Summary
Centromeres play a critical role in chromosome inheritance but are among the most difficult genomic components to analyze in multicellular eukaryotes. Here, we present a highly detailed molecular structure of a functional centromere in a multicellular organism. The centromere of the Drosophila minichromosome Dp1187 is contained within a 420 kb region of centric heterochromatin. We have used a new approach to characterize the detailed structure of this centromere and found that it is primarily composed of satellites and single, complete transposable elements. In the rest of the Drosophila genome, these satellites and transposable elements are neither unique to the centromeres nor present at all centromeres. We discuss the impact of these results on our understanding of heterochromatin structure and on the determinants of centromere identity and function.
Introduction
The centromere is a specialized chromosomal region that is essential for normal chromosome inheritance during mitosis and meiosis. The centromeric DNA is associated with the kinetochore, a structure that attaches to microtubules and helps direct chromosome movements along the spindle (Pluta et al., 1995). The centromere also plays a role in sister chromatid cohesion and separation (Miyazaki and Orr-Weaver, 1994). Centromere malfunction results in aneuploidy, that is, chromosome loss or gain, which is associated with a variety of human disorders including birth defects (e.g., Down’s syndrome) and cancer.
A complete understanding of how centromeres function in chromosome inheritance requires identifying centromeric DNA components and determining their organization in vivo. Characterization of centromeres in Saccharomyces cerevisiae and Schizosaccharomyces pombe has shown that the centromeres of the two yeast species differ greatly in size and do not share any significant sequence identity (reviewed in Clarke and Carbon, 1985; Clarke, 1990; Schulman and Bloom, 1991). In S. cerevisiae, a 125 bp sequence (CEN) encodes all the information needed in cis for full centromere function (Hegemann and Fleig, 1993). The CEN is organized into three functionally distinct elements: two sequence-specific protein-binding sites flanking an A+T-rich central sequence. In S. pombe, the centromeres span 40–100 kb and are composed of several classes of centromere-specific repetitive elements that flank a centromere-specific, nonrepetitive A+T-rich central core (Fishel et al., 1988; Chikashige et al., 1989; Clarke et al., 1993).
Unlike centromeres of the yeasts, centromeres of multicellular organisms are usually embedded in large blocks of heterochromatin (White, 1973). Heterochromatin contains many repetitive sequences that have hindered molecular-genetic studies of higher eukaryotic centromeres. In humans, numerous studies have suggested that α-satellite, an A+T-rich, ~171 bp tandem repeat (Choo et al., 1991), is associated with centromere function (Mitchell et al., 1985; Tyler-Smith et al., 1993; Brown et al., 1994; Heller et al., 1996). Alpha satellite integrated into ectopic chromosomal sites displayed some properties of centromeres (Haaf et al., 1992) but failed to provide full centromere function (Larin et al., 1994). Recent transfection studies with purified α-satellite have led to the recovery of unstable (Taylor et al., 1996) and stable (Harrington et al., 1997) extrachromosomal elements. However, since total genomic DNA was also required for the rare recovery of the stable extra-chromosomal element, additional studies are needed to determine if α-satellite alone is sufficient for centromere function.
We have been using the Drosophila minichromosome Dp1187 as a model system for characterizing the structure and function of a higher eukaryotic centromere (Le et al., 1995; Murphy and Karpen, 1995a, 1995b; Cook et al., 1997). This fully functional minichromosome is small (1.3 Mb) and is not essential for viability, allowing direct molecular and genetic manipulation (Karpen and Spradling, 1990, 1992; Tower et al., 1993). Restriction mapping of some deletion derivatives generated by γ irradiation suggested that the 1 Mb of centric heterochromatin in the minichromosome contains regions of highly repetitive satellites interspersed with islands of complex DNA sequences, corresponding to regions of low and high restriction site density, respectively (Le et al., 1995). Analyses of the transmission of different derivatives demonstrated that a 420 kb region of centric heterochromatin contains a fully functional centromere (Murphy and Karpen, 1995b). These results prompted an important question: what DNA sequences constitute this functional centromere?
The detailed mapping of a higher eukaryotic centromere is a challenging task because of the prevalence of repetitive sequences. Ideally, one could use DNA sequences that are unique to a particular centromere as entry points for molecular analyses. Unfortunately, such unique DNA sequences have rarely been identified (reviewed in Vig, 1994; Sunkel and Coelho, 1995). Cloning higher eukaryotic centromeres in yeast or bacterial vector systems such as YACs, BACs, or PACs could separate centromeres from the majority of repetitive DNA sequences present in the genome; however, no clone is known to contain a complete centromere, possibly due to the instability of highly repetitive sequences in yeasts and bacteria (Foote et al., 1992). Fluorescence in situ hybridization (FISH) is useful for localizing DNA sequences to chromosomal regions, but the resolution in general is low (at the level of hundreds of kilobases) (Lichter et al., 1990; Ried et al., 1992; Pimpinelli et al., 1995). In the case of Dp1187, previous studies of the centric heterochromatin were facilitated by using rearranged chromosomes that juxtaposed euchromatin and heterochromatin. These rearrangements made it possible to use single-copy euchromatic sequences as entry points to restriction map the adjacent centric heterochromatin (Karpen and Spradling, 1990, 1992; Le et al., 1995; Murphy and Karpen, 1995b). More distant heterochromatin, however, could not be mapped using this method or was mapped with low resolution. We report here the detailed structure of the Dp1187 centromere obtained by a more direct approach: we purified minichromosome DNA from nuclei by pulsed-field gel electrophoresis (PFGE) and used the purified DNA as a substrate for molecular analyses. We found that this centromere is predominantly composed of common satellite DNAs and transposable elements. We will discuss how these results advance our understanding of centromere function and the organization of heterochromatin.
Results
Purification of Minichromosomal DNA
Our strategy for characterizing the Dp1187 centromere was to use purified minichromosomal DNAs. The small size of Dp1187 and its derivatives enabled us to separate them from bulk genomic DNA by PFGE (Le et al., 1995; Murphy and Karpen, 1995b). Such purification eliminated the interference by repetitive sequences from the rest of the genome during Southern hybridization analyses, and allowed us to determine which known Drosophila satellites and transposable elements are present in the minichromosome and to use them as probes to map the minichromosome heterochromatin. Although these sequences are multicopy in the genome, we demonstrated that some of them map to specific regions of minichromosome heterochromatin and thus are excellent “locally single-copy” probes when applied to a purified substrate.
We first used this strategy to analyze the heterochromatin of γ1230 (Figure 1), a deletion derivative of Dp8-23 (Dp1187 with two rosy+ (ry+) P element insertions [Le et al., 1995]). γ1230 is an excellent substrate for centromere mapping, because it is stably transmitted and therefore contains a fully functional centromere (Murphy and Karpen, 1995b), yet it retains only 420 kb of the 1 Mb of centric heterochromatin present in Dp1187 (Le et al., 1995). In fact, as we will demonstrate, the entire 420 kb heterochromatin in γ1230 is necessary and sufficient for fully stable chromosome transmission. Below we will describe the detailed molecular structure of this region obtained through analyses of γ1230 and will characterize the structure of the entire Dp1187 centric heterochromatin. Finally, we will discuss the implications of these results for determining the location of the centromere within Dp1187. For each critical conclusion, we will provide examples of data from our more extensive mapping and hybridization analyses.
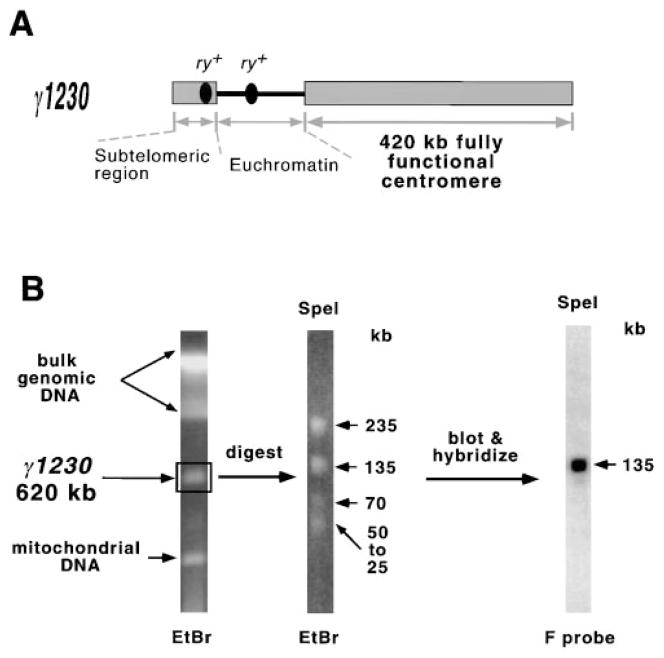
Figure 1. Strategy for Molecular Analysis of the Minichromosome Centromere.
(A) Gross structure of γ1230, the smallest stable derivative available (only 620 kb, compared to the 1.3 Mb in the parental Dp8-23 minichromosome).
(B) Purification and Southern hybridization analysis of γ1230. Minichromosomal DNA is separated from the rest of the genome by PFGE. An agarose block containing the DNA is then excised and digested with a restriction enzyme (SpeI), and restriction fragments are resolved by a second round of PFGE. The gel is blotted and probed with a Drosophila repetitive DNA sequence (F transposable element) that works as a single copy probe on the purified minichromosome DNA. Pulse conditions were 20–80 s pulses, 2 s ramp, 20 hr for resolving undigested γ1230; 1–21 s pulses, 1 s ramp, and 15 hr for resolving the SpeI fragments.
