Abstract
This study aims to examine global gene expression profiles before and after the work-shift among coke-oven workers (COW). COW work six consecutive days and then take two days off. Two blood and urine samples in each worker were collected before starting to work after two-days off and end-of-shift in the sixth-day work in 2009. Altered gene expressions (ratio of gene expression levels between end-of-shift and pre-shift work) were performed by Human OneArray expression system which probes ∼30,000-transcription expression profiling of human genes. Sixteen workers, all men, were enrolled in this study. Median urinary 1-hydroxypyrene (1OHP) levels (μmole/mole creatinine) in end-of-shift work were significantly higher than those in pre-shift work (2.58 vs. 0.29, p = 0.0002). Among the 20,341 genes which passed experimental quality control, 26 gene expression changes, 7 positive and 19 negative, were highly correlated with across-the-shift urinary 1OHP levels (end-of-shift – pre-shift 1OHP) (p-value < 0.001). The high and low exposure groups of across-the-shift urinary 1OHP levels dichotomized in ∼2.00 μmole/mole creatinine were able to be distinguished by these 26 genes. Some of them are known to be involved in apoptosis, chromosome stability/DNA repair, cell cycle control/tumor suppressor, cell adhesion, development/spermatogenesis, immune function, and neuronal cell function. These findings in COW will be an ideal model to study the relationship of PAHs exposure with acute changes of gene expressions.
Keywords: Polycyclic aromatic hydrocarbons, 1-hydroxypyrene, toxicogenomic, epidemiology
Introduction
Coke-oven workers (COW) are exposed to high concentrations of polycyclic aromatic hydrocarbons (PAHs), which are also the major hazards from cigarette smoking and traffic pollutants.1-3 Some PAHs with four or more benzene rings are considered to be human carcinogens.4 Epidemiologic studies have presented strong evidence that workers with long-term exposure to PAHs have a high incidence of cancer, especially lung and colon cancers.5,6 Besides their carcinogenesis property, PAHs can affect various organ functions, such as immunologic response and reproductive function.7-10 PAHs are present in complex mixtures of more than 100 different compounds in the vicinity of coke-oven areas.3 Our earlier studies have demonstrated that urinary 1-hydroxypyrene (1OHP), a metabolite of pyrene, is a good index of external ambient exposure to PAHs in COW.1-3
Whole genome expression microarray provides a powerful tool for biological research fields due to its ability to scan tens of thousands of genes at one time.11,12 Although microarray-related studies have increased rapidly in the fields of molecular biology and clinical medicine,13-16 few studies apply this cutting-edge technique in industrial settings.17-21 In addition, only one study used a pre- and post-exposure study design to study the gene expression changes in workers exposed to metal fumes, which can reduce the error of inter-individual variability.19 Since, to our knowledge, no one has examined the changes of whole genome expression profile before and after occupational exposure in COW which is an ideal model to study PAHs exposure, we evaluated the correlation between urinary PAHs biomarkers and the acute changes of gene expressions.
Materials & Methods
Subject
Sixteen COW who had worked in one of two coke-oven plants for at least one year in the largest steel company in Taiwan voluntarily participated in this study between July-October, 2009. COW regularly work 6 days and take two days off.3 Thus, we collected their blood and urine samples at two different time points: one was during the pre-shift work on the first day after two days off, and the second one was at the end-of-shift work on the 6th work day. Information about age and smoking status was also collected before the collection of biological specimens. This study was approved by the Institutional Review Boards of KMHU; all study subjects gave written informed consent.
Biomarkers in urine
All urine samples were stored at -68°C until analysis. The detailed analytical method is described elsewhere.1,2,22-24 Briefly, aliquot amount of thawed urine was hydrolyzed with β-glucuronidase/sulfatase (Roche Diagnostics Ltd.), purified with a Sep-Pack C18 cartridge (2 g/12 mL, BondElut® C18 HF, Varian), and condensed by dry N2 purge to obtain a 2-ml extract. The extract was analyzed by using high performance liquid chromatography (HPLC, Beckman Coulter Module 126, UK) equipped with a fluorescence detector (Jasco FP-920, Japan) to determine 1-naphthol (1NP), 2-naphthol (2NP), 9-phenanthrol (9PHE) and 1OHP levels. The linearity (expressing as R2), limit of detection (LOD), reproducibility (expressing as coefficient of variation (CV)) and mean recovery rate were 0.9982-0.9998, 1.83-47.56 ng/L, 4.02%-7.27%, and 82.97 %-107.85 % respectively. Urinary creatinine was reacted with alkaline picrate and the creatinine-picrate complex was quantified by spectrophotometry (Hitachi U-2000, Japan) using a wavelength of 520 nm. The concentrations of these four hydroxyl-PAHs were presented in units of μmol/mol creatinine.
RNA preparation
Five ml blood samples were drawn from volunteers. WelPrep RNA Stabilizer (Welgeng Biotech, Taipei, TAIWAN) was immediately added to stabilize the whole-blood total RNA in room temperature for 2 hrs and the samples were transferred to our laboratory within one hour. Then, the stabilized blood samples were stored in a -20°C freezer until extraction within 1 month. Total RNA was isolated with RNeasy® Mini Kit (Qiagen, Hilden, Germany) according to manufacturer protocol. The procedure was identical in the pre-shift and end-of-shift blood samples. The yield and quality of RNA were assessed by spectrophotometry and the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Only samples with an A260/A280 between 1.8 and 2.2 and A260/A230 ratio above 1 were eligible for the subsequent array experiment.
Oligonucleotide DNA microarrays
Human oligonucleotide DNA microarrays (Human Whole Genome OneArray™) from Phalanx Biotech Group (Hsinchu, Taiwan) were used. The Human Whole Genome OneArrayTM (HOAv4.3, Phalanx Biotech Group, Taiwan) contains 32,050 60-mer oligonucleotide probes, including 28,703 probes corresponded to the annotated genes in Unigene v175 and RefSeq database, 2,265 experimentally defined probes and 1,082 control probes.25,26 The detailed descriptions of the gene array list are available from http://www.phalanx.com.tw/tech_support/gene_lists.html.
