Abstract
Studies in long-term non-progressors (LTNP) have suggested that the quality of the CD8+ response may involve protective human leucocyte antigen (HLA) class I alleles. However, studies examining the expansion ability of different functional CD8+ T cells and their association with HLA class I alleles are lacking. LTNP, untreated typical progressors (TP) and patients successfully on highly active retroviral therapy (HAART) during 1 year (HP) were included. HLA class I typing was performed using a sequence-specific primer assay. Functional subsets of Gag- and Nef-specific CD8+ cells were analysed based on the production of macrophage inflammatory protein (MIP)-1β, tumour necrosis factor (TNF)-α and interleukin (IL)-2. Their expansion abilities were evaluated after 10-day culture in the presence of Gag and Nef human immunodeficiency virus (HIV) peptides. No differences were seen when comparing quantitative and qualitative HIV-specific CD8+ T cell responses according to the presence/absence of protective HLA alleles (B*58 and B*27 supertypes) in each group. However, LTNP with protective HLA alleles showed a higher expansion ability of Gag-specific MIP+ TNF+ IL-2+ T cells and Nef-specific MIP+ TNF+ IL-2+. HLA-B*5701+LTNP displayed a higher expansion ability of Gag and Nef-specific MIP+ TNF− IL-2+ T cells than HLA-B*5701-LTNP. This was not so for HLA-B*2705. No differences were seen in the expansion ability according to the presence/absence of protective HLA alleles in TP and HP. The expansion ability of polyfunctional CD8+ T cells is modulated by HLA class I alleles and targeted protein. LTNP with HLA class I protective alleles (mainly B*5701) display better expansion ability of polyfunctional HIV-specific CD8+ T cells than the rest, suggesting that factors other than HLA-B*5701 must contribute to the control of viral replication in other LTNP. Furthermore, these attributes of HIV-specific CD8+ T are not restored by HAART; thus, adjuvant therapies and vaccines that induce and/or normalize the expansion ability of HIV-specific T cells are required.
Keywords: CD8+ T cell response, cytokines, expansion ability, HIV, HLA class I, LTNP
Introduction
Different evidence has demonstrated that human immunodeficiency virus (HIV) infection induces a strong host cellular immune response that is critical in controlling viral replication.1,2 However, despite this cellular immune response most HIV-infected patients show clinical progression in the absence of antiretroviral therapy. Studies in long-term non-progressors (LTNPs) who remain asymptomatic for more than 10 years have suggested that the quality of cellular immune response is one of the major factors related with the spontaneous control of viral replication,3 although other characteristics of cellular immune response, such as the expansion ability of virus-specific T cells,4,5 could also be related to the control of viral replication.6,7
The expansion ability of HIV-specific CD8+ T cells is impaired in typical progressors, but maintained in LTNP.4 This ability of CD8+ T cells to proliferate is coupled to their ability for secreting perforin, which could enable them to kill HIV-infected CD4 T cells.4 In agreement with this, in our previous study5 we found a significantly higher prevalence of Gag-specific macrophage inflammatory protein (MIP)-1β+ tumour necrosis factor (TNF)-α− interleukin (IL)-2− CD8+ T cell subset expansion in LTNP, with a similar tendency for the MIP-1β+ TNF-α+ IL-2− subset. Moreover, a significant inverse correlation between HIV load and expansion ability of several subsets of Gag-specific CD8+ cells was seen in progressors, which supports an association between the ability to expand and the control of viral replication. However, the statistical association between HIV replication and HIV-specific CD8+ responses does not permit inference of what is cause and what is effect. In our previous longitudinal study, analysing the expansion ability in patients treated successfully with highly active antiretroviral therapy (HAART) for 1 year,7 neither the prevalence nor the pattern of expansion changed after achieving complete suppression of viral replication with HAART. Therefore, our results suggest that HIV replication does not significantly influence the expansion ability of HIV-specific CD8+ T cells, although we do not rule out that a high virus load before the patient was treated could have caused an irreversible change in HIV-specific CD8 T cells that impaired their ability to expand.
Conversely, HLA allele restriction drives the cellular immune response and different HLA-B alleles have been associated with a better HIV-specific CD8+ T cell response,8 essentially for both HLA-B*5701 and HLA-B*2705 alleles.9 These HLA alleles are significantly more prevalent in LTNPs compared with typical progressors, and CD8+ T cell responses restricted by these alleles exert an antiviral function mediated through cytolysis of infected cells.4 Nevertheless, the presence of these alleles does not fully explain the spontaneous control of viral replication observed in these patients.3,10
So far, there have been no studies analysing the association between HLA alleles and expansion ability of different functional subsets of HIV-specific CD8+ T cell response in LTNPs patients compared with typical progressors and with HAART-treated patients. Herein, we have analysed whether an association exists between quantitative and qualitative aspects of HIV-specific CD8+ T cells (including their expansion ability) and the HLA-B allele restriction.