Satellite DNAs and Transposable Elements Are the Primary Components of the Minichromosome Centromere
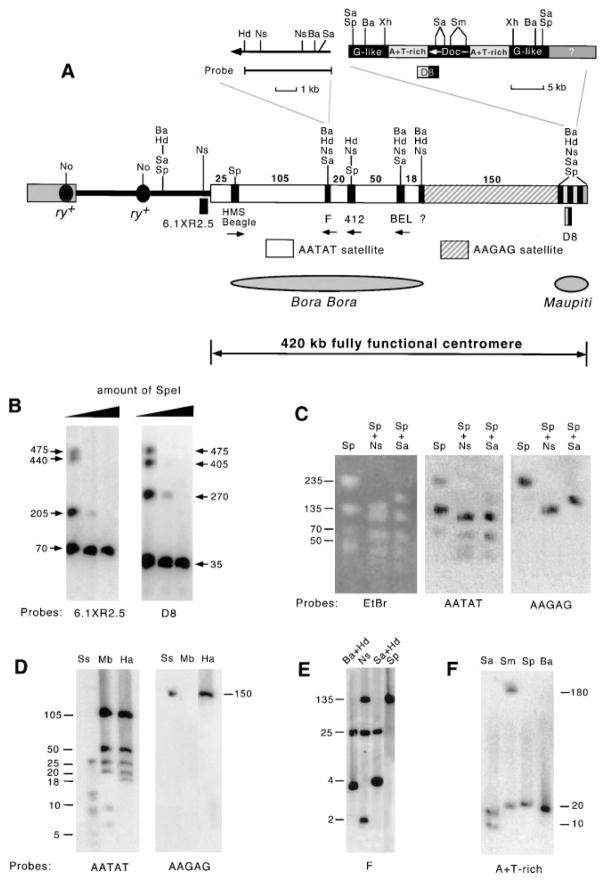
Restriction site location and frequency can help distinguish between regions of complex DNAs versus simple highly repetitive DNAs (satellites). Complex DNAs usually contain a variety of restriction sites while simple satellites can be cleaved by very few, if any, enzymes. Previous low resolution mapping indicated that there was a 220 kb island of complex DNA (termed Bora Bora; Le et al., 1995) included in the region necessary for normal transmission (Murphy and Karpen, 1995b). By partially digesting PFGE-purified γ1230 DNA and probing with two DNA sequences located close to the ends of the centromere (6.1XR2.5 and D8, Figure 2A), we were able to determine the restriction site distribution of eight enzymes with greatly improved resolution. For example, partial digestion with SpeI (Figure 2B) determined the organization of the large SpeI restriction fragments in the centromere. Our higher resolution mapping revealed a striking feature of the centromere: the restriction sites are not spread evenly across the 220 kb region (Bora Bora), as suggested previously, but are present in five widely separated small clusters (each less than 10 kb) (Figure 2A). There is also a region of complex DNA in the right terminal region (approximately 35 kb, called Maupiti). The pattern of restriction site distribution was confirmed by complete digestions with other enzymes, using probes located at different regions of the centromere (see below). This analysis indicated that the centromere is primarily composed of satellites, which are interspersed with short stretches of complex DNA.
Figure 2. Detailed Structure of the 420 kb of Centric Heterochromatin that Contains a Fully Functional Centromere.
(A) Organization of the centric heterochromatin in γ1230. Transposable elements are shown as black boxes and satellites as blank and striped boxes (numbers above the boxes show size of satellite arrays in kilobases). Orientation of transposable elements is indicated by arrows, which point to 3′ ends. Only a subset of characterized restriction sites are shown (see Experimental Procedures for a complete list of restriction enzymes used. A complete map cannot be displayed effectively here but is available upon request). Complex DNA sequence at the fifth restriction cluster (at the junction of the AATAT and AA-GAG satellites) remains to be identified (see Discussion). The region corresponding to the previously identified complex DNA island Bora Bora is indicated, as is the newly identified island Maupiti. (B) Southern hybridization of SpeI partially digested γ1230 with two probes located close to either end of this region, which localized the four SpeI fragments in the order of (from left to right) 70, 135, 235, and 35 kb (the 70 kb fragment contains the euchromatin/heterochromatin junction because it also hybridizes to the 6.1XR2.5 probe in complete digests). (C) Restriction fragments can be visualized by ethidium bromide staining and by hybridization with satellite probes. AAGAG and AATAT satellites are localized to either side of the fifth restriction site cluster by hybridizing to specific restriction fragments. For example, the AATAT satellite probe detected the 70, 135, and 235 kb SpeI fragments, but not the 150 kb SpeI/NsiI fragment (from the fifth cluster to Maupiti); the latter is the only SpeI/NsiI fragment detected by the AAGAG satellite probe. (D) Southern hybridization demonstrates high sequence homogeneity of the two satellites (see text). The blot was successively probed with the AATAT and AAGAG satellite probes. When this blot was probed with the centromeric transposons, each probe detected specific fragments that corresponded to the AATAT arrays (data not shown). For example, the 412 probe detected the 50 and 20 kb HaeIII fragments, and the BEL probe hybridized to the 50 and 18 kb fragments. Thus, the 20 kb AATAT fragment is positioned to the left of the 412 element, the 18 kb fragment is to the right of BEL, and the 50 kb fragment is in between the two elements (A). (E) Southern hybridization of F element probe to digested γ1230. This result indicates that the centromeric F is a single intact element located at the second restriction site cluster (compare the hybridization pattern with the known F map shown in [A]). (F) Southern hybridization with the A+T-rich sequence on purified γ1230. The probe detected two SalI and two SmaI fragments, indicating that homologous sequences are present at both sides of the Doc element in Maupiti (see the enlarged Maupiti structure in [A]). The same hybridization pattern was observed with the A+T-rich probe on total genomic DNA of a strain (y; ry506) that lacks the minichromosome (data not shown).
Size of restriction fragments is indicated in kilobases. Abbreviations are: EtBr, ethidium bromide; Ba, BamHI; Hd, HindIII; Mb, MboII; No, NotI; Ns, NsiI; Sa, SalI; Sm, SmaI; Sp, SpeI; Ss, SspI; Xh, XhoI. Pulse conditions were 10–50 s pulses, 1 s ramp, 17 hr for (B); 1–21 s pulses, 1 s ramp, 12 hr for (C) and (E), and 20 hr for (D); 0.5–10.5 s pulses, 0.5 s ramp, 18 hr for (F).
Transposable elements are abundant in Drosophila heterochromatin (Finnegan, 1985; Charlesworth et al., 1994; Carmena and Gonzalez, 1995; Pimpinelli et al., 1995). We probed γ1230 digests with 36 Drosophila transposable element sequences (see Experimental Procedures) and found that six elements—retrotransposons H.M.S. Beagle, 412, and BEL, and retroposons F Doc and G-like (Lindsley and Zimm, 1992)—are present in the centric heterochromatin of γ1230 (Figure 2A). These transposable elements provide excellent entry points for dissecting the centromere due to their “deep” location within the centromeric heterochromatin. Extensive mapping demonstrated that H.M.S. Beagle, F, 412, and BEL define the first four restriction site clusters (from left to right), while the Doc and the G-like sequences are present in Maupiti (Figure 2A). The DNA sequence at the fifth cluster remains to be determined.
We probed restriction enzyme–digested γ1230 DNA with nine previously cloned Drosophila satellite DNAs (see Experimental Procedures) and found that only the 1.672–38 satellite (AATAT repeats) and the 1.705–42 satellite (AAGAG repeats) (Lohe and Brutlag, 1986; Lohe et al., 1993; and references therein) are present in the centromere. Extensive restriction mapping using single and double digestions showed that the fifth restriction-site cluster separates the two satellite repeats: only AA-TAT satellite is present to the left of the fifth restriction site cluster, and only AAGAG satellite was detected between the fifth restriction-site cluster and Maupiti (Figure 2C; see also Figures 2D and 5). Neither AATAT nor AAGAG repeats were detected in the 35 kb Maupiti sequence (Figure 1A). Since satellite fragments as small as 2 kb were detected in other regions (e.g., see SspI digestion in Figure 2D), it is unlikely that Maupiti contains large amounts of any satellite.
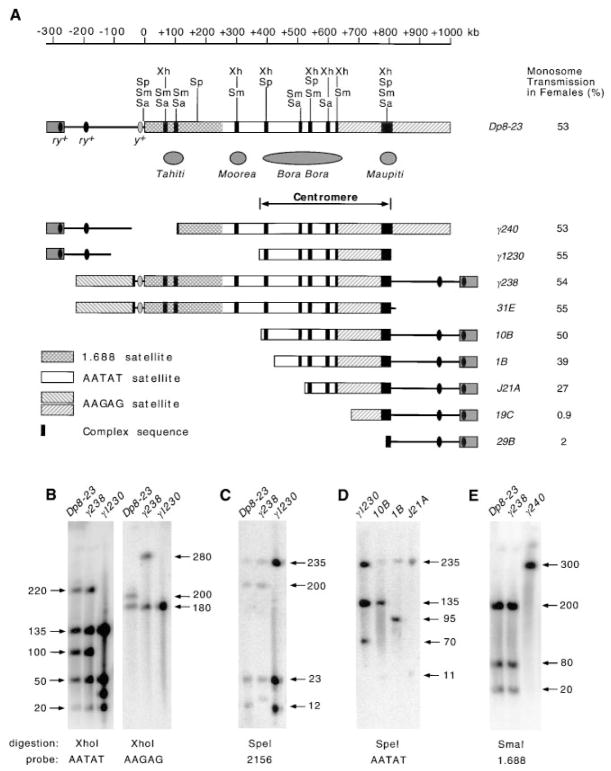
Figure 5. The Structure of Dp8-23 and Its Derivatives.