Microarray experiment
One-half μg RNA of each sample was amplified by Illumina TotalPrep RNA Amplification Kit according to the manufacturer's instructions (Ambion, Austin, TX). Then, 10 μg of fragmented biotin-labeled cRNA was hybridized on Phalanx Human OneArray™ by Phalanx hybridization buffer at 50°C in oven for 14-16 hrs using the bubble-mixing method. Each sample was hybridized in triplicate; thus, the total 96 chips were used in this study (16 workers × 2 time points × 3 experiments). After non-specific binding targets were washed, the hybridization arrays were conjugated with fluorescent detector of Strepavidin-Cy3. Finally, arrays were dried by centrifugation and scanned by DNA Microarray Scanner (Agilent Technologies, Santa Clara, US). Images from the scanned arrays were quantified using GenePix® Pro 4.0 (Molecular Devices, Sunnyvale, CA).
Qualification and normalization of microarray chips
Spots in each array with foreground median intensity of wavelength 532 nm greater than or equal to that of background median intensity plus 3 folds standard deviation of wavelength 532 nm were considered as the “Present” flag and included for further analysis. In order to evaluate the quality of each array in the entire array experiment, three evaluation steps were performed: basic, reproducible, and diagram. In the basic step, three parameters, including percentage of “Present” spots among all spots, the average intensity of “Present” spots, and coefficient of variation of intensity for control spots in the entire arrays were all considered. If any two parameters in one array were located outside the 1.5-folds interquartile range (25th-75th) of same parameters for all arrays, that array was excluded. The remaining arrays were then evaluated in reproducible steps which the repeated arrays of the same sample would pass, when their Pearson's correlation coefficient was larger than 0.95 and “2-fold percentage” was less than 15% (sFig. 1). The “2-fold percentage” was the percentage of probes among all probes in which the ratio of the same probe between two arrays exceeded 2-fold. In the final diagram step, the density plot of repeated arrays was used to examine the intensity profile of each array. An array would pass if the profile was similar to the rest of arrays in the same phenotype groups. When the arrays passed all three steps, the raw intensity of spots were log-2 transformed for subsequent analysis. To adjust the systematic variation of experiments and dye effects, global Loess normalizations were performed within repeated arrays of the same sample and between the samples. Spot was included for further analysis when it was “Present” in at least one of the qualified arrays.
Validation of microarray intensity by real-time PCR
To validate the differential expression of genes in the array experiment, we performed the quantitative real-time PCR analysis in the most significant candidate gene (MYO15B) in the 8-paired samples (pre-shift and end-of-shift samples) randomly from 16 study subjects by ABI StepOne Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The primers were 5′-tggagcaactgtgcaacaac-3′ and 5′-agcatctccaccacagcag-3′ for MYO15B and 5′-gca ccg tca agg ctg aga ac-3′ and 5′-atg gtg gtg aag acg cca gt-3′ for GAPDH (internal control). We found a high correlation of MYO15B RNA intensity analyzed by real-time PCR and microarray experiments (Spearman correlation r = 0.881, p = 0.004, n = 8) (sFig. 2).
Statistical analysis
Mann-Whitney U-test was used to compare the differences between pre-shift and end-of-shift (across-the-shift) work of urinary biomarkers. Since 1OHP in urine was recognized as the best surrogate to represent for ambient coke-oven emission exposure,1,3,27 we examined the correlation between urinary across-the-shift 1OHP levels and other biomarkers, including 1NP, 2NP, and 9PHE.
For array analysis, Spearman correlation was used to examine the relationship of across-the-shift urinary 1OHP levels with altered gene expressions (difference of log-transformed gene expression levels between end-of-shift and pre-shift work) using the Biometric Research Branch statistical program (BRB; http://linus.nci.nih.gov/BRB-ArrayTools.html).25 Significant p-value was set as < 0.001. Hierarchical cluster analysis with average linkage was also used to examine the similarity metric of pairs of samples. Distance metric used in the dendrogram was one minus Pearson correlation.28,29 Those differential expressed genes were further analyzed according to their biological process/molecular function by functional annotation clustering of DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/).
Results
Study population
All 16 COW were male and 11 (68.8%) of them were cigarette smokers. Median urinary 1OHP, 1NP, and 9PHE levels in end-of-shift work were significantly higher than those in pre-shift work (p < 0.01) (sTable 1). In contrast, no significant difference was found in 2NP. The high correlation of urinary 1OHP with 1NP (Spearman correlation coefficient r = 0.68, p = 0.0047) and 9PHE (r = 0.76, p = 0.0006), but not 2NP (r = 0.09, p = 0.7452), was noted (sFig. 3).
Array analyses
In total, 82 (85.4%) out of 96 arrays and 20,341 (70.9%) out of 28,703 genes passed the quality control of array experiments (GEO accession number GSE30504 at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30504). Each blood sample had at least two qualified arrays for analyses and means of their values were analyzed in the subsequent analyses.
Using the cut-point p-value < 0.001 of Spearman correlation statistics between across-the-shift 1OHP and altered gene expression, we identified 26 significant gene expression changes and 7 and 19 of their intensity were positively and negatively correlated with across-the-sift urinary 1OHP levels respectively (Table 1). The positive correlations ranged from 0.759∼0.847, whereas the negative correlation ranged from -0.759∼-0.912. These 26 significant genes were further classified and displayed two predominant clusters in samples in the high and low exposure groups (the cut-point level of across-the-shift urinary 1OHP levels was ∼2.00 μmole/mole creatinine) (Fig. 1). In the low across-the-shift urinary 1OHP group (0.43-1.97 μmole/mole creatinine), 19 genes were positively associated with across-the-shift 1OHP levels and 7 genes were negatively associated with 1OHP across-the-shift 1OHP levels. In contrast, among the high across-the-shift urinary 1OHP group (2.60-8.47 μmole/mole creatinine), 19 genes were negatively associated with across-the-shift 1OHP levels and 7 genes were positively associated with 1OHP across-the-shift 1OHP levels.
Table 1.
