Abstract
NOD-like receptors (NLRs) and RIG-I-like receptors (RLRs) are newly discovered pattern-recognition receptors. They detect substructures of bacterial peptidoglycan and viral RNA, respectively, thereby initiating an immune response. However, their role in eosinophil activation remains to be explored. The aim of this study was to characterize the expression of a range of NLRs and RLRs in purified human eosinophils and assess their functional importance. Expression of NOD1, NOD2, NLRP3, RIG-I and MDA-5 was investigated using real-time reverse transcription PCR, flow cytometry and immunohistochemistry. The effects of the corresponding agonists iE-DAP (NOD1), MDP (NOD2), alum (NLRP3) and poly(I:C)/LyoVec (RIG-I/MDA-5) were studied in terms of cytokine secretion, degranulation, survival, expression of adhesion molecules and activation markers, and chemotactic migration. Eosinophils expressed NOD1 and NOD2 mRNA and protein. Low levels of RIG-I and MDA-5 were found, whereas expression of NLRP3 was completely absent. In accordance, stimulation with iE-DAP and MDP was found to induce secretion of interleukin-8, up-regulate expression of CD11b, conversely down-regulate CD62 ligand, increase expression of CD69 and induce migration. The MDP also promoted release of eosinophil-derived neurotoxin, whereas iE-DAP failed to do so. No effects were seen upon stimulation with alum or poly(I:C)/LyoVec. Moreover, the NOD1-induced and NOD2-induced activation was mediated via the nuclear factor-κB signalling pathway and augmented by interleukin-5 and granulocyte–macrophage colony-stimulating factor, but not interferon-γ. Taken together, the NLR system represents a novel pathway for eosinophil activation. The responses are enhanced in the presence of cytokines that regulate T helper type 2 immunity, suggesting that the NLRs constitute a link between respiratory infections and exacerbations of allergic disease.
Keywords: allergy, cytokines, eosinophils, innate immunity
Introduction
Allergic diseases such as rhinitis and asthma have increased in prevalence during the last decades, particularly in countries with a western lifestyle.1 Eosinophils have a central role in allergic diseases, affecting airway inflammation, hyper-responsiveness and remodelling by the release of different tissue-damaging mediators. Also, their numbers are correlated with the disease severity.2–4 In addition to allergens, both bacterial and viral respiratory infections are known to promote hyper-reactive responses.5–7 The mechanisms behind infection-induced exacerbations are still far from understood, but one possible explanation is linked to the activation of airway eosinophils through different pattern-recognition receptors. Previous studies have demonstrated expression of functional Toll-like receptors (TLRs) in eosinophils.8–12 In addition, we have shown that activation of eosinophils via TLR7 and TLR9 is stronger in patients with allergic rhinitis than in healthy subjects and augmented in the presence of T helper type 2 (Th2) cytokines.11,12 These findings led us to investigate whether eosinophils express and respond to the other members of the pattern-recognition receptor family; the nucleotide-binding oligomerization domain (NOD) -like receptors (NLRs) and the retinoic acid-inducible gene-I (RIG-I) -like receptors (RLRs).
The NLR family consists of more than 20 members in humans, located intracellularly in the cytosol.13 Four subfamilies have been approved based on the N-terminal domain: NLR family, acidic domain containing (NLRA); NLR family, BIR domain containing (NLRB); NLR family, CARD domain containing (NLRC) and NLR family, pyrin domain containing (NLRP).14,15 The ligands for most NLRs, and hence their physiological functions, are poorly understood. The best characterized proteins are the NLRC members NOD1 and NOD2, and NLRP3. NOD1 recognizes the peptidoglycan substructures γ-d-glutamyl-meso diaminopimelic acid (iE-DAP), found predominantly in Gram-negative bacteria. NOD2 senses muramyldipeptide (MDP), which is the largest molecular motif common to Gram-negative and Gram-positive bacteria.13,16 NLRP3 responds to microbial products and host-derived danger signals and activates the caspase-1-dependent inflammasome.13,17 Recent studies demonstrate a role for the NLRP3 inflammasome in the recognition of aluminium adjuvants (alum), although it is unclear whether this is a direct interaction.18,19 Despite the recent discovery of NLRs, genetic studies have revealed an association between polymorphisms in the NOD1 and NOD2 genes and allergic disorders, including allergic rhinitis, atopic asthma and allergen-specific serum IgE levels.13,20–22 Moreover, we have shown that patients suffering from symptomatic allergic rhinitis exhibit a lower NOD1 and NALP3 expression than healthy controls and patients outside the pollen season.23
The RLR protein family comprises three members; RIG-I, melanoma differentiation associated gene-5 (MDA-5) and laboratory of genetics and physiology-2 (LGP-2). Unlike the anti-viral TLRs (TLR3, TLR7, TLR8 and TLR9), which are located in endosomal compartments, RLRs detect viral RNA in the cytoplasm.17,24,25 Poly(I:C)/LyoVec is a mimic of viral RNA that binds to RIG-I/MDA-5. However, no synthetic ligand is currently available for LGP-2.
