Abstract
HIV-1 relies on a myriad of interactions with host cell proteins to carry out its life cycle. Traditional biochemical approaches to probe protein–protein interactions are limited in their ability to study the spatial and dynamic interactions that take place in the context of an intact cell. However, issues such as localization and dynamics of interactions between viral and host proteins can be well addressed utilizing fluorescent imaging methods. The past decade has brought about the development of many novel fluorescent imaging techniques which have proved useful to describe the interaction of HIV-1 proteins with the host cell.
1 Introduction
Encoding only 15 proteins itself, HIV has evolved to usurp many functions of its host cell in order to provide a suitable environment in which it may complete its life cycle. A myriad of interactions exist that redirect host cell functions in order to carry out a successful infection, as do interactions that have in turn evolved to combat HIV infection. In elucidating molecular targets to manipulate therapeutically, one must then consider not only the viral components but also the host cell interactions with these components that may be exploited to derail infection. Understanding this interplay between virus and host may pave the way for novel therapies to disrupt HIV replication.
The traditional manner of both identifying and characterizing protein–protein interactions is the use of biochemical techniques. Much has been uncovered using these techniques, but as the pandemic continues, new strategies for examining these interactions are desperately needed. The biochemical methods employed to probe protein–protein interactions require removing the proteins of interest from the likely relevant milieu around them as well as chemically disrupting the cells from which they are purified, modifications that can limit the physiological relevance of these complex interactions.
Proteins found interacting in biochemical assays are meaningful only if they also interact in vivo. While a protein found in the cytoplasm may be structurally capable of interacting with a nuclear protein, such a theoretical interaction between proteins residing in different subcellular neighborhoods is physiologically irrelevant. Thus, while biochemical studies are useful for characterizing populations of interactions in vitro, novel imaging methods are required to visualize these interactions as they naturally occur in living cells. The use of live cell imaging techniques allows for minimally invasive approaches to more accurately describe the dynamics of such interactions. For not only are we interested in describing the players in these interactions, but also the stage in which they take place.
The past decade has brought forth an abundance of innovative imaging techniques and an increasing pool of knowledge of HIV-1 interactions, and this review will attempt to highlight the most promising of these techniques, how they have impacted our knowledge base, and how they will continue to reveal details unable to be seen by any other means.
2 Techniques
Before understanding how novel imaging techniques have revealed aspects of HIV biology previously unseen, it is necessary to understand the theory of these techniques. The following section will describe the methodology behind a variety of imaging techniques, with an emphasis on ways in which they are useful in the study of HIV-host cell protein interactions. Before considering novel methodological approaches, an important consideration is the type of fluorescent microscopy to be utilized. Both confocal and deconvolution microscopy allow high resolution fluorescent imaging. Laser scanning confocal microscopy utilizes lasers and pinholes to illuminate a thin layer of the sample, providing a restricted region of illumination that can decrease out of focus fluorescence, generating a clearer image. Deconvolution microscopy uses computer-based image restoration to remove the out of focus light, allowing high resolution imaging. State of the art digital cameras, with highly sensitive and fast image capture, currently allow great flexibility for live cell imaging. The lasers associated with confocal microscopy generate higher levels of phototoxicity, making it less desirable for live cell microscopy. However, the use of a spinning disk confocal addresses this issue to a large degree. In the end, each has its advantage depending on the specific application to be utilized, and for the typical laboratory, access to local equipment often dictates the choice of equipment.
2.1 Colocalization versus Interaction
A common way to demonstrate the interaction of HIV proteins with cellular proteins or compartments is to show that the localization of two or more proteins overlaps within cells. This analysis can be complicated by the fact that the interaction may be transient, and taking a steady state snapshot of the localization of the protein within cells may reveal that only a subset of the populations being visualized in a cell have overlapping expression. Such interactions can be explored further in living cells by utilizing a number of techniques which can provide important insights into the dynamics of such transient interactions. Interpreting image-based analysis can also be confounded if an HIV protein and a cellular protein have overlapping expression, caused not via direct interaction but by both being localized to the same specific cellular compartment. Therefore, the interpretation of observations relating to the interaction of HIV proteins with cellular proteins can be complex. At the same time, these issues can be resolved with controls and appropriate supporting studies.
The most common approach to determining the localization and the potentially overlapping expression of proteins within cells is immunofluorescence microscopy. Typically, the analysis of potential interactions of HIV proteins and cellular proteins begins with the fluorescent imaging of fixed cells. One strength of fixed cells is their higher resolution when compared to live cells, though the method chosen for fixation can be critical to the outcome of this type of analysis because of potential artifacts caused by certain techniques. Simply staining expressing cells with fluorescently labeled antibodies to the proteins can provide initial information. If antibodies to the proteins of interest are not available, then an expression vector for the protein of interest fused to certain small peptides allows immunostaining with antibodies specific to the peptide tag. Alternatively, an expression vector for the protein of interest fused to a fluorescent protein can be utilized. However, such fluorescent fusion proteins must be carefully validated to ensure that the relatively large tag does not perturb the localization or function of the linked protein of interest. Advantageously, such fluorescent protein fusions can be used in live cell studies to provide insights into the dynamics of potential protein interactions.
One disadvantage of the use of such tagged proteins is that their expression is not likely subject to the regulatory constraints imposed on the endogenous gene. Normal regulation of promoter function or posttranscriptional aspects of regulation are lost in typical expression vectors. Over- or underexpression of the protein under study can alter the normal physiology of virus–host protein interactions. Such issues may be addressed in the future using highly-efficient homologous recombination to introduce the desired tag into the targeted protein in the natural chromosomal location (Lombardo et al. 2007).