Significance gene changes associated with across-the-shift urinary 1-hydroxypyrene levels in 16 coke-oven workers (COW) and in 5 nonsmoking COW.
| 16 COW | 5 nonsmoking COW | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene name | Gene symbol | Location | Ra | p-value | FDRb | Ra | p-value | |
| 1 | Myosin XVB | MYO15B | 17q25.1 | -0.862 | < 1e-07 | < 1e-07 | -0.700 | 0.188 |
| 2 | Solute carrier family 25, member 34 | SLC25A34 | 1p36.21 | -0.885 | < 1e-07 | < 1e-07 | -0.500 | 0.391 |
| 3 | DAZ associated protein 1 | DAZAP1 | 19p13.3 | -0.912 | < 1e-07 | < 1e-07 | -0.900 | 0.037 |
| 4 | GTPase, IMAP family member 6 | GIMAP6 | 7 | 0.847 | 0.00001 | 0.0519 | 0.600 | 0.285 |
| 5 | Prostaglandin reductase 1 | PTGR1 | 9q31.3 | 0.835 | 0.0001 | 0.2130 | 0.900 | 0.037 |
| 6 | Zinc finger protein 335 | ZNF335 | 20q13.12 | -0.826 | 0.0001 | 0.2780 | -0.500 | 0.391 |
| 7 | Spinster homolog 3 (Drosophila) | SPNS3 | 17p13.2 | -0.826 | 0.0001 | 0.2780 | -0.800 | 0.104 |
| 8 | Homo sapiens genomic DNA, chromosome 11 clone: RP11-810P12, complete sequence | AP003733.5 | 11 | 0.818 | 0.0002 | 0.3060 | 0.500 | 0.391 |
| 9 | ADP-ribosylation factor GTPase activating protein 2 | ARFGAP2 | 11p11.2-p11. 12 | -0.818 | 0.0002 | 0.3060 | -0.700 | 0.188 |
| 10 | Additional sex combs like 2 | ASXL2 | 2p24.1 | -0.818 | 0.0002 | 0.3060 | -0.600 | 0.285 |
| 11 | Vomeronasal 1 receptor 4 | VN1R4 | 19q13.42 | -0.803 | 0.0003 | 0.5070 | -1.000 | <0.001 |
| 12 | PHD finger protein 2 | PHF2 | 9q22.31 | 0.788 | 0.0004 | 0.6630 | 0.600 | 0.285 |
| 13 | Apoptosis-inducing factor, mitochondrion-associated, 3 | AIFM3 | 22q11.21 | -0.785 | 0.0005 | 0.6630 | -0.900 | 0.037 |
| 14 | Chromosome 1 open reading frame 91 | C1orf91 | 1p36.11-p34.2 | -0.785 | 0.0005 | 0.6630 | -0.600 | 0.285 |
| 15 | Chromosome 2 open reading frame 66 | C2orf66 | 2q33.1 | -0.785 | 0.0005 | 0.6630 | -0.700 | 0.188 |
| 16 | Histone cluster 1, H2bi Solute carrier family 12 | HIST1H2BI | 6p21.3 | -0.779 | 0.0006 | 0.6750 | -0.900 | 0.037 |
| 17 | (potassium/chloride transporters), member 7 | SLC12A7 | 5p15 | -0.779 | 0.0006 | 0.6750 | -0.800 | 0.104 |
| 18 | Claudin 15 | CLDN15 | 7q11.22 | -0.776 | 0.0006 | 0.6750 | -0.700 | 0.188 |
| 19 | Endoplasmic reticulum protein 27 | ERP27 | 12p12.3 | -0.776 | 0.0006 | 0.6750 | -0.900 | 0.037 |
| 20 | Intersection 2 | ITSN2 | 2pter-p25.1 | 0.771 | 0.0007 | 0.7070 | 0.600 | 0.285 |
| 21 | E4F transcription factor 1 | E4F1 | 16p13.3 | -0.771 | 0.0007 | 0.7070 | -1.000 | <0.001 |
| 22 | Chromosome 17 open reading frame 70 | C17orf70 | 17q25.3 | -0.765 | 0.0009 | 0.7070 | -0.700 | 0.188 |
| 23 | NIPA-like domain containing 4 | NIPAL4 | 5q33.3 | 0.762 | 0.0009 | 0.7070 | 0.900 | 0.037 |
| 24 | Low density lipoprotein receptor-related protein 12 | LRP12 | 8q22.2 | -0.759 | 0.0010 | 0.7070 | -0.800 | 0.104 |
| 25 | Retinitis pigmentosa 11 | RP11-399K21. 6 | 19q13.4 | 0.759 | 0.0010 | 0.7070 | 0.200 | 0.747 |
| 26 | Hydroxyacylglutathione hydrolase-like | HAGHL | 16p13.3 | -0.759 | 0.0010 | 0.7070 | -0.800 | 0.104 |
R: Spearman correlation coefficient
FDR: false discovery rate.
Fig. 1.
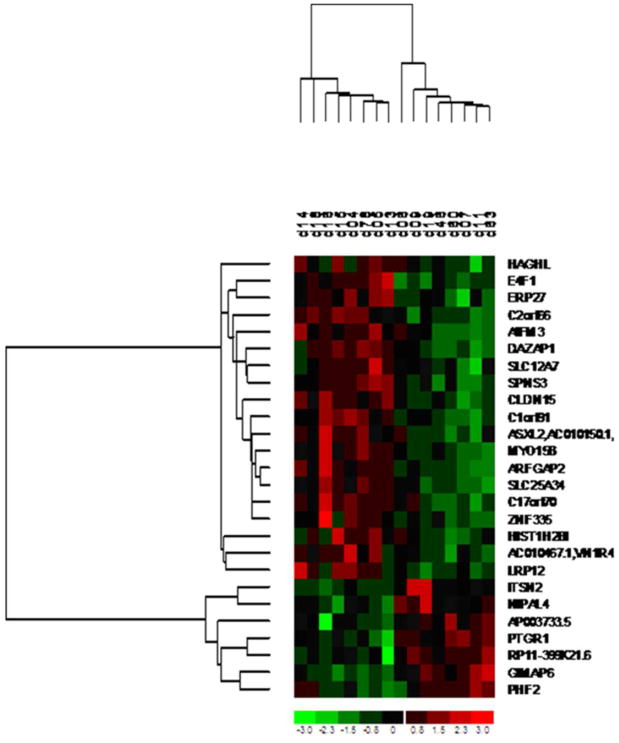
Heatmap and cluster analysis of 16 paired samples using 26 significant gene expression changes identified by Spearman correlation test with across-the-shift 1-hydroxypyrene (1OHP, μmole/mole creatinine). Each column and row represent one coke-oven worker and gene respectively.