Patients and methods
Study population
A total of 49 individuals with chronic HIV infection were identified at one HIV/acquired immune deficiency syndrome (AIDS) reference centre located in Madrid, Spain, in which approximately 2600 HIV-infected individuals are followed-up regularly. Twenty-four patients met the criteria for LTNP, meaning serologically proven HIV-1 infection lasting for over 10 years, CD4 counts always above 500 cells/μl and lack of HIV-related symptoms in the absence of any antiretroviral therapy. All these patients belonged to an already well-characterized cohort of LTNP.11 These individuals were split out into two categories according to viral load evolution during a 10-year follow-up: (i) 10 elite controllers (EC) that had maintained undetectable viral load, and (ii) 14 viraemic controllers (VC) that had persistent detectable viral load below 2000 copies RNA/ml. The remainder of the patient population comprised 13 antiretroviral-naive typical progressors (TP) and 12 subjects on HAART for at least 1 year and maintaining undetectable viral load (HP). All patients were infected with B subtype and were Caucasians. Sixty-nine per cent of typical progressors, 67% of LTNPs and 79% of patients treated with HAART were male. No significant differences were observed in the medians of age when different groups were compared [41(20), 46(7) and 47(1), P = 0·47 median of age (years) in TP, LTNPs and HP]. Table 1 shows the immunological and virological characteristics of the patient population.
Table 1.
Immunological and virological characteristics of human immunodeficiency virus (HIV)-infected patients included in the study
| Group | CD4 count (cell/μl) | Viral Load (log copies RNA/ml) | Length of infection (years) | HLA-A* | HLA-B* | HLA-C* | |||
|---|---|---|---|---|---|---|---|---|---|
| EC 1 | 1080 | 1·7 | 11 | 0205 | 3301 | 1402 | 5001 | 0602 | 0802 |
| EC 2 | 840 | 1·7 | 18 | 2402 | 3201 | 0801 | 5701 | 0602 | 0701 |
| EC 3 | 754 | 1·7 | 14 | 0201 | 2902 | 1501 | 5701 | 0401 | 0701 |
| EC 4 | 616 | 1·7 | 17 | 0101 | 1101 | 2705 | 5201 | 0202 | 5201 |
| EC 5 | 999 | 1·7 | 13 | 0301 | 2501 | 1501 | 4402 | 0303 | 0501 |
| EC 6 | 972 | 1·7 | 15 | 0201 | 6601 | 1503 | 5701 | 0602 | 1203 |
| EC 7 | 1372 | 1·7 | 10 | 0301 | 1101 | 2705 | 4402 | 0102 | 0704 |
| EC 8 | 714 | 1·7 | 15 | 2301 | 2301 | 1501 | 4901 | 0307 | 0701 |
| EC 9 | – | 1·7 | 20 | 0101 | 0201 | 2705 | 5701 | 0102 | 0602 |
| EC 10 | 570 | 1·7 | 18 | 0301 | 1101 | 0702 | 2705 | 0102 | 0702 |
| VC 1 | – | 2·50 | 13 | 0101 | 1101 | 2705 | 5701 | 0102 | 0701 |
| VC 2 | 651 | 3·08 | 10 | 0101 | 2403 | 1401 | 3801 | 0802 | 1203 |
| VC 3 | – | 3·02 | 16 | 2402 | 3002 | 1503 | 1801 | 0210 | 1203 |
| VC 4 | 600 | 2·65 | 10 | 0201 | 2501 | 1801 | 4403 | 1203 | 1601 |
| VC 5 | 986 | 2·03 | 14 | 3002 | 3101 | 2705 | 4001 | 0202 | 0304 |
| VC 6 | 592 | 3·30 | 15 | 2601 | 2601 | 1401 | 3801 | 0802 | 1203 |
| VC 7 | 609 | 3·04 | 10 | 0201 | 6601 | 5001 | 5201 | 0602 | 1202 |
| VC 8 | 594 | 3·15 | 13 | 2601 | 2601 | 3801 | 5701 | 0602 | 1203 |
| VC 9 | 1760 | 3·06 | 10 | 2402 | 6801 | 3503 | 5201 | 0401 | 1202 |
| VC 10 | 900 | 2·99 | 15 | 0101 | 1101 | 0801 | 2705 | 0102 | 0701 |
| VC 11 | 1200 | 2·04 | 10 | 1101 | 3201 | 2705 | 5101 | 0102 | 1502 |
| VC 12 | 961 | 3·34 | 10 | 0301 | 3101 | 3501 | 3901 | 0401 | 1203 |
| VC 13 | 920 | 1·91 | 13 | 0201 | 3201 | 1401 | 3503 | 0401 | 0802 |
| VC 14 | 550 | 2·51 | 15 | 0201 | 1101 | 5101 | 5701 | 0602 | 1402 |
| TP 1 | – | 3·79 | 18 | 0201 | 6801 | 1501 | 5101 | 0202 | 0303 |
| TP 2 | 594 | 4·15 | 1 | 0201 | 0301 | 1801 | 3503 | 0401 | 0501 |
| TP 3 | 60 | 5·35 | 1 | 0201 | 0301 | 0702 | 3906 | 0702 | – |
| TP 4 | 229 | 4·34 | 5 | 2402 | 3001 | 1302 | 5101 | 0602 | 1402 |
| TP 5 | 320 | 5·03 | 6 | 0201 | 2301 | 0801 | 4901 | 0701 | – |
| TP 6 | 546 | 4·78 | 10 | 2402 | 2501 | 3503 | – | 1203 | – |
| TP 7 | 728 | 3·13 | 1 | 0201 | 6801 | 3901 | 3914 | 0401 | 0702 |
| TP 8 | 390 | 4·35 | – | 0301 | 2402 | 2705 | 3501 | 0102 | 0401 |
| TP 9 | 341 | 3·93 | 3 | 0201 | 0301 | 0702 | – | 0702 | – |
| TP 10 | 780 | 4·99 | 4 | 1101 | 6801 | 1401 | 5101 | 0602 | 1502 |
| TP 11 | 648 | 2·47 | 2 | 0205 | 3301 | 1402 | 4101 | 0701 | 0802 |