(A) A single 420 kb region of centric heterochromatin is necessary and sufficient for fully stable chromosome transmission. The ruler defines the position along Dp8-23 relative to the euchromatin/heterochromatin junction (Le et al., 1995). Structure is shown along with the percent monosome transmission in females (Murphy and Karpen, 1995b). Regions corresponding to previously identified islands of complex DNA (Tahiti, Moorea, and Bora Bora) are indicated; Maupiti is the newly identified island (see Figure 2). Note that our new, more precise alignment of γ1230 with Dp8-23 has moved Bora Bora 200 kb leftward compared to the previous location (Le et al., 1995; Murphy and Karpen, 1995b). γ1230 and 10B, the two smallest derivatives that display normal monosome transmission (50%), share almost the exact same 420 kb region of centric heterochromatin. (B) Hybridization of AATAT and AAGAG satellite probes to XhoI fragments of Dp8-23, γ238, and γ1230, demonstrating that γ1230 was produced by an internal and a terminal deletion. γ1230 lacks the 220 kb XhoI fragment (from +80 to +300) detected by the AATAT satellite probe in Dp8-23 and γ238, and contains a 30 kb fragment instead of a 100 kb fragment (from +300 to +400). In addition, the 200 kb fragment (from +800 to +1000) detected by the AAGAG satellite probe in Dp8-23 is missing in γ1230. Note that the signal intensity of the 220 kb XhoI fragment (from +80 to +300) detected by the AATAT probe is approximately equal to that of the 50 kb XhoI fragment (left), indicating that AATAT satellite comprises about 50 kb of the 220 kb XhoI fragment. This maps, as shown in (A), the transition point between the 1.688 and the AATAT satellite to approximately +250 (300–50 = 250). γ238 displays a 280 kb Xho fragment instead of the 200 kb fragment in Dp8-23 (right), consistent with the inversion in γ238 that placed the 200 kb of AAGAG satellite to the left of the y+ gene. (C) Hybridization of the 2156 probe to SpeI digested Dp8-23, γ238, and γ1230. The fragment corresponding to the 12 kb SpeI fragment in Dp8-23 is either larger (in γ238) or smaller (in γ1230), indicating that the right breakpoints of γ238 and γ1230 fall into this SpeI fragment in Dp8-23. This made the size of Maupiti in γ1230, and γ238 is smaller than in Dp8-23. (D) Hybridization with the AATAT probe on SpeI digested γ1230, 10B, 1B, and J21A. Comparison of the patterns confirms that these derivatives contain progressively larger deletions of the centromere and precisely maps the positions of the terminal breakpoints. (E) Hybridization of the 1.688 satellite probe to SmaI digested Dp8-23, γ238, and γ240. The hybridization pattern indicates that the first 250 kb of centric heterochromatin is largely composed of the 1.688 satellite. Extensive mapping suggested that stretches of complex DNA are present around +100 kb and +300 kb, including a Doc at +300 kb (data not shown; see also Le et al., 1995). Only the restriction sites relevant to the hybridization results shown in (B)–(E) are shown in (A). The following pulse conditions were used for all blots shown in this figure: 1–21 s pulses, 1 s ramp, and 20 hr. Size of restriction fragments is indicated in kilobases, and abbreviations are the same as in Figure 2.
The Centromeric Satellite Arrays Display a High Degree of Sequence Homogeneity
Are there any other complex sequences hidden within the satellite arrays? How variable are the satellite sequences? To address these questions, we analyzed the structure of the satellite arrays with sixteen enzymes that recognize 4, 5, or 6 bp sites, representing a variety of recognition sequences and base compositions (see Experimental Procedures). The rationale is that failure to cleave satellite blocks with enzymes that cut complex DNA frequently (e.g., most four-cutters) would suggest that the satellite blocks are devoid of complex DNA, while the frequency of cleavage by an enzyme that recognizes a variant of the satellite would reflect the level of sequence variation within the satellite.
The nucleotide sequence variation in the centromeric AATAT satellite is very low. Enzymes that require only a single nucleotide change (MseI/TTAA and SspI/AATATT) cut the AATAT blocks into fragments up to 25 kb (see SspI digestion shown in Figure 2D). Note that production of a 5 kb fragment indicates that the single nucleotide change occurred only once in 1000 repeats. The five blocks remained intact after digestions with all the other enzymes except MboII/GAAGA, AseI/ATTAAT, and RsaI/GTAC, which cut, probably just once, in the 18, 25, and 50 kb blocks, respectively (see Figure 2D for MboII digestion). Eleven enzymes did not cut the centromeric AATAT satellite blocks; from these results, we estimate that there is only a 1% chance that we missed a 150 bp complex sequence in the 5 blocks of AATAT, which total 228 kb (see Experimental Procedures). These results indicated that the satellite blocks are homogeneous AATAT repeats with occasional nucleotide changes, suggesting that significant amounts of complex DNA are not likely to be hidden within the five AATAT satellite blocks.
The 150 kb AAGAG block appears to be even more homogeneous. The only enzymes that cut this repeat array are MboII and BbsI (see Figure 2D for examples). MboII recognizes GAAGA, which is present in every tandem repeat of AAGAG, and as expected, no AAGAG-containing fragment was detected after MboII digestion. Only a few large fragments (90, 30, 25, and 9 kb) were generated by BbsI (GAAGACNN), which requires one nucleotide change in the AAGAG repeats (data not shown). Restriction enzymes that recognize AAGAG repeat variants with two altered nucleotides (e.g., BfaI/CTAG and Sau3AI/GATC) did not cut the AAGAG array (data not shown). These results demonstrate that sequence variation in the AAGAG repeat array is even lower than in the AATAT arrays.
The Transposons in the Centromere Are Single, Intact Elements
Are the centromeric transposable elements complete or incomplete, and are they single, isolated elements, tandem repeated elements, or the scrambled clusters previously seen for heterochromatic transposons (Devlin et al., 1990; Hochstenbach et al., 1993)? Are other complex sequences present at the restriction site clusters? We addressed these questions by fine scale restriction mapping of the γ1230 transposons, using more than 30 different restriction enzymes (see Experimental Procedures). For example, the F probe detected the expected restriction fragments (Di Nocera et al., 1983; Di Nocera and Casari, 1987) within the γ1230 F element and also detected large fragments that contain both the flanking AATAT repeats and the terminal F sequences (Figure 2E, compare with the map in 2A). H.M.S. Beagle, F, 412, and BEL are probably immediately juxtaposed with the AATAT satellite, since four-cutter restriction fragments were detected by both the corresponding transposable element probes and the AATAT probe (see Figure 2 legend); very little complex DNA sequences should remain attached to a satellite block after four-cutter digestion.
The transposable elements in the centromere appear to be well conserved compared to previously characterized euchromatic transposon clones. No restriction site variations were observed, with one minor exception: a SalI site was detected in the fourth restriction site cluster in γ1230 that was not present in the published BEL sequence (Davis and Judd, 1995). The 1.55 kb 3′ end of the Doc in Maupiti (see below) is 96% identical to the published sequence of another Doc element at an euchromatic location (O’Hare et al., 1991). We conclude that each of the first four restriction site clusters corresponds to a single transposable element that is immediately flanked by AATAT satellite, and that the elements are conserved and intact in comparison to cloned or sequenced euchromatic versions. This organization of heterochromatic transposons in a satellite-rich region is unexpected and will be considered further in the Discussion.
A Novel A+T-Rich Sequence Is Present in the Right Terminus of the Centromere
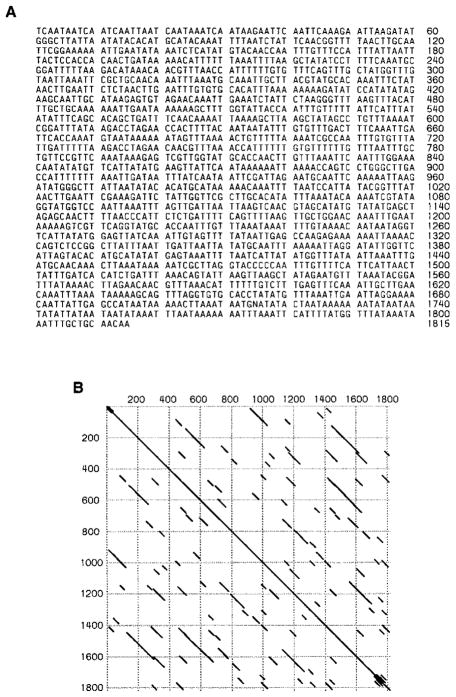
A 35 kb region of complex DNA, Maupiti, was identified to the right of the 150 kb block of AAGAG satellite, comprising the right terminal region of the centromere (Figure 2A). Molecular characterization of Maupiti was made possible by the identification of three DNA sequences present in this region: a novel A+T-rich sequence, the Doc retroposon, and a G-like sequence. The organization of these sequences is shown in Figure 2A.
The AT-rich sequence corresponds to part of the D8 clone (see Experimental Procedures) and has an A+T composition of 75% and many short stretches of internal repeats (Figure 3). The A+T-rich sequence was derived (by direct cloning) from a small part of Maupiti, but hybridization to digested γ1230 DNA showed that homologous sequences are present on both sides of the Doc element (Figure 2A). Because the A+T-rich probe detected a single 14 kb XhoI fragment that also contains nearly 5 kb of Doc, the amount of the homologous sequence cannot be more than 10 kb. Hybridization of the A+T-rich sequence to digested total genomic DNA from a strain that lacks the minichromosome gave essentially the same pattern (data not shown) as seen with digested purified γ1230 (Figure 2F), indicating that the sequences homologous to the probe are confined to one small region in the normal genome as well. The location of these sequences is most likely at the heterochromatic base of the X chromosome (see below), the origin of the centric heterochromatin in the minichromosome (Le et al., 1995). Such local distribution is unusual since all other sequences identified in the centromere so far are either middle or highly repetitive and have multiple locations in the genome (see below).
Figure 3. Analysis of the Maupiti A+T-Rich Sequence.
(A) Nucleotide sequence of the A+T-rich sequence. This sequence is followed by the 3′ end of a Doc element in the D8 clone (not shown).
(B) DotPlot of the A+T-rich sequence showing multiple short stretches of internal repeats. DNAStar program, percentage: 70; Window: 30; Min Quality: 1.