Of the 16 COW, five were nonsmokers. We also looked at the Spearman correlation between those 26 candidate gene expression changes and across-the-sift urinary 1OHP levels among these five nonsmokers. We found most values of Spearman correlation coefficients were close to the original ones in 16 COW, although only 8 genes reached the significance of p-value at 0.05 (Table 1).
Using the David program analysis, we were able to annotate 24 out of these 26 significant genes (except AP003733.5 and RP11-399K21.6) (sTable 2). Among these twenty-four genes, two main biological process/molecular functions involving in ‘metal ion binding’ and ‘transport’ were identified by David functional annotation clustering (sTable 2). Of these significant genes, 4 had false discovery rate (FDR) ≤ 0.05, including myosin XVB (MYO15B), solute carrier family 25, member 34 (SLC25A34), DAZ-associated protein 1 (DAZAP1), and GTPase, IMAP family member 6 (GIMAP6) (Fig. 2).
Fig. 2.
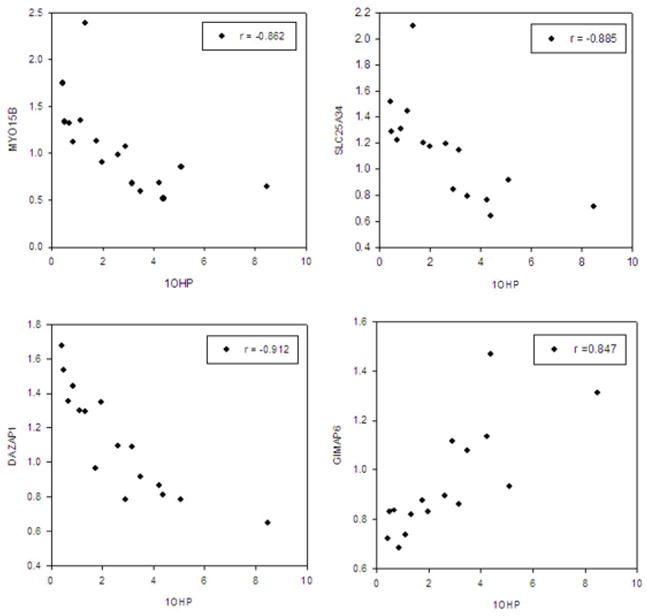
Correlation of urinary across-the-shift 1-hydroxypyrene (1OHP, μmole/mole creatinine) with gene expression changes (folds) between pre-shift work and end-of-shift work among 16 coke-oven workers. MYO15B, Myosin XVB; SLC25A34, Solute carrier family 25, member 34; DAZAP1, DAZ associated protein 1; GIMAP6, GTPase, IMAP family member 6.
Discussion
1NP and 2NP, 9PHE, and 1OHP in urine are the metabolites of naphthalene, phenanthrene, and pyrene respectively, which belong to 2-, 3-, and 4-benzene rings of PAHs (30). Urinary 1OHP is commonly used to represent ambient PAHs exposures in occupational settings, including COW.1,3,27 This study found that median urinary 1OHP, 1NP, and 9PHE levels in end-of-shift work were significantly higher than those in their pre-shift work. In addition, there were the high correlations of across-the-shift urinary 1OHP levels with across-the-shift urinary 1NP and 9PHE levels. These results suggest that the COW were occupationally exposed to a high amount of different PAHs during work. In contrast, no significant increase of urinary across-the-shift 2NP levels and no significant correlation between across-the-shift urinary 1OHP levels and 2NP levels were found. Possible explanations are: 2NP is not the final metabolite of ambient naphthalene exposure, this may be due to the contributions of smoking larger than coke-oven emissions exposure on 2NP metabolite, or the issue of collection of only one-spot urine sample is not representative for continuous coke-oven exposure.31
This is, to our knowledge, the first study to use the technique of whole-genome array to investigate the effect of coke-oven emissions, which mainly contain PAHs, on altered gene expressions in humans. Although a few studies have examined the effect of environmental and occupational hazards, such as arsenic or benzene, on whole genome expression changes in humans,17,19,32 most of the study designs compared gene function differences between exposed and non-exposed subjects, which would potentially introduce inter-individual variability bias, especially in the study of evaluating thousands of genes changes at the same time. Our study design, along with the study of Wang et al 19 to evaluate metal fume exposures, using the same subjects to examine altered gene expressions before and after work shifts, can overcome this bias.
The Human Whole Genome OneArrayTM (Phalanx Biotech Group, Taiwan) used in this study can evaluate ∼30,000 functional gene expressions. Since many genes are unknown functions or their expressions are correlated with each other, we used the p-value significance of <0.001 in Spearman statistics, rather than multiple testing adjustment, to explore the correlation between across-the-shift 1OHP and gene expression changes. Under this approach, we were able to identify 26 significant altered gene expressions, 7 positively and 19 negatively correlated with across-the-shift urinary 1OHP levels (Table 1). These positive and negative Spearman correlations were at least 0.75. Some known gene functions are involved in apoptosis (e.g., AIFM3),33,34 chromosome stability/DNA repair (e.g., C17orf70, HIST1H2BI),35,36 cell cycle control/tumor suppressor (e.g., E4F1, LRP12),37,38 cell adhesion (e.g., CLDN15),39 development/spermatogenesis (e.g., ASXL2, DAZAP1),40-42 immune function (e.g., GIMAP6),43-45 and neuronal cell function (e.g., ITSN2).46
David functional annotation clustering tool clustered two main biological process/molecular functions involved in ‘metal ion binding/cation binding’ and ‘transport’ among the 24 out of 26 significant genes (except AP003733.5 and RP11-399K21.6). At this moment, we cannot fully explain how it is that these two molecular functions related to PAH exposures are picked-up by DAVID annotation. Molecular function of 10 candidate proteins involved in ‘metal ion binding/cation binding’ may interact selectively and non-covalently with any metal ions, including vanadium, manganese, iron, copper, cobalt, nickel, molybdenum and silver (sTable 2). Two articles have observed that binding of heavy metal ions to cell membrane may affect the sorption of organic pollutants such as PAHs by modulating the structure and chemistry of cell membrane.47,48 Another molecular function of 6 candidate proteins associated with ‘transport’ is involved in the transport of amino acids, calcium ions, trace elements of copper or cobalt (a component of vitamin B12), or electrons probably related to the process of energy conversion. PAHs are well-known carcinogens, but they can also influence intracellular gap-junction communication to act as a tumor promoter in culture cells.49-50 This is an exploratory study and further research is needed to elucidate those molecular functions.