The use of fluorophore-tagged viral proteins and interactors is a widely utilized method, and with novel fluorophores continually being added to the repertoire, the investigation of the dynamic nature of interactions can be performed more completely. Though the green fluorescent protein (GFP) from the Aequorea victoria jellyfish is still routinely used 14 years after its development (Rizzuto et al. 1995), novel fluorophores with the ability to change colors at the whim of the investigator are making more questions answerable with fluorescence microscopy. Such fluorophores include photoactivatable, photo-convertible, and photo-switchable variants, the use of which allows for pulse-chase type experiments where movement and localization changes can be tracked over time, and thus are useful for studying the dynamics of interactions.
Photoactivatable fluorophores are unresponsive to a given excitation wavelength until they are activated by a brief burst of a specific wavelength of light, after which they behave similarly to their nonphotoactivatable counterparts (Patterson and Lippincott-Schwartz 2002). By selectively activating a very small region within the cell, a specific population of a protein can be tracked over time. As a virion will interact with proteins in a very particular region of the cell, researchers can concentrate on the molecules truly involved in these interactions, no matter how dynamically they move about the cell.
One drawback of photoactivatable fluorescent proteins is the inability to visualize the protein before activation. One solution to this problem is the use of photo-convertible or photo-switchable fluorescent proteins. These fluorophores possess two distinct nonoverlapping fluorescence profiles, enabling the visualization of the entire population of a protein within a cell and selective activation and tracking of a subset of that protein (Gurskaya et al. 2006). Photo-switchable fluorescent proteins have the added benefit of being able to not only switch the fluorescence profile one time, but also to reversibly switch it back to its original state (Ando et al. 2004).
Despite the advancements in fluorescent protein technology, analysis of interactions by visualization alone can be problematic. One problem with interpreting such data is that while colocalization can signify an interaction, it can also simply mean that the proteins occupy adjacent locations; resolving these discrepancies remains a problem with traditional microscopy methods.
2.2 Probing Association by Energy Transfer
Fluorescent imaging analysis of proteins brings to light a wealth of information regarding the subcellular localization and the dynamic movements of proteins, but more objective methods are necessary to definitively label this colocalization as an interaction.
Other techniques utilizing fluorescent proteins can probe the association of viral proteins in more detail. The transfer of energy from a fluorescent protein of one color to a nearby fluorescent protein of another color can reveal information about the distance separating the two proteins.
2.2.1 FRET
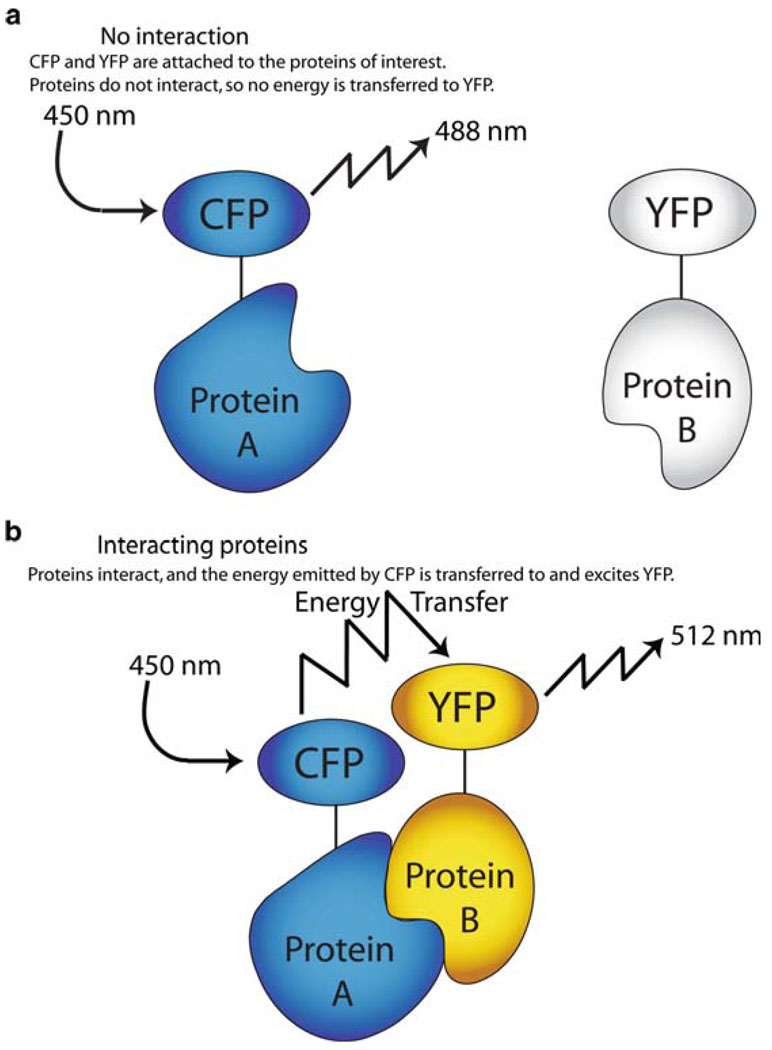
Though microscopic studies analyzing colocalization can provide much insight into viral and host cell interactions, distinct fluorescence-based methods are capable of focusing on determining real interactions. Fluorescence resonance energy transfer (FRET), for example, relies of the proximity of two proteins to produce a fluorescence signal. The distance at which a transfer of energy between two proteins can occur (<10 nm) is so miniscule that it may be interpreted as an interaction (Stryer 1978). By conjugating putative interacting proteins to a specific pair of fluorophores, an interaction can be determined. Cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) is a popular pair for FRET due to their partially overlapping fluorescence profiles. The wavelength of light emitted by CFP is the precise wavelength required to excite YFP, a fact conveniently exploited in FRET. If this pair of fluorescent proteins is sequestered in close proximity via some interaction of fusion partners, exciting the sample with the CFP wavelength will produce a YFP emission signal (Fig. 1). In addition to this popular pair, the mCherry and GFP pair is becoming more widely used as this combination is more photostable (van der Krogt et al. 2008).
Fig. 1.