The aim of the present study was to examine the expression of NOD1, NOD2, NLRP3, RIG-I and MDA-5 in purified eosinophils and assess the effects of their ligands on a range of eosinophil functions, including regulation of adhesion molecules and activation markers, chemotactic migration, cytokine release, degranulation and survival.
Materials and methods
Antibodies and reagents
Unlabelled mouse anti-human monoclonal antibodies against NOD2 (msIgG1, clone 2D9), NLRP3 (msIgG1, clone nalpy3-b) and RIG-I (msIgG2a, clone not specified), along with rabbit anti-human polyclonal antibodies against NOD1 and MDA-5 were obtained from Abcam (Cambridge, UK). The peptidoglycan-like molecules iE-DAP and MDP, the negative control compounds iE-Lys (γ-D-glutamyl-Lysine)and MDP control (d isoform of MDP) (all with endotoxin levels < 0·125 EU/ml), along with poly(I:C)/LyoVec, R-837 and celastrol were obtained from Invivogen (San Diego, CA). Aluminium hydroxide (alum) and PMA were purchased from Sigma-Aldrich (St Louis, MO), and recombinant human granulocyte–macrophage colony-stimulating factor (GM-CSF), interferon-γ (IFN-γ), interleukin-5 (IL-5), IL-8 and eotaxin-1 were purchased from R&D Systems (Minneapolis, MN).
Cell separation and cell culture
The local Ethics Committee (Stockholm, Sweden) approved the study (2008/1190–31/3), and all participants gave their informed consent. Freshly drawn peripheral blood was diluted with PBS and centrifuged using Ficoll-Paque (Amersham Bioscience, Uppsala, Sweden). The granulocyte-containing cell pellet was collected and treated with an ammonium chloride erythrocyte lysis solution (0·8% NH4Cl, 10 mm KHCO3 and 0·1 mm EDTA) for 10 min on ice. Eosinophils were isolated using magnetic antibody cell sorting separation (Eosinophil Isolation kit, Miltenyi Biotec, Cologne, Germany) as previously described in detail.11 For immunohistochemistry analysis, eosinophils were purified from fresh buffy coats obtained from the Blood Transfusion Centre, as described above. The purity of the cells was > 98%; contaminating cells were neutrophils (< 1%) and mononuclear cells (< 1%). Cell viability was 95·8 ± 0·7% as determined by propidium iodide staining. Cells were resuspended in complete medium consisting of RPMI-1640 (PAA, Pasching, Austria) supplemented with 0·3 g/l l-glutamine, 10% fetal bovine serum (PAN, Aidenbach, Germany), and 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA). Neutrophils were isolated from peripheral blood using Polymorphprep™ (Axis-Shield PoC AS, Oslo, Norway) as previously described.26 All experiments were performed with freshly isolated cells that had not been further maintained in culture. Purified cells were cultured for 3 or 24 hr at 37° in a humidified 5% CO2 air atmosphere in complete medium alone or with various stimuli and inhibitors as indicated.
RNA extraction and real-time reverse transcription-PCR
Freshly isolated eosinophils and neutrophils were lysed in RLT buffer (Qiagen, Hilden, Germany) supplemented with 1% 2-mercaptoethanol. RNA was extracted using an RNeasy Mini kit (Qiagen), and the quality and quantity of the obtained RNA was determined by spectrophotometry based on the absorbance A260/A280 ratio. The Omniscript Reverse Transcriptase kit (Qiagen) and oligo(dT)16 primer (DNA Technology A/S, Aarhus, Denmark) were used for cDNA synthesis. The samples were denatured (65° for 5 min), chilled (4° for 5 min) and amplified (37° for 1 hr) using a Mastercycler PCR machine (Eppendorf, Hamburg, Germany). Real-time PCR was performed using a stratagene Mx3000P (Agilent Technologies, Santa Clara, CA) using TaqMan Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems, Foster City, CA). Probes (FAM™ labelled) for NOD1 (assay ID Hs01036721_m1), NOD2 (Hs00223394_m1), NLRP3 (Hs00366465_m1), RIG-I (Hs01058986_m1), MDA-5 (Hs00223420_m1) and β-actin (Hs99999903_m1) were obtained from Applied Biosystems. The thermal cycler was set to perform an initial set-up (95°, 10 min) and 45 cycles of denaturation (95°, 15 seconds) followed by annealing/extension (60°, 1 min). The relative amount of mRNA for the genes of interest was determined by subtracting the threshold cycle (Ct) values for the gene from the Ct value for the internal control gene β-actin (ΔCt). Data are depicted as number of NLR/RLR molecules per 100 000 molecules of β-actin (2−ΔCt × 105).27–29
FACS analysis