The Centromeric Satellites and the Transposable Elements Are Neither Specific for nor Universal to All Drosophila Centromeres
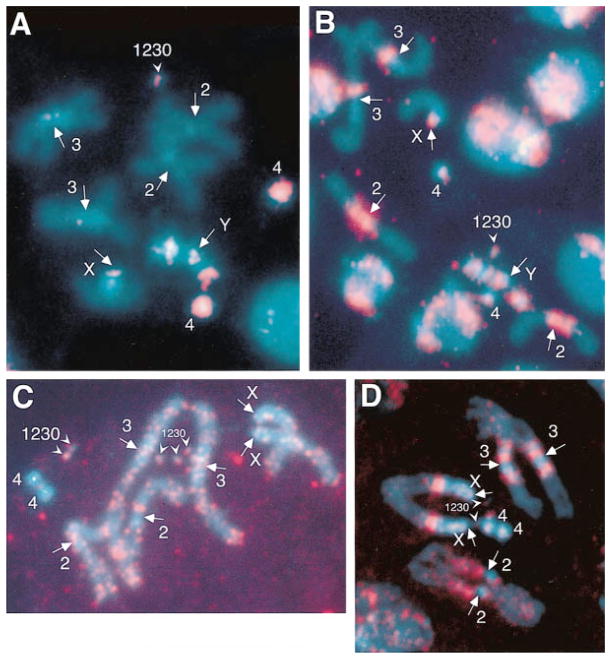
Previous studies have demonstrated that the transposable elements found in the minichromosome centromere (H.M.S. Beagle, F, 412, BEL, Doc, and G) have both euchromatic and heterochromatic locations in the Drosophila genome (Finnegan, 1985; Charlesworth et al., 1994; Carmena and Gonzalez, 1995; Le et al., 1995; Pimpinelli et al., 1995) and that the AATAT and AAGAG satellites are predominantly located in heterochromatin outside of the cytologically visible centromeres (Bonaccorsi et al., 1990; Lohe et al., 1993). Therefore, both the satellites and transposable elements identified in the Dp1187 centromere are not centromere-specific. Nevertheless, it was possible that some or all of these sequences are present in all centromeres. We evaluated the locations and amount of the centromeric sequences with FISH to mitotic chromosome spreads. The sensitivity was improved over previous analyses of Drosophila satellite DNA distribution in two ways. First, observations were made using a cooled CCD camera instead of autoradiographic methods (Bonaccorsi et al., 1990; Lohe et al., 1993). Second, we included an internal quantitation control, γ1230, in which the amount of the two satellites and the copy number of the transposable elements is known precisely. We estimated that the limits of detection in our FISH experiments were usually around 15 kb for the AAGAG satellite and 25 kb for the AATAT satellite, respectively (see Experimental Procedures). Our results demonstrated that AAGAG and AATAT are present at multiple sites in the Drosophila genome, predominantly in heterochromatic regions outside the centromeres (Figures 4A and 4B) in a pattern that is consistent with previous studies (Bonaccorsi et al., 1990; Lohe et al., 1993). Most importantly, we observed that some centromeric regions did not contain detectable amounts of either satellite. For example, no signal was observed at the second and third chromosome centromeres with the AATAT satellite probe or at the third chromosome centromere with the AAGAG satellite probe (Figures 4A and 4B). Low stringency in situ hybridization with the Maupiti A+T-rich sequence showed that no hybridization signals were observed at any centromeres, except γ1230 and the X chromosome centromeres (data no shown). With the 412 and F probes, we unequivocally detected the single copy element in γ1230 (see Figures 4C and 4D for 412 result). Both probes produced a large number of euchromatic and heterochromatic signals, which generally made it difficult to determine conclusively if a particular centromere contained these transposons. However, no hybridization was observed on the entire 4th chromosome with the 412 probe, demonstrating that there is no 412 present in at least one of the Drosophila centromeres. We conclude that all the γ1230 centromeric satellites and the transposable elements are not confined to Drosophila centromeres, and that the AATAT and AAGAG satellites, the A+T-rich sequence, and the 412 retrotransposon are not present in all Drosophila centromeres.
Figure 4. Fluorescence In Situ Hybridization (FISH) to Mitotic Metaphase Chromosomes with AATAT (A) and AAGAG (B) Satellites, and transposable elements 412 (C) and F (D).
DAPI stained (blue) neuroblast squashes are superimposed with the hybridization signals (pseudocolored FITC fluorescence in red). The normal chromosomes are indicated, as is the γ1230 minichromosome that serves as an internal quantitation control. Note that only one copy of γ1230 is present in (A) and (B) (outcrossed stocks), while five copies are seen in (C) and two in (D) (inbred stocks). None of these probes hybridizes to all centromeric regions (arrows); for example, the centromeric regions of the second chromosomes in (A) lack AATAT satellite signal, the centromeric regions of the third chromosomes in (B) are not labeled by the AAGAG probe, and the fourth chromosomes contain no 412 signal. None of these sequences are confined only to the centromeric regions.
A Single 420 kb Region of Dp1187 Is Both Necessary and Sufficient for Full Chromosome Transmission
Previous analyses in this laboratory provided a low resolution picture of the structure of the minichromosome centromere (Le et al., 1995; Murphy and Karpen, 1995b). Rearrangement derivatives generated from Dp8-23, such as γ240, γ1230, and γ238 (Figure 5A), brought different parts of the centric heterochromatin close to euchromatic sequences, allowing the adjacent heterochromatin to be “end-labeled” with euchromatic probes and mapped at low resolution (Le et al., 1995). From these analyses, the centric heterochromatin retained in γ1230 was originally thought to be derived from the right end of Dp8-23 (Le et al., 1995). A series of terminal deletion derivatives were also generated from γ238 and their in vivo stability was determined (Murphy and Karpen, 1995b) (examples are shown in Figure 5A). The smallest stable γ238 derivative is 10B, which was thought to have 220 kb overlap with γ1230 according to the original placement of γ1230 at the right end of Dp8-23. The apparently partial overlap between the two small yet fully functional derivatives led to the conclusion that the overlapping region (Bora Bora) served as an essential core of the centromere and that an additional 200 kb of satellite repeats flanking either side of the essential core was required for full function (see Figure 6A in Murphy and Karpen, 1995b). Our higher resolution analyses of γ1230, as well as the detailed structural characterization of other derivatives described below, demonstrate that the centric heterochromatin present in γ1230, like that in 10B, comes from the middle portion of the centric heterochromatin in Dp8-23, which greatly simplifies our view of the region required for full centromere function.
We generated detailed maps of the parental chromosomes Dp8-23, γ238, and some of their derivatives, using PFGE-purified minichromosomal DNAs as substrates and satellites and transposable elements as probes. Our results, as shown in Figures 5A and 5B, demonstrated that γ1230 was derived from an internal deletion (from −120 to +380) and a terminal deletion (break at +800) of Dp8-23, and that γ1230 contains essentially the same heterochromatin as 10B. The right heterochromatic breakpoints of γ1230 and γ238 (hence 10B also) appear to be very close, within a 12 kb SpeI fragment in Dp8-23 (Figure 5C), and the left heterochromatin breakpoint of γ1230 is only 15 kb to the left of the corresponding 10B breakpoint (Figures 5A and 5D). Therefore, we conclude that a nearly identical block of heterochromatin is associated with full centromere function in both γ1230 and 10B, which places the Dp1187 centromere within only one specific 420 kb part of the centric heterochromatin. Furthermore, the 1.688 satellite (~250 kb), the first 130 kb of AATAT satellite (from +250 to +380), the 200 kb of AAGAG satellite(from +800 to +1000), and some small stretches of complex DNAs can be deleted with no effect on inheritance and are not required for full centromere function.
Analyses of various deletion derivatives demonstrated that specific regions of this centromere are required for efficient chromosome inheritance. No derivatives with a right terminal deletion were recovered that lost any part of Maupiti, after irradiation of γ238 (31E was the smallest), suggesting that the right side is absolutely essential for inheritance (Murphy and Karpen, 1995b). Deletion from the left side results in a progressive decrease in transmission (Murphy and Karpen, 1995b). For example, monosomic transmission of derivative 1B, which has a 55 kb deletion at the left side of the centromere, is 39% in females, compared to 55% for γ1230. J21A deleted 150 kb of the centromere, and its transmission decreased to 27% in females (Figures 5A and 5D). Therefore, this 420 kb of centric heterochromatin is necessary for fully stable chromosome transmission.
Discussion
Here we describe the detailed molecular structure of a large region of Drosophila centric heterochromatin. We took advantage of the small size of Dp1187 and its deletion derivatives (Le et al., 1995; Murphy and Karpen, 1995b) to purify minichromosome DNA away from bulk genomic DNA. This approach overcame the difficulties inherent to studying the molecular structure of repetitive sequences present in multiple heterochromatic sites in the whole genome. We found that a single 420 kb region of the minichromosome centric heterochromatin is both necessary and sufficient for fully stable chromosome inheritance and is primarily composed of AATAT and AAGAG satellites interspersed with transposable elements. A novel A+T-rich sequence was also identified at the right end of the centromere. Deletion of centromeric sequences rendered the resulting minichromosome derivatives increasingly unstable. The following discussion will focus on how these results impact our understanding of the general organization and composition of heterochromatin, and how centromere structure is related to centromere function.
The Fine Structure of Centric Heterochromatin in Drosophila
Previous FISH and Southern analyses have demonstrated that transposons and other complex DNAs are prevalent in Drosophila centric heterochromatin (Hochstenbach et al., 1993; Carmena and Gonzalez, 1995; Le et al., 1995; Pimpinelli et al., 1995; Kurek et al., 1996). However, these studies could not address important questions about the fine structure of Drosophila heterochromatin, and the overall organization and conservation of the transposons and satellite DNAs. The results reported here reveal the fine-scale organization and composition of a large, continuous block of Drosophila centric heterochromatin. Given the extreme difficulties in cloning centric heterochromatin, these results are particularly valuable in that they allowed us to determine whether heterochromatic transposable elements are intact or deleted/rearranged and are single or clustered, and to assess the variability inherent to long satellite arrays.