Of 26 significant genes, 4, including MYO15B, SLC25A34, DAZAP1, and GIMAP6, had FDR ≤ 0.05. Gene expression changes of MYO15B, SLC25A34, and DAZAP1 were positively correlated with urinary across-the-shift 1-OHP, whereas GIMAP6 was negatively correlated with urinary across-the-shift 1-OHP. Although these 4 candidate genes were the most significant ones, their gene expression differences between pre-shift and end-of-shift works were less than 2.5-fold (Fig. 2). Since these subjects for array analyses were healthy, their gene expression changes may not be as large as the studies of examining the gene differences between cancer and normal tissues.17,51
MYO15B is a transcribed, untranslated pseudogene located at 17q25.52 Based on Bootstrap analysis, MYO15B is the most significant similarity to MYO15A which are mainly involved in muscular contraction and related to important functions of some specific cells such as melanocytes, kidney and intestinal brush border microvilli, nerve growth cones, or inner ear hair cells associated with hearing impairment.52-54 SLC25A34 is located at 1p36.21 and belongs to the SLC25 family of mitochondrial carrier proteins which function is to transport molecules over the mitochondrial membrane.55 The molecules transported by the SLC25 family protein include ATP/ADP, amino acids (glutamate, aspirate, lysine, histidine, and arginine), malate, ornithine, and citrulline.56 DAZAP1 is an RNA-binding protein located at 19p13.3. Its primary expression is in the testis and its main function involves spermatogenesis.40 Recently, Prima et al.41 also showed that DAZAP1 can translocate and fuse to myocyte enhancer factor 2D to become a fusion protein in an acute lymphoblastic leukemia cell line (TS-2), suggesting DAZAP1 may also contribute to human leukemogenesis. GIMAP6 gene encodes a protein belonging to the GTPases of the immunity-associated protein family and is located in a cluster at 7q36.1.44 Their predominated mRNA expression sites are in the immune system such as in spleen and lymph nodes. A few studies have shown that GIMAP GTPases are expressed at very low levels in diverse cancer tissues and cell lines including leukemia and lymphoma.43 Thus, GIMAP may play a role to decrease cancer growth and probably act as a tumor suppressor gene.
Except for MYO15B, the biological functions of the other three significant genes (SLC25A34, DAZAP1, and GIMAP6) selected by arrays are coincidentally consistent to the findings of our previous epidemiological studies in COW from the same large steel company.7,9 Besides well-known carcinogenic properties of some PAHs, one study of ours has shown that COW had significantly higher serum IgE and tumor necrosis factor-α levels than rolling steel workers as a comparison non-exposed group.7 In contrast, serum IgA levels were significantly lower in COW than in rolling steel workers, suggesting that PAHs exposure may alter the immune responses in COW. The mechanism of different immune-modulation by PAHs exposure is probably due to the binding activity of aryl hydrocarbon receptor or oxidative stress induction.57-59 In addition, another of our studies found a positive correlation between urinary 1OHP concentration and percentage of abnormal sperm morphology in the same COW.9 PAHs such as benzo[a]pyrene and its metabolites can accumulate in the testis and epididymis and may therefore affect androgen-dependent processes by acting as antiandrogens.51,60 PAHs may also directly influence sperm function through the DNA damage to form PAHs-DNA adduct or to bind dioxin receptor in sperm.61,62 The findings of our genome arrays may add additional information about the biological mechanisms of PAHs effect on systems of carcinogenesis, immune response, and reproductive dysfunction.
In conclusion, this study demonstrates that microarray analysis can become a useful tool to discover the potential known and novel genes or biomarkers in an ideal exposure group such as PAHs exposure in COW, although the sample size is small. Since microarray experiments will generate thousands of items of gene function information, using a repeated measure design, like ours, can reduce inter-individual variability bias. Some known gene functions identified by this study and associated with carcinogenesis, immune response, and reproductive dysfunction are consistent with our earlier epidemiological studies in the same group of COW. Future research is necessary to study the role of other significant genes in COW professionally exposed to PAHs.
Supplementary Material
sFig. 1 Scatter plots of one representative coke-oven workers with 3 qualified array assays by Pearson's correlation coefficients (r). A. Data of log2 intensities of pre-shift work; B. Data of log2 intensities of end-of-shift work.
sFig. 2 The Spearman correlation of MYO15B gene expression change (fold, end-of-shift/pre-shift work) by real-time PCR and microarray analyses in eight coke-oven workers.
sFig. 3 Correlation of urinary across-the-shift 1-hydroxypyrene (1OHP, μmole/mole creatinine) with 1-naphthol (1NP, μmole/mole creatinine) and 9-phenanthol (9PHE, μmole/mole creatinine).
Acknowledgments
We thank Yu-Hsiu Hung and Hui-Shan Chen for assisting in the collection of blood specimens. This work was supported in part by grants from the Taiwan Institute of Occupational Safety & Health (IOSH99-M303), National Science Council (NSC97-2314-B-037-MY3), Department of Health (DOH778-8876-768), Kaohsiung Medical University (KMUO-O100001), and the National Institute of Health (NIH 2 R01 ES011297-06).