Flow cytometry analyses were performed on a Coulter Epics XL flow cytometer (Beckman Coulter, Marseille, France). Live cells were gated based on their forward and side scatter properties, and 10 000–15 000 events were collected and analysed using Expo 32 ADC analysis software (Beckman Coulter). For the detection of NLRs and RLRs, intracellular staining of the cells was performed using the IntraPrep™ Permeabilization Reagent kit (Immunotech, Beckman Coulter), as previously described.23,28,30 Unlabelled monoclonal antibodies against NOD2, NLRP3 and RIG-I were used together with AlexaFluor 488 mouse IgG1 or IgG2a labelling kit from Molecular Probes (Eugene, OR). Unlabelled rabbit polyclonal antibodies against NOD1 and MDA-5 were detected using an FITC-conjugated goat anti-rabbit IgG (H&L) polyclonal antibody (Abcam). Isotype controls relevant for each antibody were used for background staining. In addition, cultured cells were analysed for the expression of cell surface markers using the following anti-human antibodies: CD45-ECD (energy-coupled dye) (J.33), CD16-ECD (3G8), CD16-PCy5 (3G8), CD11b-FITC (Bear1), CD62 ligand (CD62L) -phycoerythrin (DREG56) and CD69-ECD (TP1.55.3) from Immunotech, and IL-8RA-fluorescein and CCR3-phycoerythrin from R&D Systems. All washing and labelling steps were performed in PBS or PBS supplemented with 3% fetal bovine serum to avoid unspecific binding. For both extracellular and intracellular staining, cells were incubated with antibodies or appropriate isotype controls for 15 min at room temperature.
Quantification of cell survival
The percentage of viable cells was determined using the Vybrant™ Apoptosis Assay kit #3 (Molecular Probes). Briefly, cells were stained with Annexin V (ANXV)-FITC and propidium iodide (PI) and analysed by flow cytometry. Viable cells were quantified as ANXV− PI−.
Immunohistochemistry
Purified eosinophils were resuspended in a fixation solution containing equal amounts of 10% neutral-buffered formalin and 95% ethanol. Fixed cells were centrifuged, the supernatants were removed and the pellets were embedded in paraffin. Cell preparations were cut in 3 μm thick sections and mounted on glass slides. The NLR and RLR proteins were visualized using Dako Cytomation Envision+ System horseradish peroxidase kits (Copenhagen, Denmark) as previously described.28 Briefly, the sections were treated with xylene to remove paraffin, then ethanol for rehydration, target retrieval solution followed by 1% Triton-X to increase membrane permeability, and 0·03% hydrogen peroxide to quench endogenous peroxidase activity. The sections were incubated overnight at 4° with antibodies against NOD1, NOD2, NLRP3, RIG-I and MDA-5 (1 : 20 and 1 : 50 dilution), thereafter treated with horseradish peroxidase-labelled goat anti-mouse or goat anti-rabbit polymer for 30 min, followed by 3,3′-diaminobenzidine (DAB) substrate-chromogen for 5 min. Counterstaining was performed with Mayer's haematoxylin. Thereafter, the glass slides were dehydrated in ethanol, rinsed in xylene and mounted in Faramount Aqueous Mounting Medium (Dako). As negative controls, N-series universal negative control reagents (Dako) or antibody diluent (Dako) were used. Tris-buffered saline (pH 7·6) supplemented with 0·05% Tween-20 was used in all washing steps.
Chemotaxis assay
Eosinophils (1 × 106 cells/ml) were cultured for 16 hr in the absence or presence of iE-DAP and MDP (100 μg/ml). Thereafter, cell culture inserts (pore size 8 μm) from Becton Dickinson (Franklin Lakes, NJ) were placed in 12-well culture plates containing complete medium supplemented with IL-8 or eotaxin (10 ng/ml). The cultured cells were subsequently added to the upper wells and allowed to migrate for 4 hr. Migrated cells were counted microscopically using a Bürker chamber. For each well, the number of migrated cells in four random fields was determined. The results are expressed as chemotactic index (CI = migration with NLR ligands/spontaneous migration) as previously described.10,11
ELISA
Cell culture supernatants were assayed for IL-8 and RANTES using ELISA plates from R&D Systems and eosinophil-derived neurotoxin (EDN) from MBL (Nagoya, Japan). An IFN-α multi-subtype ELISA from PBL Biomedical Laboratories (Piscataway, NJ) was used to detect all IFN-α subunits except IFN-α F.
Statistics
Data are expressed as mean ± SEM, and analysed using paired Student's t-test (for two sets of data) or one-way repeated measures analysis of variance (for more than two sets of paired data). P-values < 0·05 were considered statistically significant.