The primary components of this region of Dp1187 centric heterochromatin are the AATAT and AAGAG satellites, making up more than 85% (370 kb) of the total 420 kb. Sequence variation of the two satellites, especially the AAGAG repeats, is very low. Restriction fragments of up to 90 kb of the AAGAG satellite or 25 kb of the AATAT satellite remained after digestion by enzymes that can cleave with only one nucleotide change in the repeat unit. Thus, these analyses allowed us to assess the organization and sequence variation of Drosophila satellites over an extensive region.
Transposable elements comprise about 10% of this region of the centric heterochromatin. Single elements are interspersed in the AATAT but not the AAGAG satellite and are also clustered in Maupiti, the right terminal region of the centromere. Frequent interruption of the AATAT satellite may reflect a preference of some transposable elements for insertion into A+T-rich sequences (Sandmeyer et al., 1990). The sequence composition at the fifth restriction site cluster is not known at this time; we do know that the restriction site distribution at this cluster is confined to a region of a few kilobases. Since the other four clusters were of similar size (5–10 kb) and found to be transposable elements embedded in the AATAT repeats, we speculate that the fifth cluster is still within the AATAT repeats and most likely represents another transposable element that simply was not included in the 30 transposon clones we utilized for these studies (see Experimental Procedures).
The conservation of the centromeric transposable elements is very high. The fine scale restriction maps of the centromeric H.M.S. Beagle, 412, F, and BEL elements and the sequence of the centromeric Doc retroposon were nearly identical to previously published elements (Snyder et al., 1982; Di Nocera et al., 1983; Yuki et al., 1986; Di Nocera and Casari, 1987; O’Hare et al., 1991; Davis and Judd, 1995). At this time, we do not know when these Dp1187 heterochromatic transposable elements were inserted during evolution and whether the elements are mobile. The sequence similarity suggests that these centric elements are recent insertions and are likely to be active, or that they are ancient insertions conserved due to selective/functional constraints.
Does the fine structure and organization defined here for the Dp1187 centromeric region apply to other parts of Drosophila centric heterochromatin? Many of our observations are unexpected, including the conservation of the structure of the heterochromatic transposons (in comparison to the corresponding euchromatic copies), their interspersion in the AATAT but not the AAGAG satellite, the extremely low sequence variation of the satellites, and the presence of a special region (Maupiti) that contains several transposons and a novel unique A+T-rich sequence. Nevertheless, the main structural pattern, satellites and interspersed single transposons, predominates in both the centromeric and the non-centromeric portions of the entire 1 Mb of minichromosome heterochromatin. Southern hybridization analyses suggest that most other satellite blocks in Drosophila heterochromatin are frequently interrupted by complex DNAs (Le et al., 1995), which most likely consist of transposons present as single elements (X. S., J. W., T. Nguyen, and G. K., unpublished data). Therefore, we postulate that the structural organization described here for Dp1187 centric heterochromatin is one of the major structural patterns displayed by centric heterochromatin throughout the Drosophila genome. However, similar fine scale analyses of other regions of heterochromatin, in Drosophila and other organisms, must be performed to determine if the interspersion pattern observed in Dp1187 is ubiquitous.
Centromeric Domains Required for Full Chromosome Transmission
A nearly identical block of heterochromatin is associated with full centromere function in both γ1230 and 10B, which localizes the Dp1187 centromere to within a specific 420 kb region of the centric heterochromatin (Figure 5). The structural analyses presented here constitute the most detailed molecular structure currently known for a multicellular eukaryotic centromere with demonstrated full in vivo function. Combining knowledge of fine structure with transmission behavior of minichromosome deletion derivatives also allows us to identify grossly sequence domains within the centromere that are necessary for chromosome transmission. Previous analyses demonstrated that no derivatives of γ238 with a right terminal deletion were recovered that lost any part of Maupiti (Murphy and Karpen, 1995b). The complete absence of this class of derivative suggests that Maupiti and/or the AAGAG block are absolutely essential for full centromere function and thus may encode kinetochore formation. However, Maupiti could be required for a noncentromeric function, such as replication initiation. In contrast, derivatives with deletions of the AATAT satellite/transposon region (Bora Bora) were recovered—they display diminished stability, with larger deletions producing lower transmission rates (Murphy and Karpen, 1995b, and Figure 5). This instability is not caused by decreased total chromosome size. J21A (580 kb) is nearly the same size as γ1230 (620 kb), and 1B (680 kb) is larger than γ1230, yet both J21A and 1B are significantly less stable than γ1230 (Figure 5A). These data suggest that the deleted Bora Bora satellites and/or transposons are required for full chromosome transmission and that partial loss of the centromeric components cannot be compensated for by simply increasing the amount of euchromatic sequences. Derivatives with deletions in Bora Bora (e.g., J21A) display high rates of sister chromatid nondisjunction and low rates of loss, suggesting that this region may be primarily responsible for sister chromatid cohesion, rather than kinetochore formation (Murphy and Karpen, 1995a; K. Cook and G. K., unpublished data; T. Murphy and G. K., unpublished data). Further investigations are required to test directly the model that Bora Bora and the AAGAG/Maupiti regions are separately responsible for sister chromatid cohesion and kinetochore formation, respectively.
Do Specific Primary Sequences Determine Dp1187 Centromere Identity and Function?
In the yeast S. cerevisiae, centromere identity and function are determined by specific primary DNA sequences that interact with particular DNA-binding proteins (for review, see Hegemann and Fleig, 1993). Do multicellular eukaryotes such as Drosophila utilize a similar mechanism to determine which region will act as a centromere and how well it will function? Our sensitive FISH analyses corroborate previous results (Bonaccorsi et al., 1990; Lohe et al., 1993) and demonstrate that the AATAT and AAGAG satellites are present at multiple places in the genome, predominantly in regions that never act as functional centromeres. In addition, we performed FISH analyses of the satellites using an internal quantitation control, a minichromosome with a known amount of satellite DNA. This method allowed more precise quantitation of the amount of the satellites in specific heterochromatic regions and demonstrated that at least some of the endogenous Drosophila centromeres cannot contain more than 15 kb of AAGAG satellite or 25 kb of AATAT satellite (Figure 4). Thus, the distribution of AA-TAT and AAGAG satellites within the genome indicates that these satellites are neither necessary nor sufficient for centromere function.
Transposable elements in Drosophila are known to be present in multiple heterochromatic and euchromatic locations throughout the genome; none have been found to be centromere-specific or present at all centromeres (Gatti and Pimpinelli, 1992; Carmena and Gonzalez, 1995; Pimpinelli et al., 1995). However, among the centromeric transposable elements identified here, the distributions of Doc, F, and G were analyzed in previously published studies, and it is unclear if these analyses were sensitive enough to identify single elements (Gatti and Pimpinelli, 1992; Carmena and Gonzalez, 1995; Pimpinelli et al., 1995). Using 412 and F elements as probes, we performed FISH on mitotic chromosome spreads containing γ1230 as an internal single element control and demonstrated that at least the fourth chromosome centromere does not contain even one copy of a 412 element. We are currently analyzing the distribution of other Dp1187 centromeric transposons. However, the abundance and broad distribution of all these elements demonstrates that none are present only at centromeres, and it is highly unlikely that they are present at all Drosophila centromeres and are specifically and absolutely required for centromere function.
Maupiti, which appears to be essential for normal chromosome transmission (see above), displays unusual structural features. In addition to a cluster of transposable elements, it contains an A+T-rich sequence—this is the only known sequence within the Dp1187 centromere that is single copy in the genome. The A+T-rich sequence could play a special role in centromere function, since many sequences unique to centromeres in other organisms are A+T-rich; these include CDEII in S. cerevisiae centromeres, the central core in S. pombe centromeres, and the α-satellite in human centromeres (Clarke and Carbon, 1985; Clarke, 1990; Schulman and Bloom, 1991; Tyler-Smith et al., 1993; Brown et al., 1994). However, the A+T-rich sequence is only present in one small region in the normal Drosophila genome (most likely the base of the X), and in situ hybridization demonstrates that this sequence is definitely not present in all the other centromeres, even at low stringency.
We conclude that none of the sequences identified to date in the Dp1187 centromere fulfill the criteria for a centromere “magic sequence” consistent with the S. cerevisiae model, that is, a specific primary DNA sequence that is both necessary and sufficient for centromere function on all chromosomes. It is unlikely that we missed a large sequence that would fulfill these criteria; our fine-scale restriction analyses demonstrate that significant amounts of sequences are not hidden within the satellite arrays and transposons. However, the structural analysis is only 95% complete, and it is possible that a short sequence is present that has the properties of a magic sequence. If specific small sequences are involved in centromere function, they most likely function as nucleating sites to recruit other nonspecific sequences during centromere assembly, since there is a clear requirement for a larger (420 kb) region of heterochromatin. It is also possible that different Drosophila centromeres contain different essential primary sequences, as observed for the central core sequences in S. pombe centromeres (Fishel et al., 1988; Chikashige et al., 1989; Clarke et al., 1993). We are currently cloning and sequencing regions of the Dp1187 centromere to test the magic sequence hypothesis.
Alternatively, particular combinations of different, common DNAs, or overall nucleotide composition, could determine centromere identity and function, rather than the primary sequences. For example, the juxtaposition of the two satellites and/or the interspersion of the transposons in the AATAT repeats may be specific to centromeres and required for function. At this time, we do not have evidence that supports this “combinatorial” model. The satellite/transposon structure is a common pattern in the Dp1187 centric heterochromatin and in Drosophila heterochromatin in general (see above), and thus is not unique to centromeres. To test directly the combinatorial model, we need to develop techniques that allow us to analyze the function of directed DNA rearrangements in vivo. For example, it would be informative to evaluate the transmission of constructs in which the centromeric satellites are replaced with another satellite, and other constructs in which the centromeric transposable elements are replaced or removed.
Do Higher Order Structure and Epigenetic Regulation Determine Centromere Identity and Function in Drosophila?