References
- 1.Wu MT, Mao IF, Ho CK, Wypij D, Lu PL, Smith TJ, Chen ML, Christiani DC. Urinary 1-hydroxypyrene concentrations in coke oven workers. Occup Environ Med. 1998;55(7):461–467. doi: 10.1136/oem.55.7.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu MT, Wypij D, Ho CK, Mao IF, Chen ML, Lu PL, Christiani DC. Temporal changes in urinary 1-hydroxypyrene concentrations in coke-oven workers. Cancer Epidemiol Biomarkers Prev. 1998;7(2):169–173. [PubMed] [Google Scholar]
- 3.Lin YC, Pan CH, Chen CJ, Wu KY, Chang-Chien GP, Ho CK, Wu TN, Chuang HY, Kuo HW, Wu MT. Associations between exposure to polycyclic aromatic hydrocarbons and temporal change of urinary 1-hydroxypyrene levels in Taiwanese coke-oven workers. J Occup Environ Med. 2006;48(9):930–936. doi: 10.1097/01.jom.0000226974.91335.5b. [DOI] [PubMed] [Google Scholar]
- 4.Boogaard PJ, van Sittert NJ. Exposure to polycyclic aromatic hydrocarbons in petrochemical industries by measurement of urinary 1-hydroxypyrene. Occup Environ Med. 1994;51(4):250–258. doi: 10.1136/oem.51.4.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger J, Manz A. Cancer of the stomach and the colon-rectum among workers in a coke gas plant. Am J Ind Med. 1992;22(6):825–834. doi: 10.1002/ajim.4700220605. [DOI] [PubMed] [Google Scholar]
- 6.Chau N, Bertrand JP, Mur JM, Figueredo A, Patris A, Moulin JJ, Pham QT. Mortality in retired coke oven plant workers. Br J Ind Med. 1993;50(2):127–135. doi: 10.1136/oem.50.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu MT, Pan CH, Wu TN, Huang YL, Chen CY, Huang LH, Ho CK. Immunological findings in a group of coke-oven workers exposed to polycyclic aromatic hydrocarbons. J Occup Environ Med. 2003;45(10):1034–1039. doi: 10.1097/01.jom.0000088876.25970.96. [DOI] [PubMed] [Google Scholar]
- 8.Karakaya A, Ates I, Yucesoy B. Effects of occupational polycyclic aromatic hydrocarbon exposure on T-lymphocyte functions and natural killer cell activity in asphalt and coke oven workers. Hum Exp Toxicol. 2004;23(7):317–322. doi: 10.1191/0960327104ht455oa. [DOI] [PubMed] [Google Scholar]
- 9.Hsu PC, Chen IY, Pan CH, Wu KY, Pan MH, Chen JR, Chen CJ, Chang-Chien GP, Hsu CH, Liu CS, Wu MT. Sperm DNA damage correlates with polycyclic aromatic hydrocarbons biomarker in coke-oven workers. Int Arch Occup Environ Health. 2006;79(5):349–356. doi: 10.1007/s00420-005-0066-3. [DOI] [PubMed] [Google Scholar]
- 10.Detmar J, Jurisicova A. Embryonic resorption and polycyclic aromatic hydrocarbons: putative immune-mediated mechanisms. Syst Biol Reprod Med. 2010;56(1):3–17. doi: 10.3109/19396360903296754. [DOI] [PubMed] [Google Scholar]
- 11.Marcucci G, Radmacher MD, Maharry K, Mrozek K, Ruppert AS, Paschka P, Vukosavljevic T, Whitman SP, Baldus CD, Langer C, Liu CG, Carroll AJ, Powell BL, Garzon R, Croce CM, Kolitz JE, Caligiuri MA, Larson RA, Bloomfield CD. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 12.Director's Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma. Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, Misek DE, Chang AC, Zhu CQ, Strumpf D, Hanash S, Shepherd FA, Ding K, Seymour L, Naoki K, Pennell N, Weir B, Verhaak R, Ladd-Acosta C, Golub T, Gruidl M, Sharma A, Szoke J, Zakowski M, Rusch V, Kris M, Viale A, Motoi N, Travis W, Conley B, Seshan VE, Meyerson M, Kuick R, Dobbin KK, Lively T, Jacobson JW, Beer DG. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14(8):822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 14.Liu ET, Kuznetsov VA, Miller LD. In the pursuit of complexity: systems medicine in cancer biology. Cancer Cell. 2006;9(4):245–247. doi: 10.1016/j.ccr.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 15.Maraqa L, Donnellan CF, Peter MB, Speirs V. Clinicians' guide to microarrays. Surg Oncol. 2006;15(4):205–210. doi: 10.1016/j.suronc.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Parikh AA, Johnson JC, Merchant NB. Genomics and proteomics in predicting cancer outcomes. Surg Oncol Clin N Am. 2008;17(2):257–277. doi: 10.1016/j.soc.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Forrest MS, Lan Q, Hubbard AE, Zhang L, Vermeulen R, Zhao X, Li G, Wu YY, Shen M, Yin S, Chanock SJ, Rothman N, Smith MT. Discovery of novel biomarkers by microarray analysis of peripheral blood mononuclear cell gene expression in benzene-exposed workers. Environ Health Perspect. 2005;113(6):801–807. doi: 10.1289/ehp.7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith MT, Vermeulen R, Li G, Zhang L, Lan Q, Hubbard AE, Forrest MS, McHale C, Zhao X, Gunn L, Shen M, Rappaport SM, Yin S, Chanock S, Rothman N. Use of ‘Omic’ technologies to study humans exposed to benzene. Chem Biol Interact. 2005;153-154:123–127. doi: 10.1016/j.cbi.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Neuburg D, Li C, Su L, Kim JY, Chen JC, Christiani DC. Global gene expression profiling in whole-blood samples from individuals exposed to metal fumes. Environ Health Perspect. 2005;113(2):233–241. doi: 10.1289/txg.7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gwinn MR, Weston A. Application of oligonucleotide microarray technology to toxic occupational exposures. J Toxicol Environ Health A. 2008;71(5):315–324. doi: 10.1080/15287390701738509. [DOI] [PubMed] [Google Scholar]
- 21.Maeda S, Yu X, Wang RS, Sakakibara H. A pilot study of gene expression analysis in workers with hand-arm vibration syndrome. Ind Health. 2008;46(2):188–193. doi: 10.2486/indhealth.46.188. [DOI] [PubMed] [Google Scholar]