Fluorescence resonance energy transfer (FRET). (a) No interaction. CFP and YFP are attached to the proteins of interest. Proteins do not interact, so no energy is transferred to YFP. (b) Interacting proteins. Proteins interact, and the energy emitted by CFP is transferred to and excites YFP
2.2.2 BiFC
An alternative to FRET is bimolecular fluorescence complementation (BiFC), a technique invaluable for probing interactions. BiFC relies on the production of a fluorescent signal when two interacting proteins attached to complementary halves of GFP come together, complementing the structure of GFP and producing fluorescence (Fig. 2). By separating GFP into halves, neither the N-terminal nor the C-terminal portion is capable of emitting radiation on its own. However, if the proteins to which they are conjugated interact, this will bring the halves close enough to each other to take on the conformation of native GFP and emit fluorescence as usual.
Fig. 2.
Bifluorescence complementation (BiFc). (a) No interaction. GFP is split into two halves which cannot fluoresce alone and attached to the proteins of interest. Proteins do not interact, so no fluorescent signal is produced. (b) Interacting proteins. Proteins interact, allowing the two halves of GFP to come together and produce a fluorescent signal
2.3 Improving Imaging Conditions by Increasing Sensitivity and Resolution
Despite the advances in fluorescence light microscopy, there are limitations of fluorescent signal detection and diffraction-limited spatial resolution. Background fluorescence and emission filters make the detection of especially weak signals problematic. Likewise, the highest resolution of light microscopy attainable by traditional optical techniques is approximately 200 nm (Richard 2003), making it impossible to determine association from proximity. It is important to define resolution in this case, and this resolution limit means that it is not possible to identify two distinct fluorescent objects unless they are separated by at least 200 nm. New subdiffraction techniques such as stimulated emission depletion (STED) and structured illumination allow imaging at much higher resolution (Klar et al. 2000 and Bailey et al. 1993), but highly specialized equipment is required. Below, we describe alternative methods allowing molecules located at subdiffraction distances to be resolved. Photoactivated localization microscopy (PALM) uses photoactivation and photobleaching to beat the limits, while total internal reflection fluorescence (TIRF) microscopy uses restricted illumination to improve axial resolution and decrease background, allowing the detection of weak signals.
2.3.1 TIRF
One notable advance in fluorescence imaging is total internal reflection fluorescence (TIRF) microscopy. Traditional epifluorescence microscopy illuminates more of the sample than an optical section will capture, creating out of focus fluorescence that causes unnecessary background. By decreasing the portion of the sample in the range of the excitation wavelength, the signal-to-noise ratio is increased, greatly increasing the sensitivity of fluorescent detection and allowing the detection of individual fluors.
TIRF exploits optical principles in order to illuminate only the area of the sample that will be analyzed. The principle works by adjusting the system so that the excitation light passing through the glass coverslip reaches a critical angle, causing all of the light to reflect back into the medium from which it came. On the other side of this medium, an evanescent wave is created that extends up to 100 nm into the sample (Fig. 3) (Axelrod et al. 1984). By focusing the light on such a thin region of the sample, this limits the amount of background fluorescence and thus creates greater sensitivity. This technique is especially useful when interest lies in a comparatively weak signal, which by traditional optical techniques may be masked by stronger signals in a region of the cell that one wishes to ignore. However, the weakness of this method is that the objects of interest must be localized within 100 nm of the coverslip.
Fig. 3.
Total internal reflection fluorescence (TIRF) microscopy. Approaching light at the critical angle undergoes total internal reflection back through the glass coverslip An evanescent wave is created on the other side of the glass, extending approximately 100 nm into the sample. As only this extremely thin region is illuminated, background fluorescence from out-of-focus light is dramatically decreased, creating greater sensitivity
The thin region of illumination by the evanescent field is adjustable by altering the critical angle. Modulating the penetration of the evanescent field allows increased resolution in the z-plane, and additional resolution in the z-plane can be obtained by switching between epifluorescence and TIRF. In this way, it is possible to determine when a signal enters or exits the evanescent field, allowing its proximity to the coverslip to be determined. While not a method designed specifically to probe interactions, the increased spatial resolution attainable with TIRF microscopy lends itself to examining colocalization of proteins more accurately.
2.3.2 PALM
A more recent approach for obtaining high-resolution images is photoactivated localization microscopy (PALM). This technique is in theory capable of resolving molecules separated by only a few nanometers, making it an incredibly powerful technique to visualize the spatial and temporal intricacies of certain interactions. In essence, PALM does not increase optical resolution, but instead allows measurement of subdiffraction distances between fluorescent signals.
Though the theory behind PALM has existed for over a decade, the ability to exploit it for high-resolution imaging of fluorescent molecules is still in its infancy. Within a specific diffraction-limited region lie thousands of individual molecules that cannot be visualized separately. PALM is capable of determining the location of a large number (105/µm2) of these molecules by the use of sequential photo-activation and subsequent photobleaching of subsets of fluorescent molecules within a specific field (Betzig et al. 2006). After this information is obtained, the probability that each molecule resides in a particular location is statistically estimated based on the point spread function of the optical path. Identifying the center of each signal allows the distance between them to be determined. Of more interest to studies of interactions, a dual-color version of PALM has been developed capable of providing superresolution images of two labeled proteins. One notable achievement with this technology is the demonstration that proteins viewed as clearly colocalized by conventional microscopic techniques have been revealed by PALM as very close but distinct clusters of molecules (Shroff et al. 2007). Such a technique should prove valuable for deciphering putative interactions between viral and host cell proteins.
Going even further down the road towards the goal of nanometer resolution is the amalgamation of PALM and single-particle tracking into a method entitled sptPALM (Manley et al. 2008). While single-particle tracking can provide information on the dynamic nature of molecules, it is still subject to diffraction-limited resolution. When using this method to study events occurring on the plasma membrane, the proximity of clusters of molecules renders the interpretation of these paths uncertain. The great spatial resolution of sptPALM, on the other hand, is capable of enabling visualization of the dynamic nature of the plasma membrane by tracking the behavior of thousands of individual molecules. Moreover, the ability to selectively photoactivate populations of molecules allows the determination of multiple trajectories of individual molecules within a very small region of the cell.