Centromere function may be achieved by formation of a specific higher order structure (HOS) (Zinkowski et al., 1991; Vig, 1994; Sunkel and Coelho, 1995), an overall three-dimensional organization that results from special DNA architectures (e.g., DNA bending) and/or DNA–protein interactions in the centromere. Different DNA sequences could function as centromeric components in different organisms, or even in chromosomes of the same organism (as in flies [this study] and in S. pombe [Clarke et al., 1993]), as long as they facilitate the formation of the appropriate HOS. Repetitive, A+T-rich DNA seems to be a common feature of centromeric DNAs in different organisms (e.g., see Rattner, 1991; Alfenito and Birchler, 1993; Clarke et al., 1993); the AATAT and AA-GAG satellites found in the Dp1187 centromere may be examples of sequences that can facilitate the formation of a centromere-specific HOS. The transposable elements may contribute to formation of this HOS; alternatively, the transposable elements simply could be tolerated by the centromere if they do not disrupt the HOS.
Formation and propagation of a centromere-specific HOS may be epigenetically regulated. Epigenetic mechanisms have been proposed to account for heritable changes in gene function that cannot be explained by changes in DNA sequence (for review, see Russo et al., 1996). Epigenetics and the concept of higher order structure emphasize the importance of centromere structure beyond the primary sequence level, which can account for the numerous cases of centromere plasticity reported for mammals, flies, and pombe. First, centromeric DNA is not always sufficient for centromere function. In fission yeast, it has been shown that centromeric DNA can be associated with two functionally different states (Steiner and Clarke, 1994). Similarly, the global genomic distribution of the AATAT and AAGAG satellites and the inactivation of one of the centromeres in stable dicentric chromosomes in humans and flies (Hsu et al., 1975; Earnshaw and Migeon, 1985; Ault and Lyttle, 1988; Page et al., 1995; Sullivan and Schwartz, 1995) demonstrate that the same type of sequences at different places in the genome may or may not be associated with centromere function. Secondly, centromeric DNA is not always necessary for centromere function; centromere activity can be associated with normally noncentromeric DNAs. Human marker chromosomes are stable in transmission even though they do not contain centromere-associated alphoid repeats (Voullaire et al., 1993; Ohashi et al., 1994; Sacchi et al., 1996; du Sart et al., 1997). Such “neocentromere activity” of normally noncentromeric DNA has also been demonstrated to account for the transmission of structurally acentric derivatives of the Dp1187 minichromosome (Murphy and Karpen, 1995b; Williams et al., 1998).
Formation of a centromere-specific higher order structure by nonspecific sequences may be the underlying mechanism for the observed neocentromere activation observed in humans and flies. However, we also need to account for the fact that once a functional centromere is present, it is stably propagated at that site through multiple DNA replications and cell divisions by a mechanism that seems to be independent of the underlying DNA sequence. Epigenetic regulation provides a plausible explanation for these observations. Centromere identity and function may be self-propagating and may be inherited by DNA and/or protein marking (e.g., DNA methylation and histone modification or even centromere-specific histones) (Ekwall et al., 1997, this issue of Cell). In other words, centromere identity may be determined by the fact that a region functioned as a centromere in the previous cell division. However, experiments are required to test directly the role of epigenetic regulation in normal centromere function in higher eukaryotes. For example, if preassembly of an HOS or DNA marking is an important component of centromere regulation, we expect that purified, intact γ1230 chromosomes would be transmitted better than deproteinized, naked γ1230 DNA when introduced into cells or embryos. Similarly, we also need to determine if proteins that modify chromatin structure or HOS (e.g., histone acetylases) affect centromere function, as has been suggested for S. pombe (Ekwall et al., 1997).
In summary, the studies reported here are an important step toward understanding heterochromatin and centromere structure and function, and provide useful information and tools for future investigations. Knowledge of the fine structure of a fully functional centromere will help elucidate the biochemical nature of DNA architectures and DNA–protein interactions at the centromere, which will be critical to our understanding of centromere function and the efficient construction of artificial chromosomes in Drosophila and other eukaryotes, including humans. Our data suggest that the majority of the centromeric sequences are not specific to centromeres. If there are specific sequences involved in centromere function, they must comprise a minor portion of the regions required for full function and may even differ among individual centromeres. Alternatively, centromere function in this metazoan, and perhaps other multicellular eukaryotes, may be provided by a specific three-dimensional higher order structure, which may be under the control of epigenetic mechanisms. This model is consistent with our current knowledge of centromere structure and function and can most easily account for both centromere plasticity and stability. However, the roles of specific sequences, combinations of nonspecific sequences, and epigenetics in regulating centromere function in multicellular eukaryotes may not be mutually exclusive and must be evaluated with direct experimental tests.
Experimental Procedures
Drosophila Stocks and Culture
Stocks of Dp8-23 and its derivatives were described previously (Le et al., 1995; Murphy and Karpen, 1995b).
Purification and Southern Hybridization Analysis of Minichromosome DNA
Preparation of high molecular weight DNA from embryos was described previously (Le et al., 1995). DNA from Dp8-23 or its derivatives was separated from bulk genomic DNA by PFGE (1% agarose gel with 60–120 s pulses, 2 s ramp, and 28 hr run time). After ethidium bromide staining and visualization on a longwave UV illuminator, a block of agarose gel containing the minichromosomal DNA was excised and stored in TE buffer at 4°C. For a typical restriction enzyme reaction, half an agarose block was equilibrated with restriction buffer for 30 min at room temperature and incubated overnight at the appropriate temperature with 10–20 U of a restriction enzyme in 350 μl buffer containing 0.1 mg/ml BSA. The agarose block containing digested DNA was then washed with 0.5× TBE buffer and loaded for a second round of PFGE. Pulsed-field gels were blotted and hybridized as described previously (Le et al., 1995). Various pulse conditions were used depending on the DNA fragment sizes separated (see figure legends). Hybridization was carried out in Church and Gilbert buffer (Church and Gilbert, 1984) at 55°C and 48°C for the Maupiti A+T-rich sequence and the AATAT satellite probe, respectively, and at 68°C in QuickHyb buffer (Stratagene) for all other probes.
Partial restriction digestion of PFGE-purified, intact γ1230 DNA was performed in the same way as the complete digestions described above, except that varying amounts of enzyme were used (e.g., 0.2, 0.8, and 20 U of SpeI were used for Figure 2B). Complete NotI digestion was included in partial digestion reactions to remove the subtelomeric region and part of the euchromatic sequences. This allowed the euchromatic probe 6.1XR2.5 to be used as a probe to map the centric heterochromatin (see Figure 2A for the location of the NotI sites and the 6.1XR2.5 probe).
For fine scale mapping of transposable elements in the centromeric heterochromatin, we used restriction enzymes AccI, ApaI, ApoI, AvaI, BamHI, BglI, BglII, ClaI, DraI, DraIII, EcoRV, EcoRI, HaeII, HincII, HindIII, HpaI, KpnI, NaeI, NdeI, NsiI, PstI, PvuI, PvuII, SacI, SacII, SalI, ScaI, SmaI, SpeI, SphI, StyI, XbaI, XhoI, and XmaI. In addition, we used the following enzymes/recognition sites to analyze the sequence homogeneity of the AATAT and AAGAG satellites: AluI/AGCT, AseI/ATTAAT, AvaII/GGACC, BbsI/GAAGACNN, BfaI/CTAG, DraI/TTTAAA, HaeIII/GGCC, MboI/GATC, MboII/GAAGA(N)8, MseI/TTAA, MspI/CCGG, NlaIII/CATG, NlaIV/GGNNCC, Sau3AI/GATC, SspI/AATATT and RsaI/GTAC. Monte Carlo simulations were used to calculate the distribution of restriction fragment sizes expected for random sequences of specific GC contents (from 0% to 100%) when cut with the collection of thirteen 4 bp cutter restriction enzymes (excluding TTAA, AATATT, and GAAGA, which cut the AATAT or AAGAG satellites). These calculations indicated that there is a 99% probability of at least one restriction site in a 149 bp random sequence of random GC content. Random sequences of 20% GC content contained the lowest frequency of restriction sites but still have a 99% probability of at least one restriction site in 260 bp of random sequence.
In Situ Hybridization
Neuroblast squashes were prepared on microscope slides essentially as described in “protocol 2” (Gatti et al., 1994). An 8 min hypotonic incubation was used, without colchicine treatment. Satellite in situ probes were made by 3′ tailing synthetic oligonucleotides with biotin-14-dCTP (Gibco/BRL) using terminal deoxynucleotidyl transferase (Promega). A 9 kb Bam fragment containing the 412 element and a 4.4 kb fragment of the F element were digested with restriction enzymes to yield fragments with an average length of 50 bp, and then each was labeled, as were the satellite oligos (Dernburg et al., 1996). Hybridization was performed using a modified version of published methods (Pardue, 1986). The labeled AAGAG 30-mer was hybridized at 26°C, the labeled AATAT 35-mer at 18°C, and the labeled 412 or F transposon probe at 37°C. Separate images were captured for DAPI signal and FITC signal using a cooled CCD camera (Princeton Instruments), and were merged and analyzed using IP-Lab Spectrum (Signal Analytics) and Photoshop (Adobe) programs. Limits of detectable amount of satellites were estimated by comparing the intensity of smallest visible fluorescence signals to the signals on γ1230 that contains about 220 kb of AATAT and 150 kb of AAGAG satellites.