Results
Expression of NOD1, NOD2, NLRP3, RIG-I and MDA-5 in eosinophils
The expression of NOD1, NOD2, NLRP3, RIG-I and MDA-5 transcripts was quantified in purified eosinophils and neutrophils using real-time reverse transcriptase-PCR. Eosinophils expressed NOD1, NOD2 and RIG-I, whereas the whole range of receptors was present in neutrophils (Fig. 1).
Figure 1.

Expression of nucleotide-binding oligomerization domain (NOD) -like receptor (NLR) and retinoic acid-inducible gene-I (RIG-I) -like receptor (RLR) mRNA. NOD1, NOD2, NLR-family, pyrin domain containing (NLRP3), RIG-I and melanoma differentiation associated gene-5 (MDA-5) mRNA expression in purified eosinophils (n = 10) and neutrophils (n = 10) determined by real-time reverse transcriptase-PCR. Data are presented in relation to the internal control gene β-actin as 2−ΔCt × 105 and depicted as mean ± SEM.
To verify the presence of the receptors, FACS analysis was carried out on freshly isolated eosinophils. Expression of NOD1 and NOD2 was seen in eosinophils, along with a less distinct expression of RIG-I and MDA-5. NLRP3 could not be detected (Fig. 2a). In line with mRNA data, all receptors were present in neutrophils (Fig. 2b). Also, the expression levels were quantified by determining the relative mean fluorescence intensity (rMFI = MFIantibody/MFIisotype control), which confirmed the generally higher expression seen in neutrophils (Fig. 2c).
Figure 2.

Expression of nucleotide-binding oligomerization domain (NOD) -like receptor (NLR) and retinoic acid-inducible gene-I (RIG-I) -like receptor (RLR) proteins. (a) NOD1, NOD2, NLR-family, pyrin domain containing (NLRP3), RIG-I and melanoma differentiation associated gene-5 (MDA-5) protein expression in purified eosinophils, and (b) neutrophils. Cells were stained intracellularly with FITC-conjugated NOD1, NOD2, NLRP3, RIG-I and MDA-5 (open histograms) and analysed by flow cytometry. Appropriate isotype controls (grey histograms) were used for background staining. Data show one out of three independent experiments. (c) Data are presented as relative mean fluorescence intensity (rMFI = MFIantibody/MFIisotype control) and depicted as mean ± SEM (n = 3). (d) Immunohistochemical staining of eosinophils with antibodies against NOD1, (e) NOD2, (f) NLRP3, (g) RIG-I, (h) MDA-5 (all diluted 1 : 20), and (i) N-series universal negative control reagent. Slides were visualized using 3,3′-diaminobenzidine (brown). All slides were counterstained with haematoxylin (blue) and analysed by microscopy; magnification 1000 ×.
Immunohistochemical staining of purified eosinophils with antibodies against the receptors further demonstrated the presence of NOD1 and NOD2 (at dilutions 1 : 20 and 1 : 50) (Fig. 2d,e). A weak positive immunostaining was seen with RIG-I and MDA-5 (dilution 1 : 20) (Fig. 2g,h), whereas the staining was negative for NLRP3 (Fig. 2f). Replacement of the primary specific antibodies with N-series universal negative control reagent resulted in a complete loss of staining (Fig. 2i).
iE-DAP and MDP induce cytokine secretion
To investigate whether the receptors exhibit functional activity, the responsiveness of purified eosinophils to different concentrations of iE-DAP (NOD1), MDP (NOD2), alum (NLRP3) and poly(I:C)/LyoVec (RIG-I/MDA-5) was assessed. The negative control compounds iE-Lys and MDP control were used to rule out unspecific activation of NOD1 and NOD2. Purified cells were cultured for 3 and 24 hr in the absence or presence of the ligands followed by analysis of IL-8, IFN-α and RANTES levels in the supernatants by ELISA. After 24 hr, an increase in the production of IL-8 was seen with iE-DAP and MDP, whereas no effects were seen with alum or poly(I:C)/LyoVec (Fig. 3a). The control compounds iE-Lys and MDP control (white bars) did not affect cytokine secretion, whereas R-837 functioned as the positive control. A 3-hr culture period revealed that the IL-8 induction was both concentration-dependent and time-dependent (Fig. 3b,c). Levels of IFN-α were below the detection limit of the assay both in the absence and presence of the ligands (n = 6, data not shown). There were large inter-individual differences in the release of RANTES, but despite normalizing the values to the untreated control, effects of the ligands were seen after neither 3 nor 24 hr of stimulation (n = 15, data not shown). Interestingly, the differences in RANTES levels between donors were found to be related to their allergic status. The variation was marked among atopic patients with seasonal allergic rhinitis outside the pollen season, whereas it was almost negligible among healthy non-allergic controls. This phenomenon was not seen for IL-8 (Fig. 3d,e).