| TP 12 | 406 | 4·81 | 2 | 0301 | 2402 | 4402 | 4901 | 0501 | 0701 |
| TP 13 | 432 | 4·72 | 1 | 0201 | 2902 | 4402 | 4403 | 0501 | – |
| HP 1 | 1480 | 1·7 | 3 | 2402 | 2902 | 1517 | 4403 | 0701 | 1601 |
| HP 2 | 440 | 1·7 | – | 0302 | 2601 | 0702 | 5101 | 0702 | 1502 |
| HP 3 | 532 | 1·7 | 4 | 2902 | 3002 | 1801 | 4403 | 0501 | 1601 |
| HP 4 | 494 | 1·7 | 6 | 0101 | 3001 | 0801 | 1302 | 0602 | 0701 |
| HP 5 | 434 | 1·7 | 1 | 3002 | 6802 | 1503 | 1801 | 0210 | 0501 |
| HP 6 | – | 1·7 | – | 2301 | 6802 | 4501 | 5801 | 0718 | 1601 |
| HP 7 | 228 | 1·7 | 1 | 2402 | 2902 | 0801 | 5001 | 0602 | 0701 |
| HP 8 | 575 | 1·7 | 9 | 0301 | 3101 | 2702 | 4403 | 0202 | 1601 |
| HP 9 | 240 | 1·7 | – | 2902 | 3101 | 3502 | 4501 | 0401 | 1601 |
| HP 10 | 782 | 1·7 | 11 | 0101 | 2301 | 0702 | 4403 | 0401 | 0702 |
| HP 11 | 792 | 1·7 | 7 | 0101 | 3301 | 1402 | 1517 | 0701 | 0802 |
| HP 12 | 987 | 1·7 | 2 | 0201 | 0301 | 0702 | 1501 | 0303 | 0702 |
HLA: human leucocyte antigen; HP: subjects on HAART for at least 1 year and maintaining undetectable viral load; TP: typical progressors; VC: viraemic controllers; EC: elite controllers.
Samples from patients were kindly provided by the HIV BioBank integrated into the Spanish AIDS Research Network (RIS). Written informed consent for the examinations conducted in this study was obtained from all individuals, and the study protocol was evaluated and approved by the hospital Ethical Committee.
Clinical specimens
All studies were performed in cryopreserved peripheral blood mononuclear cells (PBMC). Ethylenediamine tetraacetic acid (EDTA)-anticoagulated blood was obtained by venipuncture; PBMC were isolated immediately by density gradient centrifugation using Ficoll-Hypaque (Sigma Chemical Co., St Louis, MO) and frozen in fetal calf serum (FCS) plus 10% dimethyl sulphoxide (DMSO). Viability of thawed PBMC was always greater than 85%.
HIV peptides
A total of 66 optimally defined HIV-specific CTL epitopes from 9 to 11 amino acids in length derived from Gag (45 peptides) and Nef (21 peptides) HIV proteins were selected on the basis of the published list of HIV epitopes at the HIV Molecular Immunology Database 2000 available at Los Alamos National Laboratory (Table S1). They were synthesized as free acids more than 98% pure (Peptide Synthesis Facility; Autonomous University, Barcelona, Spain). Lyophilized peptides were resuspended in DMSO (Sigma, St Louis, MO), mixed into two pools (Gag and Nef), aliquoted and stored at −80°. The concentration of each individual peptide in the pools was 1 mg/ml, and the final culture concentration was 10 μg/ml.
HLA typing
HLA class I typing was carried out at Centro de Transfusiones de la CAM, Spain, using a sequence specific primers (SSP) assay. Table 1 shows class I HLA alleles for each patient.
Functional profile of HIV-specific CD8+ T cells
Production of MIP-1β, IL-2 and TNF-α (the most relevant soluble mediators secreted by HIV-specific CD8+ T cells in differentiating between progressors and non-progressors) was examined simultaneously in response to HIV-Gag and Nef peptide pools using multiparameter flow cytometry, as described previously.5 Briefly, a million PBMC were incubated with each peptide pool in the presence of CD28 and CD49d monoclonal antibodies for 6 hr at 37°. The secretion inhibitor Brefeldin A was added to the cultures during the second hour of incubation. Control conditions included stimulation with medium alone as negative control, and with phorbol myristate acetate (PMA) plus ionomycin as positive control. After incubation, cells were harvested, washed with phosphate-buffered saline (PBS) and incubated with anti-CD8α-energy-coupled dye (ECD) (Beckmann–Coulter, Fullerton, CA) for 30 min at 4°. A commercial kit was used to permeabilize cells (Cytofix/Cytoperm Plus; Pharmingen, San Diego, CA). Permeabilized cells were then incubated with anti-MIP-1β-fluorescein isothiocyanate (FITC) (R&D Systems, Minneapolis, MN), anti-IL-2-phycoerythrin (PE) (Beckmann-Coulter) and anti-TNF-α-biotin (BD Biosciences, San José, CA). Cells were washed and incubated with streptavidin-PEcy5 (BD Biosciences). Gating was performed on CD8bright cells and a minimum of 50 000 of these cells were analysed using an FC 500 cytometer (Beckman Coulter).