DNA Probes
The D8 clone was directly cloned from γ1230 DNA. Purified γ1230 DNA was partially digested with Sau3AI and ligated to BamHI digested λFixII vector (Stratagene). D8 contains two types of sequences: the 3′ end of a Doc transposable element and an A+T-rich sequence. The H.M.S. Beagle probe is a 5 kb sequence to the left of the EcoRI site and lacks the long terminal repeats (LTR) ([Amy Csink, personal communication; X. S. et al, unpublished data]. See Lindsley and Zimm, 1992, for a restriction map—note that the SalI site in the published map should be SacI [Snyder et al., 1982]). The F probe lacks the sequence 3′ to the HindIII site (Pimpinelli et al., 1995) (Figure 2A). The 412 probe contains the entire element (Charlesworth et al., 1994), including the two LTRs at its ends that are almost identical in sequence. The BEL probe is a 1.2 kb sequence at the 5′ terminus, which includes the 361 bp LTR sequence (Amy Csink, personal communication; Davis and Judd, 1995). The G-like sequence was cloned from 2156, a Drosophila genomic clone containing different middle repetitive DNA sequences (Charlesworth et al., 1994; X. S. et al., unpublished data). Only the left terminal 500 bp of 2156 hybridized to γ1230; this region is 70% identical to part of the reverse transcriptase ORF of the G retroposon (Di Nocera, 1988). The 6.1XR2.5 probe and the satellite probes (1.672–38 [AA-TAT], 1.705–42 [AAGAG], and 1.688 [359 bp repeats]) were described previously (Le et al., 1995). The following satellites and transposable elements were tested and found not to be present in the minichromosome heterochromatin. Satellites: 1.672–181 (AATAC), 1.672–453 (AATAAAC), 1.705–34 (AAGAGAG), 1.686–198 (AAGAC), 1.686–171 (AATAACATAG), and dodeca (Lohe and Brutlag, 1986; Abad et al., 1992; Lohe et al., 1993). Transposable elements: 17.6, 297, 2158 (Charlesworth et al., 1994), 2198 (Charlesworth, personal communication), 2219 (Charlesworth, personal communication), 2244 (Charlesworth, personal communication), 3S18, aurora (Shevelyov, 1993), blood, BS, circe (Losada, personal communication), copia, coral (Csink, personal communication), gate (Gvozdev, personal communication), gypsy, HeT-A (Danilevskaya et al., 1994), Hoppel (Kurenova et al., 1990), I, jockey, Kermit/flea, mdg1, mdg3, micropia, NEB, opus, Pogo, roo, S, sancho2, and springer (see Lindsley and Zimm, 1992 for references unless specified). Clones of most of these satellites and transposable elements are available upon request.
Acknowledgments
We thank Thai Nguyen for analyzing the structure of Maupiti, and Hiep Le and Dan Entrikin for technical assistance. We thank Kevin Cook, Kenneth Dobie, Kumar Hari, Martin Latterich, Keith Maggert, Terence Murphy, and Barbara Wakimoto for stimulating discussions and critical reading of the manuscript. We are grateful to the following people for providing us with transposable element and satellite clones: Sonsoles Campuzano, Maki Chaboissier, Brian Charlesworth, Thomas Cline, Amy Csink, Olga Danilevskaya, Thomas Eickbush, Rosa de Frutos, David Finnegan, Vladimir Gvozdev, David Kuhn, Dirk Lankenau, Bob Levis, Allan Lohe, Ana Losada, Howard Lipshitz, Vladic Mogila, Kevin O’Hare, Renato Paro, Sergio Pimpinelli, Michael Simmons, and Alfredo Villasante.
Footnotes
GenBank Accession Number
The accession number for the nucleotide sequence reported in this paper is AF036950.
References
- Abad JP, Carmena M, Baars S, Saunders RD, Glover DM, Ludena P, Sentis C, Tyler-Smith C, Villasante A. Dodeca satellite: a conserved G+C-rich satellite from the centromeric heterochromatin of Drosophila melanogaster. Proc Natl Acad Sci USA. 1992;89:4663–4667. doi: 10.1073/pnas.89.10.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfenito MR, Birchler JA. Molecular characterization of a maize B chromosome centric sequence. Genetics. 1993;135:589–597. doi: 10.1093/genetics/135.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ault JG, Lyttle TW. A transmissible dicentric chromosome in Drosophila melanogaster. Chromosoma. 1988;97:71–79. [Google Scholar]
- Bonaccorsi S, Gatti M, Pisano C, Lohe A. Transcription of a satellite DNA on two Y chromosome loops of Drosophila melanogaster. Chromosoma. 1990;99:260–266. doi: 10.1007/BF01731701. [DOI] [PubMed] [Google Scholar]
- Brown KE, Barnett MA, Burgtorf C, Shaw P, Buckle VJ, Brown WR. Dissecting the centromere of the human Y chromosome with cloned telomeric DNA. Hum Mol Genet. 1994;3:1227–1237. doi: 10.1093/hmg/3.8.1227. [DOI] [PubMed] [Google Scholar]
- Carmena M, Gonzalez C. Transposable elements map in a conserved pattern of distribution extending from beta-heterochromatin to centromeres in Drosophila melanogaster. Chromosoma. 1995;103:676–684. doi: 10.1007/BF00344228. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Jarne P, Assimacopoulos S. The distribution of transposable elements within and between chromosomes in a population of Drosophila melanogaster. III. Element abundances in heterochromatin. Genet Res. 1994;64:183–197. doi: 10.1017/s0016672300032845. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O, Yanagida M. Composite motifs and repeat symmetry in S. pombe centromeres: direct analysis by integration of NotI restriction sites. Cell. 1989;57:739–751. doi: 10.1016/0092-8674(89)90789-7. [DOI] [PubMed] [Google Scholar]
- Choo KH, Vissel B, Nagy A, Earle E, Kalitsis P. A survey of the genomic distribution of alpha satellite DNA on all the human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res. 1991;19:1179–1182. doi: 10.1093/nar/19.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L. Centromeres of budding and fission yeasts. Trends Genet. 1990;6:150–154. doi: 10.1016/0168-9525(90)90149-z. [DOI] [PubMed] [Google Scholar]
- Clarke L, Carbon J. The structure and function of yeast centromeres. Annu Rev Genet. 1985;19:29–55. doi: 10.1146/annurev.ge.19.120185.000333. [DOI] [PubMed] [Google Scholar]
- Clarke L, Baum M, Marschall LG, Ngan VK, Steiner NC. Structure and function of Schizosaccharomyces pombe centromeres. Cold Spring Harb Symp Quant Biol. 1993;58:687–695. doi: 10.1101/sqb.1993.058.01.076. [DOI] [PubMed] [Google Scholar]
- Cook KR, Murphy TD, Nguyen TC, Karpen GH. Identification of trans-acting genes necessary for centromere function in Drosophila melanogaster using centromere-defective mini-chromosomes. Genetics. 1997;145:737–747. doi: 10.1093/genetics/145.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya O, Slot F, Pavlova M, Pardue ML. Structure of the Drosophila HeT-A transposon: a retrotransposon-like element forming telomeres. Chromosoma. 1994;103:215–224. doi: 10.1007/BF00368015. [DOI] [PubMed] [Google Scholar]
- Davis PS, Judd BH. Nucleotide sequence of the transposable element, BEL, of Drosophila melanogaster. Drosoph Inf Serv. 1995;76:134–136. [Google Scholar]
- Dernburg AF, Daily DR, Yook KJ, Corbin JA, Sedat JW, Sullivan W. Selective loss of sperm bearing a compound chromosome in the Drosophila female. Genetics. 1996;143:1629–1642. doi: 10.1093/genetics/143.4.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin RH, Bingham B, Wakimoto BT. The organization and expression of the light gene, a heterochromatic gene of Drosophila melanogaster. Genetics. 1990;125:129–140. doi: 10.1093/genetics/125.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nocera PP. Close relationship between non-viral retroposons in Drosophila melanogaster. Nucleic Acids Res. 1988;16:4041–4052. doi: 10.1093/nar/16.9.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nocera PP, Casari G. Related polypeptides are encoded by Drosophila F elements, I factors, and mammalian L1 sequences. Proc Natl Acad Sci USA. 1987;84:5843–5847. doi: 10.1073/pnas.84.16.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nocera PP, Digan ME, Dawid IB. A family of oligo-adenylate-terminated transposable sequences in Drosophila melanogaster. J Mol Biol. 1983;168:715–727. doi: 10.1016/s0022-2836(83)80071-0. [DOI] [PubMed] [Google Scholar]
- du Sart D, Cancilla MR, Earle E, Mao JI, Saffery R, Tainton KM, Kalitsis P, Martyn J, Barry AE, Choo KH. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Migeon BR. Three related centromere proteins are absent from the inactive centromere of a stable isodi-centric chromosome. Chromosoma. 1985;92:290–296. doi: 10.1007/BF00329812. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. this issue. [DOI] [PubMed] [Google Scholar]
- Finnegan DJ. Transposable elements in eukaryotes. Int Rev Cytol. 1985;93:281–326. doi: 10.1016/s0074-7696(08)61376-5. [DOI] [PubMed] [Google Scholar]
- Fishel B, Amstutz H, Baum M, Carbon J, Clarke L. Structural organization and functional analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol. 1988;8:754–763. doi: 10.1128/mcb.8.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote S, Vollrath D, Hilton A, Page DC. The human Y chromosome: overlapping DNA clones spanning the euchromatic region. Science. 1992;258:60–66. doi: 10.1126/science.1359640. [DOI] [PubMed] [Google Scholar]
- Gatti M, Pimpinelli S. Functional elements in Drosophila melanogaster heterochromatin. Annu Rev Genet. 1992;26:239–275. doi: 10.1146/annurev.ge.26.120192.001323. [DOI] [PubMed] [Google Scholar]
- Gatti M, Bonaccorsi S, Pimpinelli S. Looking at Drosophila mitotic chromosomes. Methods Cell Biol. 1994;44:371–391. doi: 10.1016/s0091-679x(08)60924-3. [DOI] [PubMed] [Google Scholar]
- Haaf T, Warburton PE, Willard HF. Integration of human α-satellite DNA into simian chromosomes: centromere protein binding and disruption of normal chromosome segregation. Cell. 1992;70:681–696. doi: 10.1016/0092-8674(92)90436-g. [DOI] [PubMed] [Google Scholar]