3 Examples
Innovative imaging techniques have been used to probe many stages of the HIV life cycle and, as novel methods arise, they will undoubtedly be adapted to visualize interactions that currently seem impossible. A selection of key studies that have utilized these techniques will be highlighted below using examples progressively delving into each stage of the viral life cycle (Fig. 4). As there is an immense wealth of innovative studies of this topic, it is impossible to cite every contribution.
Fig. 4.
HIV-1 Life Cycle
3.1 Entry
HIV enters a target CD4+ T cell after the envelope glycoprotein gp120 binds the CD4 receptor, allowing gp120 to interact with either a CCR5 or CXCR4 coreceptor. After a conformational change exposes the hydrophobic fusion peptide, fusion between the viral and host cell membranes occurs. Upon fusion of the viral envelope to the plasma membrane of the host cell, the viral core is deposited into the host cell cytoplasm. The capsid is lost from the virion in the uncoating process, and reverse transcription converts the viral RNA genome into double-stranded DNA. The preintegration complex containing newly synthesized DNA along with several viral and host cell proteins enters the nucleus and integrates into the host cell genome, establishing its everlasting presence (Freed 2001). This process of entry relies on numerous interactions with host cell proteins to achieve the goal of gaining access to the host cell cytoplasm and eventually nuclear DNA. Information regarding the localization of these interactions can be readily gained by exploiting the fluorescence methods described in the previous section.
3.1.1 Mobile Receptors
Normally the HIV entry receptors form locally enriched domains separated by approximately 10 nm, a distance small enough to appear colocalized by simply monitoring their locations by traditional fluorescence microscopy. The envelope gp120 induces interactions between the receptors, but the resolution of light microscopy limits our ability to see any difference. FRET has been used to explore the interaction of receptors due to the presence of virus. Yi et al. (2006) took advantage of the sensitivity of FRET to evaluate the effect of gp120 on interactions between CD4 and CCR5. FRET enables the determination of very closely adjacent or truly interacting molecules, clearing up this ambiguity.
While the receptors normally form separate microdomains on the plasma membrane, complexes including gp120 and the receptors form during HIV entry. Previous studies had inconclusively implicated lipid rafts in entry complex formation, with fluorescence microscopy studies offering conflicting evidence on whether CD4 and CCR5 localization to lipid rafts is important for entry complex formation and infection. A complication in visualizing these interactions is that lipid rafts are dynamic structures less than 70 nm in diameter, well below the resolution of light microscopy. Utilizing FRET to examine interactions between CD4 and CCR5 enabled Yi et al. to assess the importance of lipid rafts on receptor interactions. Using CD4-YFP and CCR5-CFP as a FRET pair, they found that gp120 can bring CD4 and CCR5 together on the plasma membrane of live cells. Because FRET will occur only at miniscule distances, this eliminates complicated interpretations of colocalization and gives a more objective measure of association. Additionally, the authors found that chemically disrupting lipid rafts blocked this gp120-induced FRET signal, and that adding back cholesterol restored it. Studies such as these not only have the potential to visualize an interaction, but to quantify the impact of inhibitory compounds on interactions between virus and host.
3.1.2 Hitching a Ride
The revolution in HIV imaging began with the ability to visualize a virion interacting with living cells. The exploitation of live cell imaging within the HIV field started with the creation of a fluorescently labeled virus whose entry into the host cell and early interactions inside it could be followed under the microscope.
The first observation of HIV in living cells was made possible by attaching a fluorescent protein to the HIV accessory protein Vpr. In 2002, McDonald et al. observed HIV entering cells and hijacking the microtubule network to enable its voyage towards the nucleus (McDonald et al. 2002; Fig. 5). As well as establishing a key method enabling the visualization of HIV entry, this study also highlighted the dependence on the host cell cytoskeleton in order to reach the viral genome’s ultimate destination in the nucleus.
Fig. 5.
GFP-Vpr can be used to visualize HIV-1 trafficking within living cells
Though this remains a valuable method for studying interactions, several limitations are inherent in such an approach. First, Vpr is apparently lost before nuclear entry, limiting the portion of the HIV life cycle that can be visualized with this label. Second, it is not clear whether all virions within the cell have entered via fusion with the cell membrane or have been nonspecifically endocytosed. Finally, the problem of interpreting colocalization cannot be resolved without complementary biochemical techniques.
3.1.3 Discriminating Entry from Endocytosis
One improvement in labeling virions is the use of the 15 N-terminal residues of Src (S15), a signal incorporated into virions that is lost after fusion but not after nonproductive receptor-independent endocytosis. S15 targets the plasma membrane, and this sequence is incorporated into virions (Campbell et al. 2007). Using a fluorescently tagged version of S15 (S15-mCherry) together with GFP-Vpr allows a double label that provides a clear picture of a virion that has entered the host cell via fusion. Being able to visualize a productive entry event allows interactions within the cell not relevant to productively entered virions to be discriminated and ignored.
This dual-labeled system was used again by Campbell et al. (2008) to visualize the retroviral restriction factor rhTRIM5α interacting with cytoplasmic HIV complexes. The virions observed associating with TRIM5α complexes were specific to GFP-Vpr-labeled particles that had lost the S15 membrane label, indicating that they had entered the host cell cytoplasm by fusion. By being able to identify viral complexes that had entered the cytoplasm after fusion, it was possible to validate that the interaction with TRIM5α had the appropriate specificity.