On the basis of expression of each of the three molecules examined, seven different unique subsets of CD8+ T cells were defined. For each subset, a level of 0·05% CD8+ cells (after background subtraction) was considered as the threshold for a positive response. In addition, all CD8+ T cells producing one (MIP-1β+, TNF-α+ or IL-2+) or two different cytokines (MIP-1β+ TNF-α+, MIP-1β+ IL-2+, or TNF-α+ IL-2+) were analysed further. Finally, the number of functions expressed by CD8+ T cells (defined as number of different cytokines produced by the same cell) was analysed.
Expansion ability of HIV-specific CD8+ T cells
The capacity of HIV-specific CD8+ T cells to proliferate and expand in vitro after peptide stimulation was tested using a million PBMC. In vitro cultures were conducted for 10 days in the presence of HIV Gag or Nef peptide pools and IL-7 (25 ng/ml). As control for unspecific proliferation, a separate aliquot of cells was cultured in the absence of HIV peptides. At day 3 and every 2 days thereafter, IL-2 (40 UI/ml) was added to the culture. At the end of culture, cells were recovered from the plate, washed and restimulated with peptide pools in a standard 6-hr assay as described above. Gag- and Nef-stimulated cells in the 10-day culture were restimulated with Gag and Nef, respectively, whereas cells cultured in the absence of peptides were split into two aliquots and restimulated with Gag and Nef peptides as controls of unspecific expansion. After the 6-hr culture, cells were washed, permeabilized and stained with the same panel of monoclonal antibodies mentioned above. The used gating strategy is shown in Fig. S1. The level of Gag- and Nef-specific expansion for each of the seven different CD8+ T cell subsets was expressed as the ratio between the level of each subset in the stimulated cells and the one observed in unstimulated cells.
Statistical methods
The prevalence of HLA alleles in each group of patients was analysed using HLA graphing (HLA Analysis Tools; HIV molecular immunology database at Los Alamos Laboratory, http://www.hiv.lanl.gov/content/immunology) and to compare HLA frequencies between two groups HLA comparison was used (HLA Analysis Tools). The remaining statistical analyses were performed using the spss version 15 software (SPSS Inc, Chicago, IL). For each CD8+ T cell subset analysed, the prevalence (expressed as the proportion of patients having a detectable response for that particular subset), the level and the contribution to the global CD8+ response (expressed as the proportion of the global response due to that particular subset) were calculated. Levels and contribution for each subset are expressed as median (interquartile range) as well as levels of expansion. Mann–Whitney U- or Kruskall–Wallis tests were used to compare the levels and contribution of each CD8 T cell subset in the different groups, and the Wilcoxon signed-rank test was used to compare Gag and Nef responses within the same group of patients. The level of expansion of CD8+ T cells was compared using Student's t-test. The potential association between the different subsets of CD8+ T cells, plasma HIV-RNA and CD4 counts was examined using Spearman's or Pearson's correlation coefficients, as appropriate.
Results
Prevalence of class I HLA alleles
According to the presence or absence of protective class I alleles (HLA-B*27, B*58 supertypes and B*1302, B*5101, B*8101), 75% of LTNPs presented protective class I HLA-B alleles. However, only 46% of TP and 58% of HP patients had these alleles. When the group of LTNPs was separated into EC and VC, 80% of EC and 71% of VC presented protective class I HLA-B alleles.
Interestingly, the protective HLA-B*5701 allele was present only in LTNPs (29%), with a similar prevalence in the subgroup of EC compared to the group of VC (40% versus 21%; P = 0·3). The prevalence of protective HLA-B*2705 allele was significantly higher in LTNPs compared to TP and HP patients (33%, 7% and 0%, respectively, P = 0·02) and its prevalence was similar in EC compared to VC (40% versus 29%, P = 0·3).
Lastly, no significant differences were observed in viral load when TP and LTNPs were stratified according to the presence/absence of protective HLA*B alleles and also when LTNPs were stratified according to the presence/absence of either HLA*B-5701 or B-2705.
Association between HLA-B alleles and polyfunctional HIV-specific CD8+ T cells
According to the presence/absence of protective class I HLA-B* alleles, no significant differences in the levels and contribution of different functional Gag- and Nef-specific CD8+ T cell subsets were observed in any of the groups analysed (Fig. 1a). However, when the association between the presence of particular protective HLA-B* alleles and quantitative and qualitative profiles of Gag- and Nef-specific CD8+ T cell responses in each group was analysed, only LTNPs with HLA-B*5701 presented a slightly better HIV-specific CD8+ T cell response than those without this allele. LTNPs with HLA-B*5701 allele had a higher level of Gag-specific MIP-1β+ TNF− IL-2− CD8+ T cells than LTNPs without this allele [3.3%(2.96) and 0.24%(078), P = 0·012], although no significant differences were noted in the contribution profile of Gag-specific CD8+ T cell response in both groups of LTNPs (Fig. 1b).