Figure 3.

Nucleotide-binding oligomerization domain 1(NOD1) and NOD2 activation induces cytokine secretion. (a) Purified eosinophils (1 × 106 cells/ml) were cultured for 24 hr in the absence or presence of γ-d-glutamyl-meso diaminopimelic acid (iE-DAP), muramyl dipeptide (MDP), alum (10 and 100 μg/ml), poly(I:C)/LyoVec (PLV; 0·1 and 1 μg/ml) and R-837 (10 μg/ml) (grey bars), and the control compounds iE-Lys and MDP control (10 and 100 μg/ml) (white bars). Thereafter, the cell-free culture supernatants were analysed for levels of interleukin-8 (IL-8) by use of ELISA (n = 11). (b–c) IL-8 secretion by iE-DAP and MDP after 3 and 24 hr (n = 4). (d–e) Release of IL-8 and RANTES by eosinophils from atopic patients with seasonal allergic rhinitis outside the pollen season (n = 7) and healthy non-allergic controls (n = 8). Data are presented as mean ± SEM. *P < 0·05, **P < 0·01.
Eosinophil degranulation
To study eosinophil degranulation, release of EDN was measured in the cell culture supernatants after 3 and 24 hr of culture. MDP gave rise to an increase in EDN levels after 3 hr compared with the untreated control, whereas remaining ligands were unable to induce degranulation (Fig. 4a). PMA (1 ng/ml) was used as a positive control. Moreover, the basal EDN release was significantly higher after a 24-hr culture period, and consequently, the MDP-induced EDN secretion was less distinct (Fig. 4b).
Figure 4.

Muramyl dipeptide (MDP) induces eosinophil degranulation. Purified cells (1 × 106 cells/ml) were cultured for 3 and 24 hr with or without γ-d-glutamyl-meso diaminopimelic acid (iE-DAP), MDP, alum (10 and 100 μg/ml), poly(I:C)/LyoVec (0·1 and 1 μg/ml) and PMA (1 ng/ml). Thereafter, levels of eosinophil-derived neurotoxin (EDN) were determined in the cell-free culture supernatants by ELISA. (a) EDN release after 3 hr (n = 6). (b) EDN release by MDP after 3 and 24 hr (n = 6). Data are presented as mean ± SEM. #P < 0·05, ##P < 0·01 (black square compared with white circle); **P < 0·01 (compared with untreated control).
Stimulation via NOD1 and NOD2 affects eosinophil survival
The ability of the ligands to prolong eosinophil survival was investigated by staining cultured cells with ANXV and PI followed by FACS analysis. After 24 hr of culture, the percentage of viable cells was slightly, but significantly higher in the presence of iE-DAP (10 μg/ml) and MDP (both concentrations). No effects on viability were seen with iE-Lys, MDP control, alum or poly(I:C)/LyoVec. However, the increase in response to the NOD1 and NOD2 ligands was modest compared with R-837 (Table 1).
Table 1.
Effects on eosinophil survival
| Control | iE-DAP (μg/ml) | iE-Lys (μg/ml) | MDP (μg/ml) | MDP control (μg/ml) | Alum (μg/ml) | PLV (μg/ml) | R-837 (μg/ml) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 10 | 100 | 10 | 100 | 10 | 100 | 10 | 100 | 10 | 100 | 0·1 | 1 | 10 | |
| Viable cells(%) | 37·6 ± 3·9 | 41·2 ± 4·2* | 39·8 ± 4·4 | 38·2 ± 9·7 | 37·8 ± 8·5 | 43·0 ± 3·7*** | 42·1 ± 3·7** | 35·7 ± 8·9 | 35·5 ± 8·6 | 35·3 ± 5·4 | 40·8 ± 4·4 | 36·4 ± 3·7 | 35·5 ± 3·8 | 54·9 ± 5·1*** |
Data are presented as the percentage of live (ANXV− PI−) cells and depicted as mean ± SEM (n = 7).
P < 0·05
P < 0·01
P < 0·001.
iE-DAP, γ-d-glutamyl-meso diaminopimelic acid; iE-Lys, γ-d-glutamyl-Lysine; MDP, muramyl dipeptide; PLV, poly(I:C)/LyoVec.