3.1.4 Journey to the Center of the Nucleus
Despite the utility of GFP-Vpr labeling, one severe limitation of this approach is that it is only possible to visualize early postentry events, leaving any nuclear movements and interactions of the intracellular HIV complex to remain a mystery. One step in the direction of looking further into the nuclear aspects of the HIV life cycle has been achieved with the labeling of integrase. Arhel et al. (2006) reported a FlAsH-tagged integrase that enables the visualization of intranuclear movements. FlAsH (fluorescein arsenical hairpin) is a reagent that specifically binds to a small tetracysteine sequence that may be inserted into a target protein. One virtue of the FlAsH system compared to traditional fluorescent protein labeling is that the sequence added is much smaller than a fluorescent protein like GFP, and as such may be less prone to perturbing the structure and function of the protein to which it is added. The use of this tag on the HIV integrase protein allowed the observation of the kinetics of movement towards the nucleus, supporting both microtubule- and actin-dependent movement. With this system, HIV complexes apparently located in the nucleus exhibited restrained diffuse movement.
Potentially improving on the FlAsH integrase tag to visualize virions trafficking within the nucleus, a subsequent study by Albanese et al. (2008) utilized a fluorophore-tagged integrase (IN) in trans in order to achieve this goal. Because inserting sequences within the proviral sequence often causes processing problems, they exploited the fact that Vpr is incorporated into virions by attaching Vpr to a fluorescently tagged IN. This was done in such a way that, in the presence of HIV protease, cleavage separating Vpr from IN-EGFP will occur. Thus, the Vpr-IN-EGFP was expressed in trans with a proviral construct containing an IN deletion, allowing the production of a virion with fluorescently labeled IN.
Rather than investigate interactions of HIV with host cell proteins, Albanese et al. examined the relationship with host cell DNA. The study of integration sites has revealed that HIV has a preference to integrate into actively transcribed genes. One open question is what determines this preference. Their findings reveal that virions tend not to travel far after entering the nucleus, and that a preference is seen for areas of decondensed chromatin. This work not only begins to reveal what areas of the genome HIV associates with, but opens many doors to further investigate the interactions occurring within the nucleus.
3.2 Exit
After integrating its reverse transcribed DNA into the genome of its host cell, this provirus acts as the template from which RNA is transcribed. Viral proteins are synthesized and virions are assembled at the plasma membrane, where these immature virions bud from the infected cell (Freed 2001) with the help of the host cell’s endosomal sorting complex required for transport (ESCRT) machinery. The nascent virions undergo maturation and then go on to infect other target cells. Imaging techniques have provided important insights into the interactions and dynamics leading to new virion assembly.
3.2.1 Correlative Imaging: The Best of Both Worlds
As fluorescently tagged virions enable the study of entry, other methods must be employed to visualize egress of budding virions. One innovative technique used to resolve this difficulty is correlating high-resolution electron microscopy with fluorescent imaging capable of visualizing molecules within the interior of a cell. This approach was used by Larson et al. (2005) in order to visualize retroviral budding. The authors utilized live cell multiphoton laser scanning microscopy (MPM) of transfected Gag and subsequent scanning electron microscopy (SEM) in order to combine the strengths of both methods. Namely, SEM can resolve single budding structures but can only be used for visualizing bulges on the surface, and fluorescence imaging can look within the cell but can only infer budding from the disappearance of a fluorescent signal. As both imaging methods are powerful techniques with their own strengths and weaknesses, the information gained by the combination is invaluable.
Studies of single particles entering the cell have been carried out, but this had not yet been accomplished for budding. Larson et al. visualized real-time budding of single HIV-1 or Rous Sarcoma Virus (RSV) virus-like particles (VLPs) from live cells and correlated their fluorescent results to those seen by SEM. A fraction of punctate Gag spots seen by fluorescent microscopy was found to correspond to budding structures on the plasma membrane as visualized by SEM, confirming the interpretation as sites of budding. Such a method of correlating the same budding structures as seen by two different methods should prove useful in future studies.
3.2.2 ESCRTing Through Microdomains
The use of basic fluorescence microscopy has aided in evaluating the role of previously noted biochemical interactions. For example, while characterization of highly purified HIV particles had already suggested a potential interaction between the tetraspanin CD63 and HIV Gag, the functional importance of this interaction was not understood. Simply staining for CD63 showed that only a small portion of CD63 was present on the plasma membrane, while the majority was located on intracellular membranes (Nydegger et al. 2003). The next step in investigating this interaction was to selectively stain surface CD63, which revealed that it was present in clustered microdomains rather than being uniformly distributed throughout the plasma membrane (Nydegger et al. 2006).
Aside from traditional microscopy, Nydegger et al. also used electron microscopy (EM) to complement their light microscopy findings. Moreover, by combining the real-time fluorescent imaging of live cells with high-resolution immuno-EM, the size of these microdomains could be appraised. Immuno-EM also enabled the authors to see that tetraspanin-enriched microdomains (TEMs) formed near clathrin-coated areas and cytoskeletal elements. Furthermore, they showed that ESCRT1 components Tsg101 and Vps28, already shown to be required for budding, are recruited to TEMs colocalizing with Gag.
3.2.3 Shared Machinery
Though the budding of HIV-1 virions has been overwhelmingly examined structurally, there is certainly a role for fluorescence microscopy. One demonstration of the strength of traditional colocalization studies is the discovery of shared cellular machinery for viral budding and cytokinesis. Carlton and Martin-Serrano (2007), spurred on by yeast two-hybrid studies revealing interactions of components of the ESCRT and cytokinesis machinery, used fluorescence microscopy to study the localization of these putative interactions.
ESCRT proteins Tsg101 and Alix both bind the centrosomal protein Cep55 involved in cytokinesis. When Cep55 expression is disrupted with siRNA, Tsg101 and Alix no longer exhibit their usual localization. They went on to map the residues of Tsg101 responsible for binding Cep55 and found that, upon deletion, the same aberrant localization occurred. After finding this striking example of a virus usurping the cellular machinery of a function as basic as cell division, they discovered that the Tsg101-Cep55 interaction is required for cytokinesis but not for viral budding. By studying the differential requirements of components involved in requisite cellular functions and exploited for viral replication, it may be possible to separate out specific interactions that may be disrupted for antiviral therapy while preserving necessary cellular functions.