Figure 1.
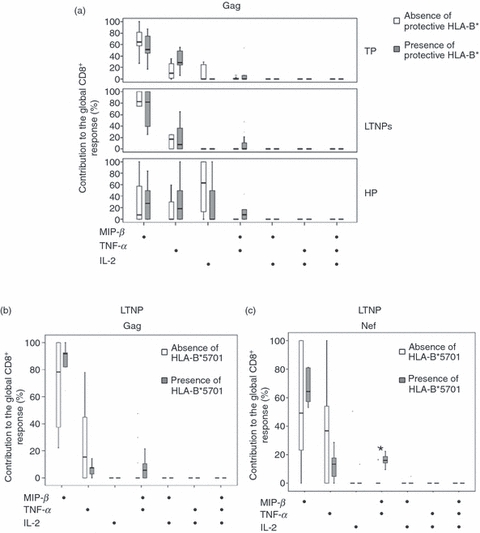
Contribution profile of different subsets to the global Gag-specific CD8+ response, according to the absence/presence of protective class I human leucocyte antigen (HLA)-B* alleles in typical progressors (TP), long-term non-progressors (LTNPs) and subjects on HAART for at least 1 year and maintaining undetectable viral load (HP) (a). Contribution profile of different subsets to the global (b) Gag- and (c) Nef-specific CD8+ T responses, according to the absence/presence of HLA-B*5701 in LTNPs.
Moreover, LTNPs with HLA-B*5701 presented higher levels of different Nef-specific functional CD8+ T cell subsets: MIP+ TNF− IL-2− [0·16%(0·3) and 0·6%(1·2), P = 0·016] and MIP+TNF+IL-2− [0·18%(0·27) and 0%(0·03) P = 0·002]. In addition, in LTNPs with HLA-B*5701 allele a higher contribution of the MIP+ TNF+ IL-2− Nef-specific CD8+ T cell subset was observed [0%(8) and 16%(8); P = 0·02 for LTNPs without and with HLA-B*5701] (Fig. 1c).
Association between HLA-B alleles and expansion ability of HIV-specific CD8+ T cells
Another functional aspect of HIV-specific CD8+ T cells analysed in this study was the ability of these cells to expand in the presence of Gag and Nef peptides, and the association of this ability with the presence of protective HLA-B* alleles was explored. First, HIV-specific CD8 T cells of LTNPs expressing protective alleles presented a higher expansion ability in the next functional subsets: Gag-specific MIP+ TNF+ IL-2+ subset [0·8(18·20) and 0; P = 0·04], Nef-specific MIP+ TNF+ IL-2+ subset [2·18(12·6) and 0; P = 0·02], Nef-specific MIP+ TNF− IL-2+ subset [2(7) and 0; P = 0·02] and Nef-specific MIP+ TNF− IL-2− subset [4·98(0·13) and 2·06(3·5); P = 0·03] than LTNPs without protective alleles (Fig. 2a,b). Interestingly, in LTNPs not expressing protective HLA alleles, all these subsets of Gag- and Nef-specific CD8+ cells (except the Nef-specific MIP+ TNF− IL-2− subset) did not present expansion ability (Fig. 2a,b). In contrast to LTNPs, the presence of protective HLA-B* alleles did not make any difference in the expansion ability of different functional Gag- and Nef-specific CD8 subsets in TP and HP patients (Fig. 2a,b).
Figure 2.
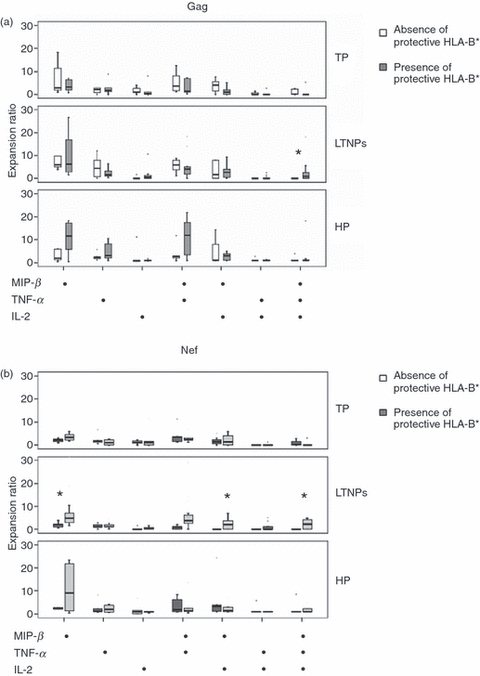
Expansion ability of different subsets to the global (a) Gag- and (b) Nef-specific CD8+ T responses, according to the absence/presence of protective class I human leucocyte antigen (HLA)-B* alleles in typical progressors (TP), long-term non-progressors (LTNPs) and subjects on HAART for at least 1 year and maintaining undetectable viral load (HP).