iE-DAP and MDP regulate cell surface expression of CD11b, CD62L and CD69 and induce eosinophil migration
Next, the influence of the ligands on a range of cell surface antigens was assessed, including the adhesion molecules CD11b and CD62L, and the eosinophil activation marker CD69. Alum and poly(I:C)/LyoVec did not affect cytokine secretion, degranulation or cell survival so the following experiments included only iE-DAP and MDP. A constitutive expression of CD11b was found on eosinophils, and a 3-hr culture period with the NOD1 and NOD2 agonists gave rise to a significant up-regulation in terms of receptor MFI (Fig. 5a,d). In contrast, for CD62L two distinct cell populations were present. NLR stimulation gave rise to a clear reduction in MFI (Fig. 5b,e), and in accordance the percentage of CD62L-positive cells decreased upon stimulation with iE-DAP (10 μg/ml: 79·1 ± 4·6%, 100 μg/ml: 74·5 ± 4·1%, P = 0·04) and MDP (10 μg/ml: 67·4 ± 5·4%, 100 μg/ml: 55·3 ± 8·1%, P = 0·006) compared with untreated control (80·5 ± 3·8%). CD69 was absent on untreated eosinophils, but induction was seen upon stimulation with iE-DAP and MDP (Fig. 5c,f). None of the surface markers were affected by the inactive equivalents iE-Lys and MDP control (Fig. 5g–i). R-837, on the other hand, regulated the MFI of CD11b (R-837: 22·1 ± 0·9 versus control: 15·5 ± 0·7, P = 0·004), CD62L (R-837: 3·2 ± 0·4 versus control: 7·2 ± 0·8, P = 0·03) and CD69 (R-837: 1·6 ± 0·1 versus control: 1·2 ± 0·1, P = 0·004) compared with the untreated control.
Figure 5.

Nucleotide-binding oligomerization domain 1 (NOD1) and NOD2 activation regulates the expression of CD11b, CD62L and CD69 and induces migration. Purified eosinophils (1 × 106 cells/ml) were cultured for 3 hr in the absence or presence of γ-d-glutamyl-meso diaminopimelic acid (iE-DAP), iE-Lys, muramyl dipeptide (MDP) and MDP control. Thereafter, cells were stained with antibodies against CD16 in combination with CD11b, CD62L and CD69 and analysed by flow cytometry. (a–c) Histograms showing unstimulated control (filled histograms) and cells stimulated with 100 μg/ml iE-DAP and MDP (open histograms). (d–i) Mean fluorescence intensity (MFI) of CD11b, CD62L and CD69 (n = 6). (j) Eosinophils were cultured for 16 hr with or without iE-DAP and MDP (100 μg/ml). Thereafter, the cell suspensions were transferred to the upper wells of cell culture inserts placed in a 12-well culture plate, while complete medium supplemented with interleukin-8 (IL-8; 10 ng/ml) or eotaxin (10 ng/ml) was added to the lower wells. The cells were allowed to migrate for 4 hr before counting. Migrated cells are expressed as chemotactic index (migration with NLR ligands/spontaneous migration) (IL-8, n = 5; eotaxin, n = 4). Data are shown as mean ± SEM. *P < 0·05, **P < 0·01, ***P < 0·001.
Moreover, because an activation of the eosinophil adhesion system indicates an increase in the migratory ability of the cells, a chemotaxis assay was carried out in which cells pre-treated with iE-DAP and MDP for 16 hr were allowed to migrate towards the chemoattractants IL-8 and eotaxin. Results showed that both ligands facilitated the chemotactic migration (Fig. 5j). To study whether this increase was a result of an up-regulated expression of the corresponding chemokine receptors, the expression of the IL-8 and eotaxin receptor; IL-8RA/CXCR1 and CCR3, respectively, was determined by flow cytometry. However, the expression levels remained unchanged (n = 5, data not shown).
iE-DAP and MDP induce nuclear factor-κB activation
Purified eosinophils were pre-treated for 45 min with vehicle (DMSO) or the nuclear factor-κB (NF-κB) inhibitor celastrol, further incubated with iE-DAP and MDP for 24 hr and thereafter analysed for IL-8 secretion. Celastrol completely abolished the increase in IL-8 secretion induced by the NOD1 and NOD2 agonists (Fig. 6). To ensure that the inhibitory effect of celastrol was not the result of unspecific mechanisms, eosinophils were cultured with the inhibitor in the absence of iE-DAP and MDP. No effects were seen, suggesting that the ability of iE-DAP and MDP to activate eosinophils was mediated via the NF-κB signalling pathway.
Figure 6.

Nuclear factor-κB (NF-κB) -dependent nucleotide-binding oligomerization domain (NOD1) and NOD2 activation of eosinophils. Purified cells (1 × 106 cells/ml) were pre-treated for 45 min with celastrol (10 μg/ml), further incubated with γ-d-glutamyl-meso diaminopimelic acid (iE-DAP) and muramyl dipeptide (MDP) (100 μg/ml) for 24 hr and thereafter analysed for secretion of interleukin-8 by ELISA (n = 6). *P < 0·05.