3.2.4 Dynamics of Budding VLPs
Another method for the visualization of assembly and egress is TIRF. Jouvenet et al. (2008) combined TIRF imaging with FRET and fluorescence recovery after photobleaching (FRAP), more objective methods of determining interactions, in order to explore viral budding. TIRF allowed the authors to visualize budding of Gag VLPs in real-time, a difficult task with epifluorescence as the more prominent signal from cytoplasmic Gag overpowers the weak signal from budding VLPs. As TIRF images regions of the sample within approximately 100 nm of the coverslip, a study of budding from the plasma membrane can benefit greatly from this approach.
In an attempt to characterize the kinetics of budding, the authors first noticed two populations of Gag: slowly appearing and rapidly appearing/disappearing. In order to explore the relevance of these two populations, the authors looked at CD63 as an endosomal marker and correlated these results to the speed at which Gag puncta appeared in order to focus only on real assembly events taking place at the plasma membrane. They found that the majority of the rapidly appearing/disappearing population stained positively for both CD63 and clathrin, implying that this population is associated with endosomes. Thus, they chose to focus on the slowly appearing population representative of budding at the plasma membrane. This use of TIRF allowed the authors to characterize kinetically variant populations based on previously characterized interactions, a feat unthinkable by traditional fluorescence. They also used FRET to interpret the population of slowly emerging particles as Gag molecules moving closer together, and thus undergoing assembly into VLPs. A similar FRET-based assay has been previously utilized to study oligomerization of Gag during the process of assembly, both within living cells and VLPs (Derdowski et al. 2004). Though the Jouvenet study focused primarily on the dynamics of budding, it should prove to be a promising approach to characterize interactions occurring during this process, such as the incorporation of cellular proteins and viral RNA into budding virions.
3.2.5 Superresolution Budding
Betzig et al. (2006) reported a use of PALM in which localization on the scale of nanometers was achieved in high-resolution imaging of Gag at the plasma membrane. PALM combined with TIRF is well suited for the study of proteins at the plasma membrane, such as budding Gag VLPs. This study combined PALM with TIRF microscopy in order to decrease autofluorescence and thus increase the resolution. Though this method requires up to 12 h to obtain a superresolution image, the detail obtained may outweigh the lengthy process for certain questions. As a dual-color version of PALM has been shown to provide resolution on the scale of 20–30 nm, the utilization of such a technique in the future seems to be quite valuable for probing interactions between virion and host cell.
The combination of PALM with single-particle tracking provides the means to investigate the dynamics of molecules in high resolution, opening up many doors to visualize interactions and determine how they are affected in real time at the single molecule level. Manley et al. (2008) use this combined sptPALM technique to track Gag molecules at the plasma membrane at an incredibly detailed resolution, as well as to follow the motion of many subsets of particles by selectively photoactivating them. One interesting finding from this study is that an immobile portion of Gag appeared stuck on the plasma membrane, an observation that the authors postulate may be due to interactions with tetraspanin-enriched microdomains (TEMs). Thus, differences in the dynamics of Gag may be further explored by the use of dual-colored PALM in order to track interactions with host cell proteins.
3.2.6 Connecting Exit and Entry
Dendritic cells have the ability to bind HIV without becoming productively infected, a strange phenomenon that both contributes to an immune response and at the same time is yet another host cell function that has been pirated by HIV. Aside from binding and degrading HIV virions and producing an antibody response, dendritic cells can also sequester virions and transfer them to T cells in a process called trans-infection, and thus contribute to establishing a systemic infection. One unresolved question has been where exactly the dendritic cell stores the virus before passing it off to a T cell. The exosome model proposes storage of virions in a protected compartment within the cell, with trans-infection occurring only when fusion with the plasma membrane brings the virions back to the surface. Recent results demonstrated an opposing model in which internalized virions are degraded by the lysosome and only surface-accessible virions are able to cause trans-infection (Cavrois et al. 2007).
While evidence existed supporting both models, Yu et al. (2008) used live cell microscopy to resolve the discrepancies between these seemingly contradictory models. First the authors characterized the subcellular components localized within the internal HIV compartment, finding that the compartment colocalizes with actin, and subsequently that formation of it requires the actin cytoskeleton. In order to characterize this compartment, they stained for various cell surface and intracellular markers, finding that tetraspanin CD81 was recruited from its uniform distribution on the cell surface to form a concentration in the compartment. Aside from determining subcellular localization, they used fluorescence microscopy to investigate the time span of HIV sequestration within the compartment, finding that it correlates with the length of time in which trans-infection can be detected. The use of the combined Vpr-S15 virion label was used in order to determine which virions had entered the cytoplasm and thus to visualize trans-infection. The virion compartment was accessible to both a surface-applied inhibitor and a fluid phase marker, indicating that this compartment was intracellular but nevertheless connected to the surface. Their surprising conclusion was that virions are stored in an internal compartment, but that this compartment is a pocket-like structure contiguous with the plasma membrane that remains surface-accessible. The use of simple antibody staining, combined with the power of live cell microscopy to study the dynamics of a subcellular compartment, allowed a more complete view of opposing models of trans-infection.
A similar mechanism has been observed for the transfer of HIV from an infected cell to a target cell. This configuration is called the virological synapse (reviewed in Jolly and Sattentau 2004). Imaging techniques have played an essential role in revealing the function of the virological synapse and addressing the long standing mystery of why infected cells are much more efficient at transmitting HIV relative to cell-free particles. Such interactions can be sites of direct contact of the plasma membranes of the two cells, or mediated by membrane projections such as cytonemes and nanotubes (Sherer et al. 2007; Sowinski et al. 2008). In all cases, the increase in infectivity is stimulated by concentrating the virus from the infected cell in close proximity with the receptors required for fusion with the target cell. It is difficult to imagine how this important aspect of HIV biology could be studied without the assistance of the investigative power of imaging techniques.