Secondly, when LTNPs were stratified according to the presence/absence of particular HLA-B* alleles, only the presence of HLA-B*5701 allele was associated with a higher level of expansion for the next functional subsets: Gag-specific MIP+ TNF− IL-2+ cells [5(35·20) and 1·18)8); P = 0·03], Nef-specific MIP+ TNF− IL-2+ subset [4·7(7·1) and 0; P = 0·03] and Nef-specific MIP+ TNF− IL-2− subset [7·15(28) and 2·7(10); P = 0·02] (Fig. 3a,b).
Figure 3.
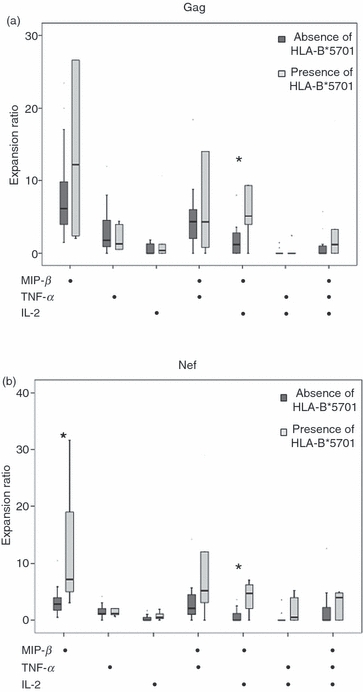
Expansion ability of different subsets to the global (a) Gag- and (b) Nef-specific CD8+ T responses, according to the absence/presence of human leucocyte antigen (HLA)-B*5701 alleles in long-term non-progressors (LTNPs).
Finally, according to the presence/absence of HLA-B*2705, no differences were observed in the expansion ability of different subsets of Gag-specific CD8+ cells in LTNPs. Interestingly, the expansion ability of the MIP+ TNF+ IL-2+ Nef-specific CD8 T cell subset in LTNPs with this allele was lower than in LTNPs without this allele [2·18(4·2) and 4·8(8·6); P = 0·014] (Fig. 4a,b).
Figure 4.
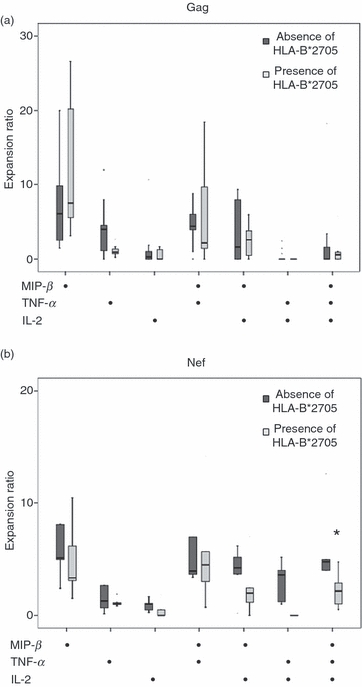
Expansion ability of different subsets to the global (a) Gag- and (b) Nef-specific CD8+ T responses, according to the absence/presence of human leucocyte antigen (HLA)-B*2705 alleles in long-term non-progressors (LTNPs).
Discussion
Due to the recent failure of Merck vaccine, designed to induce protective cellular responses, the need for a more detailed knowledge of which are the attributes of cellular immune response that are responsible for an efficient control of viral replication is required. There is no doubt that the identification of immune correlates of viral replication is of paramount importance for the rational design of new HIV vaccine approaches. In the present study, we have analysed the association between the presence of protective HLA-B* alleles and different aspects of HIV Gag- and Nef-specific CD8+ T cell responses such as the ability to produce soluble mediators and to expand in the presence of HIV peptides. These potential associations have been tested in HIV-infected patients who are able to spontaneously control HIV replication compared to those with HAART-mediated suppression of viral replication and those who are not able to control HIV replication.
Different studies have shown an increase in the prevalence of certain HLA-I alleles in LTNPs, especially HLA-B*57 and HLA-B*27,8,12–14 suggesting that HLA-B genotype is one of the host factors involved in HIV replication control. Furthermore, LTNPs presenting HLA-B*5701 alleles have higher levels of Gag-specific CD8+ T cells producing IFN-γ than those without this allele,8 and the Gag-specific CD8+ T cell response has been associated with lower levels of viral load.15 However, in studies analysing more than one function of HIV-specific CD8+ T cells, these findings have not been confirmed.3,16 In fact, although our LTNP cohort has a higher prevalence of protective HLA-B alleles than the general population of HIV-infected patients from Spain,17 we did not find important significant differences in the functional profile of Gag- and Nef-specific CD8+ T cell responses when HIV-infected patients were stratified according to the presence of protective HLA-B allele, and particularly according to the presence of HLA-B*5701 and HLA-B*2705 alleles. This same finding is observed in HP and TP. Thus, our results suggest that the functional profiles of both Gag- and Nef-specific CD8+ T cell responses mediated by the production of MIP-1β, TNF-α and IL-2 are not merely directed by the HLA-B genotype. Moreover, it has been described recently that there was no apparent relationship between protective HLA-B alleles and the capacity of HIV-specific CD8+ T cells to express perforin after stimulation,18 although we do not discount that other additional functions of antigen-specific CD8+ T cells that we have not analysed, such as cytotoxicity, could be influenced by protective HLA.19
In addition to the ability of secreting soluble mediators, antigen-specific CD8+ T cells present the ability to proliferate in the presence of cognate antigen. This ability of HIV-specific T cells may be related to the control of viral replication, as a deficiency in the in vitro expansion of these cells has been reported previously20,21 and the preservation of the ability to expand has been associated with LTNP status and with the secretion of perforin mediated by HIV-specific CD8+ T cells.4 In both HP and TP, the expansion ability of different functional subsets of HIV-specific CD8+ T cells was similar according to the presence of protective HLA-B* alleles, suggesting that in these groups of HIV-infected patients this functional aspect of HIV-specific CD8+ T cells is not influenced by HLA-B alleles. Moreover, as there were no differences between TP and HP patients, our data suggest that the capacity to expand is not restored by HAART, as has been reported previously by our own group and by others.7,22