- 22.Kim H, Cho SH, Kang JW, Kim YD, Nan HM, Lee CH, Lee H, Kawamoto T. Urinary 1-hydroxypyrene and 2-naphthol concentrations in male Koreans. Int Arch Occup Environ Health. 2001;74(1):59–62. doi: 10.1007/s004200000193. [DOI] [PubMed] [Google Scholar]
- 23.Elovaara E, Vaananen V, Mikkola J. Simultaneous analysis of naphthols, phenanthrols, and 1-hydroxypyrene in urine as biomarkers of polycyclic aromatic hydrocarbon exposure: intraindividual variance in the urinary metabolite excretion profiles caused by intervention with beta-naphthoflavone induction in the rat. Arch Toxicol. 2003;77(4):183–193. doi: 10.1007/s00204-003-0436-0. [DOI] [PubMed] [Google Scholar]
- 24.Lee MS, Eum KD, Zoh KD, Kim TS, Pak YS, Paek D. 1-hydroxypyrene as a biomarker of PAH exposure among subjects living in two separate regions from a steel mill. Int Arch Occup Environ Health. 2007;80(8):671–678. doi: 10.1007/s00420-007-0178-z. [DOI] [PubMed] [Google Scholar]
- 25.Cheng WY, Hsiang CY, Bau DT, Chen JC, Shen WS, Li CC, Lo HY, Wu SL, Chiang SY, Ho TY. Microarray analysis of vanillin-regulated gene expression profile in human hepatocarcinoma cells. Pharmacol Res. 2007;56(6):474–482. doi: 10.1016/j.phrs.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 26.Campeau PM, Rafei M, Boivin MN, Sun Y, Grabowski GA, Galipeau J. Characterization of Gaucher disease bone narrow mesenchymal stromal cells reveals an altered inflammatory secretome. Blood. 2009;114(15):3181–3190. doi: 10.1182/blood-2009-02-205708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen AM, Mathiesen L, Pedersen M, Knudsen LE. Urinary 1-hydroxypyrene (1-HP) in environmental and occupational studies--a review. Int J Hyg Environ Health. 2008;211(5-6):471–503. doi: 10.1016/j.ijheh.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 28.Xu X, Zhao Y, Simon R. Gene Set Expression Comparison kit for BRB-ArrayTools. Bioinformatics. 2008;24(1):137–9. doi: 10.1093/bioinformatics/btm541. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y, Simon R. BRB-ArrayTools Data Archive for human cancer gene expression: a unique and efficient data sharing resource. Cancer Inform. 2008;6:9–15. doi: 10.4137/cin.s448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campo L, Rossella F, Pavanello S, Mielzynska D, Siwinska E, Kapka L, Bertazzi PA, Fustinoni S. Urinary profiles to assess polycyclic aromatic hydrocarbons exposure in coke-oven workers. Toxicol Lett. 2010;192(1):72–78. doi: 10.1016/j.toxlet.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 31.Campo L, Buratti M, Fustinoni S, Cirla PE, Martinotti I, Longhi O, Cavallo D, Foà V. Evaluation of exposure to PAHs in asphalt workers by environmental and biological monitoring. Ann N Y Acad Sci. 2006;1076:405–420. doi: 10.1196/annals.1371.013. [DOI] [PubMed] [Google Scholar]
- 32.Wu MM, Chiou HY, Ho IC, Chen CJ, Lee TC. Gene expression of inflammatory molecules in circulating lymphocytes from arsenic-exposed human subjects. Environ Health Persp. 2003;111(11):1429–1438. doi: 10.1289/ehp.6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urbano A, Lakshmanan U, Choo PH, Kwan JC, Ng PY, Guo K, Dhakshinamoorthy S, Porter A. AIF suppresses chemical stress-induced apoptosis and maintains the transformed state of tumor cells. EMBO J. 2005;24(15):2815–2826. doi: 10.1038/sj.emboj.7600746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie Q, Lin T, Zhang Y, Zheng J, Bonanno JA. Molecular cloning and characterization of a human AIF-like gene with ability to induce apoptosis. J Biol Chem. 2005;280(20):19673–19681. doi: 10.1074/jbc.M409517200. [DOI] [PubMed] [Google Scholar]
- 35.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125(4):703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 36.Xue Y, Li Y, Guo R, Ling C, Wang W. FANCM of the Fanconi anemia core complex is required for both monoubiquitination and DNA repair. Hum Mol Genet. 2008;17(11):1641–1652. doi: 10.1093/hmg/ddn054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garnis C, Coe BP, Zhang L, Rosin MP, Lam WL. Overexpression of LRP12, a gene contained within an 8q22 amplicon identified by high-resolution array CGH analysis of oral squamous cell carcinomas. Oncogene. 2004;23(14):2582–2586. doi: 10.1038/sj.onc.1207367. [DOI] [PubMed] [Google Scholar]
- 38.Le Cam L, Linares LK, Paul C, Julien E, Lacroix M, Hatchi E, Triboulet R, Bossis G, Shmueli A, Rodriguez MS, Coux O, Sardet C. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell. 2006;127(4):775–788. doi: 10.1016/j.cell.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 39.Tamura A, Kitano Y, Hata M, Katsuno T, Moriwaki K, Sasaki H, Hayashi H, Suzuki Y, Noda T, Furuse M, Tsukita S, Tsukita S. Mega intestine in claudin-15-deficient mice. Gastroenterology. 2008;134(2):523–534. doi: 10.1053/j.gastro.2007.11.040. [DOI] [PubMed] [Google Scholar]
- 40.Tsui S, Dai T, Roettqer S, Schempp W, Salido EC, Yen PH. Identification of two novel proteins that interact with germ-cell-specific RNA-binding proteins DAZ and DAZL1. Genomics. 2000;65(3):266–273. doi: 10.1006/geno.2000.6169. [DOI] [PubMed] [Google Scholar]
- 41.Prima V, Gore L, Caires A, Boomer T, Yoshinari M, Imaizumi M, Varella-Garcia M, Hunger SP. Cloning and functional characterization of MEF2D/DAZAP1 and DAZAP1/MEF2D fusion proteins created by a variant t(1;19)(q23;p13.3) in acute lymphoblastic leukemia. Leukemia. 2005;19(5):806–813. doi: 10.1038/sj.leu.2403684. [DOI] [PubMed] [Google Scholar]
- 42.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138(2):389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Döhner K, Brown J, Hehmann U, Hetzel C, Stewart J, Lowther G, Scholl C, Fröhling S, Cuneo A, Tsui LC, Lichter P, Scherer SW, Döhner H. Molecular cytogenetic characterization of a critical region in bands 7q35-q36 commonly deleted in malignant myeloid disorders. Blood. 1998;92(11):4031–5. [PubMed] [Google Scholar]