IL-5 and GM-CSF augment the NOD1-induced and NOD2-induced eosinophil activation
Next, we wanted to explore if the NOD1-induced and NOD2-induced activation of eosinophils is affected by a Th1- and Th2-dominated micro-environment. Purified cells were primed with IL-5, GM-CSF and IFN-γ for 45 min before the addition of iE-DAP and MDP. After another 24-hr incubation period, IL-8 and RANTES release was measured in the cell culture supernatants. There were large inter-individual variations in RANTES levels so the results are depicted in relation to the untreated control. The experiments revealed that neither IL-5 nor IFN-γ alone affected the cytokine release. GM-CSF, on the other hand, significantly induced secretion of IL-8. Moreover, the effects of both iE-DAP and MDP were synergistically enhanced in the presence of IL-5 and GM-CSF in terms of IL-8 and RANTES secretion (Fig. 7a,b), whereas no effects were seen with IFN-γ (Fig. 7c). To investigate the mechanisms behind this augmentation in NLR responses, expression of NOD1 and NOD2 was monitored after stimulation with IL-5 and GM-CSF for 45 min and 24 hr, corresponding to the priming and cultivation periods, respectively. However, the NLR expression levels were unaffected (n = 4, data not shown). Instead, the effects of IL-5 and GM-CSF on eosinophil survival were investigated after 24 hr of stimulation. Both cytokines were found to significantly increase the percentage of viable cells (Fig. 8).
Figure 7.

Priming with interleukin-5 (IL-5) and granulocyte–macrophage colony-stimulating factor (GM-CSF) augments the nucleotide-binding oligomerization domain 1- (NOD1) and NOD2-induced eosinophil activation. Purified cells (1 × 106 cells/ml) were pre-treated with (a) 10 ng/ml IL-5 (n = 6), (b) 10 ng/ml GM-CSF (n = 7), and (c) 500 U/ml interferon-γ (IFN-γ; n = 6) for 45 min before the addition of γ-d-glutamyl-meso diaminopimelic acid (iE-DAP) and muramyl dipeptide (MDP). After 24 hr, levels of IL-8 and RANTES were measured in the cell-free culture supernatants by ELISA. Data are presented as mean ± SEM. *P < 0·05, **P < 0·01, ***P < 0·001 (compared with untreated control); #P < 0·05, ##P < 0·01, ###P < 0·001 (filled bar compared with white bar).
Figure 8.
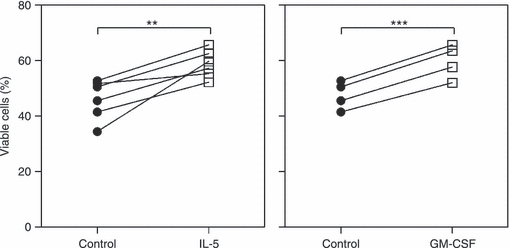
Interleukin-5 (IL-5) and granulocyte–macrophage colony-stimulating factor (GM-CSF) prolong eosinophil survival. Purified eosinophils (1 × 106 cells/ml) were cultured in the absence or presence of 10 ng/ml IL-5 (n = 6) and GM-CSF (n = 4). After 24 hr of stimulation, the viability was analysed by staining the cells with Annexin V (ANXV) and propidium iodide (PI) followed by FACS analysis. Viable cells were quantified as ANXV− PI−. Data are presented as mean ± SEM. **P < 0·01, ***P < 0·001 (compared with untreated control).
Discussion
We have previously demonstrated the TLR system to be a pathway for eosinophil activation that might link respiratory infections with exacerbations of allergic disease.11,12 Therefore, we hypothesized that the NLRs and RLRs exert similar functions. The present study shows that purified human eosinophils express NOD1 and NOD2, and that activation by iE-DAP and MDP induces cytokine secretion, regulates the expression of adhesion molecules and activation markers, and facilitates the chemotactic migration. In addition, NOD2 stimulation enhances degranulation, and priming with cytokines that regulate Th2 immunity augments the NLR-induced inflammatory response. Moreover, the effects exerted by iE-DAP and MDP are dependent on activation of the NF-κB pathway. A weak expression of RIG-I and MDA-5 is found, although the receptors seem to be functionally inactive. NLRP3 is completely absent, and no effects are seen upon stimulation with alum.