A previous study in our laboratory showed no major differences in the functional profile of HIV-specific CD8+ T cells comparing a relatively large group of LTNP and typical progressors, except for a greater prevalence of expansion of HIV-specific CD8+ T cells in LTNP.5 The similar profile of HIV-specific CD8+ T cells we saw in LTNP and progressors is in contrast with observations from others,19 who concluded that LTNP maintain highly functional HIV-specific CD8+ T cells when compared to progressors. However, that study examined a very small group of LTNP. In fact, our results regarding production of MIP-1β, TNF-α and IL-2 are similar to those obtained for these three cytokines by these authors. In the present study, we analysed if the expansion ability of the HIV-specific CD8+ T cell response is associated with particular HLA-B genotypes in different groups of chronically HIV-infected patients. Those LTNPs expressing protective HLA-B alleles had a higher expansion capacity in different HIV-specific CD8 subsets, and interestingly the majority of these subsets secreted IL-2, a cytokine that is involved in the regulation of the proliferation ability of antigen-specific CD8+ T cells.16,23 Because we did not use an irrelevant peptide as control in expansion assay, we cannot rule out a non-specific proliferation. However, giving that the HIV peptides used in our study were synthesized as free acids more than 98% pure, and moreover lyophilized peptides were resuspended in DMSO, the final concentration of DMSO in the cultures being as low as 1%, a non-specific proliferation of CD8+ T cells would be negligible. Conversely, a higher proliferation of HIV-specific CD8+ T cells has been associated with higher levels of CD4 counts;24 however, we did not observe association between CD4 count, protective HLA-B* alleles and expansion ability.
Moreover, the elevated expansion ability observed for MIP+ TNF+ IL-2+ Gag-specific CD8+ T in LTNPs is probably associated with HLA alleles others than HLA-B*5701, as when LTNPs were stratified according to this allele no differences were observed in the expansion ability from the MIP+ TNF+ IL-2+ subset. However, those LTNPs presenting HLA-B*5701 had a higher expansion ability in MIP+ TNF− IL-2+ CD8+ cells for Gag, and MIP+ TNF− IL-2+ and MIP+ TNF− IL-2− CD8+ subsets for Nef. Thus, the nature of the peptide bound by the MHC class I molecule may play an important role in control of HIV replication.25 Interestingly, when LTNPs were stratified according to the presence/absence of HLA-B*2705 those LTNPs with HLA-B*2705 allele controversially presented a lower expansion ability for the Nef-specific MIP+ TNF+ IL-2+ subset, suggesting that these cells in the context of HLA-B*2705 had dysfunctional expansion ability and are not related with the control of viral replication.
Altogether, our findings demonstrate that the quality of HIV-specific CD8+ T cell responses mediated by the production of MIP-1β, TNF-α and IL-2 is not influenced significantly by the presence of protective HLA class I alleles. However, the expansion ability of these polyfunctional CD8+ T cell subsets is modulated by HLA class I alleles and the targeted protein. LTNPs expressing HLA class I protective alleles (especially HLA-B*5701) display a better expansion ability of polyfunctional HIV-specific CD8+ T cells than the rest. However, the presence of protective HLA-B alleles does not totally explain the control of viral replication observed in these patients, as not all LTNPs express these alleles. In these patients, factors other than the presence of protective HLA class I alleles must contribute to the control of viral replication. Moreover, our results demonstrate that the secretion of soluble molecules and expansion ability of HIV-specific CD8+ T cells is not recovered by HAART. Thus, the search for new therapies and vaccines aimed to induce and/or normalize the expansion ability of HIV-specific T cells seems reasonable.
Disclosures
This work was supported in part by grants from FIPSE, Foundation IES, FIS (ISCIII-RETIC RD06/006) and European Union 6th Framework programme (NEAT, number: LSHP-CT-2006-037570).
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Representative flow plots showing expansion ability of non-specific CD8+ T as negative control (a) and expansion ability of Gag-specific CD8 T cells producing macrophage inflammatory protein (MIP)-1β, tumour necrosis factor (TNF)-α and interleukin (IL)-2 in an elite controller (b).