- Harrington JJ, Bokkelen GV, Mays RW, Gustashaw K, Willard HF. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- Hegemann JH, Fleig UN. The centromere of budding yeast. Bioessays. 1993;15:451–460. doi: 10.1002/bies.950150704. [DOI] [PubMed] [Google Scholar]
- Heller R, Brown KE, Burgtorf C, Brown WR. Minichromosomes derived from the human Y chromosome by telomere directed chromosome breakage. Proc Natl Acad Sci USA. 1996;93:7125–7130. doi: 10.1073/pnas.93.14.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstenbach R, Potgens A, Meijer H, Dijkhof R, Knops M, Schouren K, Hennig W. Partial reconstruction of the lampbrush loop pair nooses on the Y chromosome of Drosophila hydei. Chromosoma. 1993;102:526–545. doi: 10.1007/BF00368346. [DOI] [PubMed] [Google Scholar]
- Hsu TC, Pathak S, Chen TR. The possibility of latent centromeres and a proposed nomenclature system for total chromosome and whole arm translocations. Cytogenet Cell Genet. 1975;15:41–49. doi: 10.1159/000130497. [DOI] [PubMed] [Google Scholar]
- Karpen GH, Spradling AC. Reduced DNA polytenization of a minichromosome region undergoing position-effect variegation in Drosophila. Cell. 1990;63:97–107. doi: 10.1016/0092-8674(90)90291-l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Spradling AC. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics. 1992;132:737–753. doi: 10.1093/genetics/132.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurek R, Trapitz P, Bunemann H. Strukturdifferenzierungen in Y-chromosom von Drosophila hydei: the unique morphology of the Y chromosomal lampbrush loops threads results from “coaxial shells” formed by different satellite-specific subregions within megabase-sized transcripts. Chromosome Res. 1996;4:87–102. doi: 10.1007/BF02259701. [DOI] [PubMed] [Google Scholar]
- Kurenova EV, Leibovich BA, Bass IA, Bebikhov DV, Pavlova MN, Danilevskaia ON. Hoppel-family of mobile elements of Drosophila melanogaster, flanked by short inverted repeats and having preferential localization in the heterochromatin regions of the genome. Genetika. 1990;26:1701–1712. [PubMed] [Google Scholar]
- Larin Z, Fricker MD, Tyler-Smith C. De novo formation of several features of a centromere following introduction of a Y alphoid YAC into mammalian cells. Hum Mol Genet. 1994;3:689–695. doi: 10.1093/hmg/3.5.689. [DOI] [PubMed] [Google Scholar]
- Le MH, Duricka D, Karpen GH. Islands of complex DNA are widespread in Drosophila centric heterochromatin. Genetics. 1995;141:283–303. doi: 10.1093/genetics/141.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter P, Tang CJ, Call K, Hermanson G, Evans GA, Housman D, Ward DC. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- Lindsley DL, Zimm GG. The Genome of Drosophila melanogaster. San Diego, CA: Academic Press, Inc; 1992. [Google Scholar]
- Lohe AR, Brutlag DL. Multiplicity of satellite DNA sequences in Drosophila melanogaster. Proc Natl Acad Sci USA. 1986;83:696–700. doi: 10.1073/pnas.83.3.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe AR, Hilliker AJ, Roberts PA. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics. 1993;134:1149–1174. doi: 10.1093/genetics/134.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AR, Gosden JR, Miller DA. A cloned sequence, p82H, of the alphoid repeated DNA family found at the centromeres of all human chromosomes. Chromosoma. 1985;92:369–377. doi: 10.1007/BF00327469. [DOI] [PubMed] [Google Scholar]
- Miyazaki WY, Orr-Weaver TL. Sister-chromatid cohesion in mitosis and meiosis. Annu Rev Genet. 1994;28:167–187. doi: 10.1146/annurev.ge.28.120194.001123. [DOI] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH. Interactions between the nod+ kinesin-like gene and extracentromeric sequences are required for transmission of a Drosophila minichromosome. Cell. 1995a;81:139–148. doi: 10.1016/0092-8674(95)90378-x. [DOI] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH. Localization of centromere function in a Drosophila minichromosome. Cell. 1995b;82:599–609. doi: 10.1016/0092-8674(95)90032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hare K, Alley MR, Cullingford TE, Driver A, Sanderson MJ. DNA sequence of the Doc retroposon in the white-one mutant of Drosophila melanogaster and of secondary insertions in the phenotypically altered derivatives white-honey and white-eosin. Mol Gen Genet. 1991;225:17–24. doi: 10.1007/BF00282637. [DOI] [PubMed] [Google Scholar]
- Ohashi H, Wakui K, Ogawa K, Okano T, Niikawa N, Fukushima Y. A stable acentric marker chromosome: possible existence of an intercalary ancient centromere at distal 8p. Am J Hum Genet. 1994;55:1202–1208. [PMC free article] [PubMed] [Google Scholar]
- Page SL, Earnshaw WC, Choo KH, Shaffer LG. Further evidence that CENP-C is a necessary component of active centromeres: studies of a dic(X; 15) with simultaneous immunofluorescence and FISH. Hum Mol Genet. 1995;4:289–294. doi: 10.1093/hmg/4.2.289. [DOI] [PubMed] [Google Scholar]
- Pardue ML. In: In situ hybridization to DNA of chromosomes and nuclei. Roberts DB, editor. Oxford: IRL Press, Ltd; 1986. [Google Scholar]
- Pimpinelli S, Berloco M, Fanti L, Dimitri P, Bonaccorsi S, Marchetti E, Caizzi R, Caggese C, Gatti M. Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc Natl Acad Sci USA. 1995;92:3804–3808. doi: 10.1073/pnas.92.9.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta AF, Mackay AM, Ainsztein AM, Goldberg IG, Earnshaw WC. The centromere: hub of chromosomal activities. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- Rattner JB. The structure of the mammalian centromere. Bioessays. 1991;13:51–56. doi: 10.1002/bies.950130202. [DOI] [PubMed] [Google Scholar]
- Ried T, Baldini A, Rand TC, Ward DC. Simultaneous visualization of seven different DNA probes by in situ hybridization using combinatorial fluorescence and digital imaging microscopy. Proc Natl Acad Sci USA. 1992;89:1388–1392. doi: 10.1073/pnas.89.4.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo VEA, Martienssen RA, Riggs AD. Cold Spring Harbor Monograph Series 32. Plainview, NY: Cold Spring Harbor Laboratory Press; 1996. Epigenetic mechanisms of gene regulation. [Google Scholar]
- Sacchi N, Magnani I, Fuhrman-Conti AM, Monard SP, Darfler M. A stable marker chromosome with a cryptic centromere: evidence for centromeric sequences associated with an inverted duplication. Cytogenet Cell Genet. 1996;73:123–129. doi: 10.1159/000134322. [DOI] [PubMed] [Google Scholar]
- Sandmeyer SB, Hansen LJ, Chalker DL. Integration specificity of retrotransposons and retroviruses. Annu Rev Genet. 1990;24:491–518. doi: 10.1146/annurev.ge.24.120190.002423. [DOI] [PubMed] [Google Scholar]
- Schulman I, Bloom KS. Centromeres: an integrated protein/DNA complex required for chromosome movement. Annu Rev Cell Biol. 1991;7:311–336. doi: 10.1146/annurev.cb.07.110191.001523. [DOI] [PubMed] [Google Scholar]
- Shevelyov YY. Aurora, a non-mobile retrotransposon in Drosophila melanogaster heterochromatin. Mol Gen Genet. 1993;239:205–208. doi: 10.1007/BF00281619. [DOI] [PubMed] [Google Scholar]
- Snyder MP, Kimbrell D, Hunkapiller M, Hill R, Fristrom J, Davidson N. A transposable element that splits the promoter region inactivates a Drosophila cuticle protein gene. Proc Natl Acad Sci USA. 1982;79:7430–7434. doi: 10.1073/pnas.79.23.7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner NC, Clarke L. A novel epigenetic effect can alter centromere function in fission yeast. Cell. 1994;79:865–874. doi: 10.1016/0092-8674(94)90075-2. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Schwartz S. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum Mol Genet. 1995;4:2189–2197. doi: 10.1093/hmg/4.12.2189. [DOI] [PubMed] [Google Scholar]
- Sunkel CE, Coelho PA. The elusive centromere: sequence divergence and functional conservtion. Curr Opin Genet Dev. 1995;5:756–767. doi: 10.1016/0959-437x(95)80008-s. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Larin Z, Tyler-Smith C. Analysis of extra-chromosomal structures containing human centromeric alphoid satellite DNA sequences in mouse cells. Chromosoma. 1996;105:70–81. doi: 10.1007/BF02509516. [DOI] [PubMed] [Google Scholar]
- Tower J, Karpen GH, Craig N, Spradling AC. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics. 1993;133:347–359. doi: 10.1093/genetics/133.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler-Smith C, Oakey RJ, Larin Z, Fisher RB, Crocker M, Affara NA, Ferguson-Smith MA, Muenke M, Zuffardi O, Jobling MA. Localization of DNA sequences required for human centromere function through an analysis of rearranged Y chromosomes. Nat Genet. 1993;5:368–375. doi: 10.1038/ng1293-368. [DOI] [PubMed] [Google Scholar]
- Vig BK. Do specific nucleotide bases constitute the centromere? Mutat Res. 1994;309:1–10. doi: 10.1016/0027-5107(94)90036-1. [DOI] [PubMed] [Google Scholar]
- Voullaire LE, Slater HR, Petrovic V, Choo KH. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am J Hum Genet. 1993;52:1153–1163. [PMC free article] [PubMed] [Google Scholar]
- White MJD. Animal Cytology and Evolution. Cambridge: Cambridge University Press; 1973. [Google Scholar]
- Williams B, Murphy TM, Goldberg M, Karpen GH. Neocentromere activity of structurally acentric minichromosomes in Drosophila. Nat Genet. 1998 doi: 10.1038/ng0198-30. in press. [DOI] [PubMed] [Google Scholar]
- Yuki S, Inouye S, Ishimaru S, Saigo K. Nucleotide sequence characterization of a Drosophila retrotransposon, 412. Eur J Biochem. 1986;158:403–410. doi: 10.1111/j.1432-1033.1986.tb09767.x. [DOI] [PubMed] [Google Scholar]
- Zinkowski RP, Meyne J, Brinkley BR. The centromere-kinetochore complex: a repeat subunit model. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]