- 44.Krücken J, Schroetel RM, Müller IU, Saïdani N, Marinovski P, Benten WP, Stamm O, Wunderlich F. Comparative analysis of the human GIMAP gene cluster encoding a novel GTPase family. Gene. 2004;341:291–304. doi: 10.1016/j.gene.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Shiao YM, Chang YH, Liu YM, Li JC, Su JS, Liu KJ. Dysregulation of GIMAP genes in non-small cell lung cancer. Lung Cancer. 2008;62(3):287–94. doi: 10.1016/j.lungcan.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 46.Seifert M, Ampofo C, Mehraein Y, Reichrath J, Welter C. Expression analysis of human intersecting 2 gene (ITSN2) minor splice variants showing differential expression in normal human brain. Oncol Rep. 2007;17(5):1207–1211. [PubMed] [Google Scholar]
- 47.Qu X, Wang X, Zhu D. The partitioning of PAHs to egg phospholipids facilitated by copper and proton binding via cation-pi interactions. Environ Sci Technol. 2007;41(24):8321–8327. doi: 10.1021/es0718117. [DOI] [PubMed] [Google Scholar]
- 48.Xiao L, Qu X, Zhu D. Biosorption of nonpolar hydrophobic organic compounds to Escherichia coli facilitated by metal and proton surface binding. Environ Sci Technol. 2007;41(8):2750–2755. doi: 10.1021/es062343o. [DOI] [PubMed] [Google Scholar]
- 49.Blaha L, Kapplova P, Vondracek J, Upham B, Machala M. Inhibition of gap-junctional intercellular communication by environmentally occurring polycyclic aromatic hydrocarbons. Toxicol Sci. 2002;65(1):43–51. doi: 10.1093/toxsci/65.1.43. [DOI] [PubMed] [Google Scholar]
- 50.Baird WM, Hooven LA, Mahadevan B. Carcinogenic polycyclic aromatic hydrocarbon-DNA adducts and mechanism of action. Environ Mol Mutagen. 2005;45(2-3):106–114. doi: 10.1002/em.20095. [DOI] [PubMed] [Google Scholar]
- 51.Vinggaard AM, Hnida C, Larsen JC. Environmental polycyclic aromatic hydrocarbons affect androgen receptor activation in vitro. Toxicology. 2000;145(2-3):173–183. doi: 10.1016/s0300-483x(00)00143-8. [DOI] [PubMed] [Google Scholar]
- 52.Boger ET, Sellers JR, Friedman TB. Human myosin XVBP is a transcribed pseudogene. J Muscle Res Cell Motil. 2001;22(5):477–483. doi: 10.1023/a:1014507705858. [DOI] [PubMed] [Google Scholar]
- 53.Redowicz MJ. Myosins and deafness. J Muscle Res Cell Motil. 1999;20(3):241–248. doi: 10.1023/a:1005403725521. [DOI] [PubMed] [Google Scholar]
- 54.Hilgert N, Smith RJ, Van Camp G. Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics? Mutat Res. 2009;681(2-3):189–196. doi: 10.1016/j.mrrev.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haitina T, Lindblom J, Renström T, Fredriksson R. Fourteen novel human members of mitochondrial solute carrier family 25 (SLC25) widely expressed in the central nervous system. Genomics. 2006;88(6):779–790. doi: 10.1016/j.ygeno.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 56.Palmieri F. The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch. 2004;447(5):689–709. doi: 10.1007/s00424-003-1099-7. [DOI] [PubMed] [Google Scholar]
- 57.Takenaka H, Zhang K, Diaz-Sanchez D, Tsien A, Saxon A. Enhanced human IgE production results from exposure to the aromatic hydrocarbons from diesel exhaust: direct effects on B-cell IgE production. J Allergy Clin Immunol. 1995;95(1 Pt 1):103–15. doi: 10.1016/s0091-6749(95)70158-3. [DOI] [PubMed] [Google Scholar]
- 58.Tsien A, Diaz-SanchezD MaJ, Saxon A. The organic component of diesel exhaust particles and phenanthrene, a major polyaromatic hydrocarbon constituent, enhances IgE production by IgE-secreting EBV-transformed human B cells in vitro. Toxicol Appl mPharmacol. 1997;142(2):256–263. doi: 10.1006/taap.1996.8063. [DOI] [PubMed] [Google Scholar]
- 59.Hertz-Picciotto I, Park HY, Dostal M, Kocan A, Trnovec T, Sram R. Prenatal exposures to persistent and non-persistent organic compounds and effects on immune system development. Basic Clin Pharmacol Toxicol. 2008;102(2):146–541. doi: 10.1111/j.1742-7843.2007.00190.x. [DOI] [PubMed] [Google Scholar]
- 60.Ramesh A, Inyang F, Hood DB, Archibong AE, Knuckles ME, Nyanda AM. Metabolism, bioavailability, and toxicokinetics of benzo(alpha)pyrene in F-344 rats following oral administration. Exp Toxicol Pathol. 2001;53(4):275–290. doi: 10.1078/0940-2993-00192. [DOI] [PubMed] [Google Scholar]
- 61.Revel A, Raanani H, Younglai E, Xu J, Han R, Savouret JF, Casper RF. Resveratrol, a natural aryl hydrocarbon receptor antagonist, protects sperm from DNA damage and apoptosis caused by benzo(a)pyrene. Reprod Toxicol. 2001;15(5):479–486. doi: 10.1016/s0890-6238(01)00149-6. [DOI] [PubMed] [Google Scholar]
- 62.Khorram O, Garthwaite M, Jones J, Golos T. Expression of aryl hydrocarbon receptor (AHR) and aryl hydrocarbon receptor nuclear translocator (ARNT) mRNA expression in human spermatozoa. Med Sci Monit. 2004;10(5):BR135–138. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
sFig. 1 Scatter plots of one representative coke-oven workers with 3 qualified array assays by Pearson's correlation coefficients (r). A. Data of log2 intensities of pre-shift work; B. Data of log2 intensities of end-of-shift work.
sFig. 2 The Spearman correlation of MYO15B gene expression change (fold, end-of-shift/pre-shift work) by real-time PCR and microarray analyses in eight coke-oven workers.
sFig. 3 Correlation of urinary across-the-shift 1-hydroxypyrene (1OHP, μmole/mole creatinine) with 1-naphthol (1NP, μmole/mole creatinine) and 9-phenanthol (9PHE, μmole/mole creatinine).