Table S1. Best-defined human immunodeficiency virus (HIV)-specific CD8+ T cell epitopes used in this study.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Borrow P, Lewicki H, Hahn B, Shaw G, Old-stone M. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary HIV type 1 infection. J Virol. 1994;68:6103–10. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koup R, Safrit J, Cao Y, Andrews C, McLeod G, Borkowsky W, Farthing C, Ho D. Temporal association of cellular immune responses with the initial control of viremia in primary HIV type 1 syndrome. J Virol. 1994;68:4650–5. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betts MR, Nason MC, West SM, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T-cells. Blood. 2006;107:4781–9. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Migueles S, Laborico A, Shupert W, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in non-progressors. Nat Immunol. 2002;3:1061–8. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 5.López M, Soriano V, Lozano S, et al. No major differences in the functional profile of HIV Gag and Nef-specific CD8+ responses between long-term nonprogressors and typical progressors. AIDS Res Hum Retroviruses. 2008;24:1185–95. doi: 10.1089/aid.2008.0006. [DOI] [PubMed] [Google Scholar]
- 6.Horton H, Frank I, Baydo R, et al. Preservation of T cell proliferation restricted by protective HLA alleles is critical for immune control of HIV-1 infection. J Immunol. 2006;177:7406–15. doi: 10.4049/jimmunol.177.10.7406. [DOI] [PubMed] [Google Scholar]
- 7.López M, Soriano V, Rallón N, Cascajero A, González-Lahoz J, Benito JM. Suppression of viral replication with highly active antiretroviral therapy has no impact on the functional profile of HIV-specific CD8+ T cells. Eur J Immunol. 2008;38:1548–58. doi: 10.1002/eji.200738054. [DOI] [PubMed] [Google Scholar]
- 8.Migueles SA, Sabbaghian MS, Shupert WL, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci USA. 2000;97:2709–14. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Philip J, Goulder R, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619–30. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emu B, Sinclair E, Hatano H, Ferre A, Shacklett B, Martin JN, McCune JM, Deeks SG. HLA class I-restricted t-cell responses may contribute to the control of human immunodeficiency virus infection, but such responses are not always necessary for long-term virus control. J Virol. 2008;82:5398–407. doi: 10.1128/JVI.02176-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodés B, Toro C, Paxinos E, et al. Differences in disease progression in a cohort of long-term non-progressors after more than 16 years of HIV-1 infection. AIDS. 2004;18:1–8. doi: 10.1097/00002030-200405210-00004. [DOI] [PubMed] [Google Scholar]
- 12.Kaslow RA, Carrington M, Apple R, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–11. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 13.Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–51. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 14.Limou S, Le Clerc S, Coulonges CD, et al. Genomewide Association Study of an AIDS nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02) J Infect Dis. 2009;199:419–26. doi: 10.1086/596067. [DOI] [PubMed] [Google Scholar]
- 15.Kiepiela P, Ngumbela K, Thobakgale C, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 16.Harari A, Cellerai C, Enders FB, et al. Skewed association of polyfunctional antigen-specific CD8 T cell populations with HLA-B genotype. Proc Natl Acad Sci USA. 2007;104:16233–8. doi: 10.1073/pnas.0707570104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arrizabalaga J, Rodriguez-Alcántara F, Castañer JL, et al. Prevalence of HLA-B*5701 in HIV-infected patients in Spain (results of the EPI Study) HIV Clin Trials. 2009;10:48–51. doi: 10.1310/hct1001-048. [DOI] [PubMed] [Google Scholar]
- 18.Hersperger AR, Pereyra F, Nason M, et al. Perforin expression directly ex vivo by HIV-specific CD8 T-cells is a correlate of HIV elite control. PLoS Pathog. 2010;6:e1000917. doi: 10.1371/journal.ppat.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betts MR, Krowka JF, Kepler TB, et al. HIV type 1-specific cytotoxic T lymphocyte activity is inversely correlated with HIV type 1 viral load in HIV type 1-infected long-term survivors. AIDS Res Hum Retrovir. 1999;15:1219–28. doi: 10.1089/088922299310313. [DOI] [PubMed] [Google Scholar]
- 20.Betts M, Ambrozak D, Douek D, et al. Analysis of total HIV-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J Virol. 2001;75:11983–91. doi: 10.1128/JVI.75.24.11983-11991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray C, Lawrence J, Schapiro J, et al. Frequency of HLA-restricted, anti-HIV CD8+ T-cells in individuals receiving highly active antiretroviral therapy (HAART) J Immunol. 1999;162:1780–8. [PubMed] [Google Scholar]
- 22.Migueles SA, Weeks KA, Nou E, et al. Defective human immunodeficiency virus-specific CD8+ T-Cell polyfunctionality, proliferation, and cytotoxicity are not restored by antiretroviral therapy. J Virol. 2009;83:11876–89. doi: 10.1128/JVI.01153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kan-Mitchell J, Bajcz M, Schaubert KL, et al. Degeneracy and repertoire of the human HIV-1 Gag p1777-85 CTL response. J Immunol. 2006;176:6690–701. doi: 10.4049/jimmunol.176.11.6690. [DOI] [PubMed] [Google Scholar]
- 24.Kalams SA, Buchbinder SP, Rosenberg ES, et al. Association between virus-specific cytotoxic T-lymphocyte and helper responses in HIV type 1 infection. J Virol. 1999;73:6715–20. doi: 10.1128/jvi.73.8.6715-6720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loffredo JT, Sidney J, Bean AT, et al. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J Immunol. 2009;182:7763–75. doi: 10.4049/jimmunol.0900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