3.3 Regulatory and Accessory Proteins
As well as the structural components of HIV that interact with host cell molecules to exert an effect on infection, the regulatory and accessory proteins of HIV play their own roles in creating a more hospitable environment. Several examples describing the visualization of these interactions are mentioned below.
3.3.1 Tat
HIV depends on regulation of transcription to maintain the viral reservoir, a population of cells with integrated provirus that do not actively produce HIV. The transactivator of transcription (Tat) protein of HIV boosts production of viral RNA synthesis through its interaction with the trans-activation-responsive region (TAR) of the viral RNA, as well as interactions with cellular components. Tat carries out this role through its interaction with positive transcription elongation factor P-TEFb. Tat has been shown to interact with P-TEFb kinase component cyclin T1 in a complex with TAR at the HIV promoter. While the interaction between Tat and cyclin T1 was first recognized biochemically, little was known of its specifics and consequences. A method such as FRET is ideal for visualizing putative biochemical interactions and determining their physiological relevance, the use of which enabled Marcello et al. (2001) to find that Tat influences the subcellular localization of cyclin T1.
Cyclin T1 normally localizes to nuclear foci, while Tat typically resides diffusely throughout both the nucleoplasm and the nucleolus. By simply expressing fluorescently tagged versions of both proteins, they found that a reorganization of both proteins occurs when coexpressed. The authors found that Tat induced cyclin T1 to relocalize from its characteristic accumulations in the nucleus to the sites of transcription and, reciprocally, Tat lost its diffuse localization to accumulate at these same locations. The combination of analyzing changing subcellular location along with FRET to ascertain whether proteins are interacting gives insight into the function of this interaction. Not only do they note that two proteins are located in proximal locations, but this localization is only seen when they are interacting.
Though the interaction of Tat with cyclin T1 and its requisite role in activating transcription were already known, a model had not yet been described to resolve current data. The question was whether P-TEFb remained in association with the transcription complex after initiation of transcription, or if it dissociated prior to elongation.
Delving into the dynamics of this interaction with an interesting method of visualizing specific RNAs within the cell, Molle et al. (2007) explore the kintetics of the association of Tat and cyclin T1. In order to visualize HIV-1 transcription sites in live cells, they tagged the HIV-1 proviral construct with binding sites for phage MS2, and expressed this along with GFP-tagged MS2. Only when the HIV-1 proviral construct associates with GFP-tagged MS2 will any fluorescence be seen. This system allowed them to visualize the site of transcription as well as to the monitor the localization of Tat and cyclin T1 at this site. They found that Tat must interact with cyclin T1 in order to accumulate at transcription sites. Additionally, the use of FRAP allowed them to see that Tat recovers slowly at transcription sites, bringing up the possibility that Tat may dissociate before complete transcription. The strength of this study is that it used dynamic data from living cells in order to resolve conflicting models.
3.3.2 Nef Oligomerization
The HIV regulatory protein Nef is required for HIV infection to progress into AIDS. Nef is involved in downregulation of CD4 and MHC-I from the cell surface and enhanced infectivity (Freed 2001), roles it carries out through its interactions with various cellular proteins. Its interaction with Hck, a protein-tyrosine kinase, appears to be involved in pathogenesis of HIV. Ye et al. (2004) use BiFC to show that Nef oligomerizes in living cells, a prerequisite for in vivo activation of Hck. Just as BiFC has been used in this study to investigate interactions between Nef molecules that contribute to interactions with cellular molecules, it should be a promising method to reveal interactions between Nef with host cell proteins. Other studies have used BiFC to explore the potential interaction between retroviral Gag and actin (Chen et al. 2007).
4 Conclusions
There is currently a revolution taking place in the identification of cellular factors involved with HIV replication. Using libraries of siRNAs and shRNAs to knock-down cellular factors, multiple groups have identified hundreds of new factors that may interact with HIV proteins and play an essential role in HIV biology. This list will only get longer in the near future. The variety of techniques currently used to visualize interactions between HIV-1 and cellular proteins can provide an extraordinary wealth of information with regard to the functionality of such interactions. Imaging has the potential to reveal subtleties not visible by other techniques, such as the localization and dynamics of interactions. Fluorescent imaging provides a subset of techniques complementary to the standard biochemical methods, and when combined they present a powerful means to unmask complex questions. With high resolution imaging improving quickly and with different techniques suitable to probe different stages of the viral life cycle, today’s scientists are well equipped to answer questions of how HIV-1 proteins relate to the host cell environment, setting up the basis to utilize this knowledge therapeutically.
References
- Albanese A, Arosio D, Terreni M, Ceresoto A. HIV-1 pre-integration complexes selectively target decondensed chromatin in the nuclear periphery. PLoS ONE. 2008;3(6):e2413. doi: 10.1371/journal.pone.0002413. doi:10.1371/journal.pone.0002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- Arhel N, Genovesio A, Kim K, Miko S, Perret E, Olivo-Marin J, Shorte S, Charneau P. Quantitative four-dimensional tracking of cytoplasmic and nuclear HIV-1 complexes. Nat Methods. 2006;3:817–824. doi: 10.1038/nmeth928. [DOI] [PubMed] [Google Scholar]
- Axelrod D, Burghardt TP, Thompson NL. Total internal reflection fluorescence. Annu Rev Biophys Bioeng. 1984;13:247–268. doi: 10.1146/annurev.bb.13.060184.001335. [DOI] [PubMed] [Google Scholar]
- Bailey B, Farkas DL, Taylor DL, Lanni F. Enhancement of axial resolution in fluorescence microscopy by standing-wave excitation. Nature. 1993;366:44–48. doi: 10.1038/366044a0. [DOI] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Campbell EM, Perez O, Melar M, Hope TJ. Labeling HIV-1 virions with two fluorescent proteins allows identification of virions that have productively entered the target cell. Virology. 2007;360:286–293. doi: 10.1016/j.virol.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EM, Perez O, Anderson JL, Hope TJ. Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5α. J Cell Biol. 2008;180:549–561. doi: 10.1083/jcb.200706154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- Cavrois M, Neidleman J, Kreisberg JF, Greene WC. In vitro derived dendritic cells trans-infect CD4 T cells primarily with surface-bound HIV-1 virions. PLoS Pathog. 2007;3(1):e4. doi: 10.1371/journal.ppat.0030004. doi:10.1371/journal.ppat.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Jin J, Rubin M, Huang L, Sturgeon T, Weixel KM, Stolz DB, Watkins SC, Bamburg JR, Weisz OA, Montelaro RC. Association of gag multimers with filamentous actin during equine infectious anemia virus assembly. Curr HIV Res. 2007;5:315–323. doi: 10.2174/157016207780636542. [DOI] [PubMed] [Google Scholar]
- Derdowski A, Ding L, Spearman P. A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates gag-gag interactions. J Virol. 2004;78:1230–1242. doi: 10.1128/JVI.78.3.1230-1242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO. HIV-1 replication. Somat Cell Mol Genet. 2001;26:13–33. doi: 10.1023/a:1021070512287. [DOI] [PubMed] [Google Scholar]
- Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- Jolly C, Sattentau QJ. Retroviral spread by induction of virological synapses. Traffic. 2004;5:643–650. doi: 10.1111/j.1600-0854.2004.00209.x. [DOI] [PubMed] [Google Scholar]
- Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar TA, Jakobs S, Dyba M, Egner A, Hell SW. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc Natl Acad Sci USA. 2000;97:8206–8210. doi: 10.1073/pnas.97.15.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Johnson MC, Webb WW, Vogt VM. Visualization of retrovirus budding with correlated light and electron microscopy. Proc Natl Acad Sci USA. 2005;102:15453–15458. doi: 10.1073/pnas.0504812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, Holmes MC, Naldini L. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, Betzig E, Lippincott-Schwartz J. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- Marcello A, Cinelli RAG, Ferrari A, Signorelli A, Tyagi M, Pellegrini V, Beltram F, Giacca M. Visualization of in vivo direct interaction between HIV-1 TAT and human cyclin T1 in specific subcellular compartments by fluorescence resonance energy transfer. J Biol Chem. 2001;276:39220–39225. doi: 10.1074/jbc.M104830200. [DOI] [PubMed] [Google Scholar]
- McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle D, Maiuri P, Boireau S, Bertrand E, Knezevich A, Marcello A, Basyuk E. A real-time view of the TAR:Tat:P-TEFb complex at HIV-1 transcription sites. Retrovirology. 2007;4:36. doi: 10.1186/1742-4690-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nydegger S, Foti M, Derdowski A, Spearman P, Thali M. HIV-1 egress is gated through late endosomal membranes. Traffic. 2003;4:902–910. doi: 10.1046/j.1600-0854.2003.00145.x. [DOI] [PubMed] [Google Scholar]
- Nydegger S, Khurana S, Krementsov DN, Foti M, Thali M. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J Cell Biol. 2006;173:795–807. doi: 10.1083/jcb.200508165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Richard D. Near-field microscopy: throwing light on the nanoworld. Philos Trans R Soc Lond A. 2003;361:2843–2857. doi: 10.1098/rsta.2003.1282. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Brini M, Pizzo P, Murgia M, Pozzan T. Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr Biol. 1995;5:635–642. doi: 10.1016/s0960-9822(95)00128-x. [DOI] [PubMed] [Google Scholar]
- Sherer NM, Lehmann MJ, Jimenez-Soto LF, Horensavitz C, Pypaert M, Mothes W. Retro-viruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat Cell Biol. 2007;9:310–315. doi: 10.1038/ncb1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff H, Galbraith CG, Galbraith JA, White H, Gillette J, Olenych S, Davidson MW, Betzig E. Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc Natl Acad Sci USA. 2007;104:20308–20313. doi: 10.1073/pnas.0710517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowinski S, Jolly C, Berninghausen O, Purbhoo MA, Chauveau A, Köhler K, Oddos S, Eissmann P, Brodsky FM, Hopkins C, Onfelt B, Sattentau Q, Davis DM. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat Cell Biol. 2008;10:211–219. doi: 10.1038/ncb1682. [DOI] [PubMed] [Google Scholar]
- Stryer L. Fluorescence energy transfer as a spectroscopic ruler. Annu Rev Biochem. 1978;47:819–846. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
- van der Krogt GN, Ogink J, Ponsioen B, Jalink K. A comparison of donor-acceptor pairs for genetically encoded FRET sensors: application to the Epac cAMP sensor as an example. PLoS ONE. 2008;3(4):e1916. doi: 10.1371/journal.pone.0001916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Choi H, Poe J, Smithgall TE. Oligomerization is required for HIV-1 Nef-induced activation of the Src family protein-tyrosine kinase, Hck. Biochemistry. 2004;43:15775–15784. doi: 10.1021/bi048712f. [DOI] [PubMed] [Google Scholar]
- Yi L, Fang J, Isik N, Chim J, Jin T. HIV gp120-induced interaction between CD4 and CCR5 requires cholesterol-rich microenvironments revealed by live cell fluorescence resonance energy transfer imaging. J Biol Chem. 2006;281:35446–35453. doi: 10.1074/jbc.M607302200. [DOI] [PubMed] [Google Scholar]
- Yu HJ, Reuter MA, McDonald D. HIV traffics through a specialized, surface-accessible intracellular compartment during trans-infection of T cells by mature dendritic cells. PLoS Pathog. 2008;4(8):e1000134. doi: 10.1371/journal.ppat.1000134. doi:10.1371/journal.ppat.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]