The expression of NLRs and RLRs and their functions in various immune cells are being increasingly clarified, although they are still far from understood. Expression of functionally active NOD1 and NOD2 has been demonstrated in monocytes, dendritic cells and epithelial cells,13,31–33 whereas a role for NLRP3 has been reported in monocytes and macrophages.19,34,35 A recent study from our laboratory shows expression of NOD2 and NLRP3 in neutrophils and that MDP and alum promote activation.27 We have also demonstrated the presence of a range of NLRs in human peripheral and/or tonsillar B and T lymphocytes and that iE-DAP and MDP are capable of inducing activation upon concomitant B-cell receptor and T-cell receptor triggering.28,30 The latter report also demonstrates expression of RIG-I, MDA-5 and LGP-2 in tonsillar T cells and that poly(I:C)/LyoVec stimulates proliferation of tonsillar mononuclear cells activated via CD3 and CD28.30 In addition, there is one paper showing that the genes for RIG-I, MDA-5 and LGP-2 are up-regulated in human monocyte-derived DCs infected with the recombinant canarypox virus ALVAC.36 However, information about the expression of NLRs or RLRs in eosinophils is to date limited, with one study showing that murine eosinophils lack NLRP3.37 The present report shows for the first time a clear mRNA and protein expression of NOD1 and NOD2 in eosinophils, and so complements previous studies on TLRs in eosinophils in terms of innate immune receptors and pathogen recognition. Moreover, the expression profile in eosinophils differed from that in neutrophils, both quantitatively and qualitatively. Neutrophils expressed high levels of all receptors, which might reflect their important role in first-line defence by responding to a broad repertoire of pathogens.
After having characterized the expression profile of the receptors, the effects of the corresponding agonists iE-DAP, MDP, poly(I:C)/LyoVec and alum were studied on a range of eosinophil functions. The NOD1 and NOD2 ligands gave rise to an increased secretion of IL-8, a small yet significant prolongation of eosinophil survival, an altered expression of CD11b and CD62L along with a corresponding increase in migration, and an up-regulation of CD69. Moreover, even though low levels of RIG-I and/or MDA-5 mRNA and proteins were found in eosinophils, no effects were seen upon stimulation with poly(I:C)/LyoVec. In line with the absence of NLRP3 expression, eosinophils did not respond to alum. It should also be mentioned that mRNA and protein expression do not always concur, which can be related to differences in mRNA and protein half-life, transcriptional control and protein consumption, as previously reported for NLRs and RLRs in human neutrophils, B cells and T cells.27,28,30 This underscores the importance of analysing receptor functionality in addition to mRNA and protein occurrence.
It was also found that MDP increased secretion of the cytotoxic granule protein EDN after a 3-hr culture period, whereas iE-DAP failed to do so. However, after 24 hr the fold induction was less prominent (1·2-fold) compared with 3 hr of stimulation (1·4-fold) as a result of a higher basal EDN release. This phenomenon is most likely a reflection of cell death as many of the cells are dead and consequently have released their toxic contents. Similarly, we have previously shown an induction in EDN release in response to CpG stimulation after both 3 and 24 hr with a less pronounced relative increase at the later time-point.11
Stimulation of NOD1 and NOD2 has previously been described to cause recruitment of the adaptor protein RICK/RIP2, which in turn results in activation of the transcription factor NF-κB, which regulates the production of pro-inflammatory cytokines and type I IFNs.38–40 Accordingly, inhibition of NF-κB resulted in a complete attenuation of the iE-DAP and MDP-induced IL-8 secretion in eosinophils.
Pre-treatment of eosinophils with IL-5 and GM-CSF was found to augment the NLR-induced eosinophil activation without concomitantly affecting the expression levels of NOD1 and NOD2. In accordance, we have previously reported that priming with IL-5 enhances the R-837-induced and CpG-induced responses without affecting the expression of TLR7 and TLR9,11 and Nagase et al.8 have shown that treatment with IL-4 and IL-5 does not alter the TLR mRNA levels. Instead, the augmentation in NOD1 and NOD2 responses might be related to the ability of IL-5 and GM-CSF to prolong survival. It is well-known that IL-5 and GM-CSF have significant functional homology and contribute to the regulation of Th2 immunity by, for example, stimulating eosinophil growth, differentiation and survival.41,42 However, it should be mentioned that GM-CSF is also involved in diseases that are not considered Th2-driven.43 Moreover, the lack of responsiveness upon co-culture with IFN-γ is in line with its role in promoting Th1-type and inhibiting Th2-type responses.41
Some of the responses to iE-DAP and MDP are fairly small in magnitude, which raises questions as to the biological versus the statistical significance. However, as eosinophils were found to be completely unresponsive to the control compounds iE-Lys and the d-isoform of MDP at equivalent concentrations, the biological relevance of these results is strengthened and the risks of off-target effects and the possible contribution of contaminants are minimized. Also, the fact that the cells responded in a synergistic manner to iE-DAP and MDP in the presence of IL-5 and GM-CSF shows that the effects are not a result of differences in survival or cell fitness in untreated and treated cells, but are truly mediated via NOD1 and NOD2. Moreover, culturing purified eosinophils with specific NLR ligands creates a monoculture and an isolated system that does not mirror the natural biological environment in which different cell types and mediators interact and cooperate. Efficient detection and clearance of a bacterial infection by the immune system is enabled by the fact that several signals and events are triggered simultaneously. This is reflected by the cytokine-induced augmentation in the responsiveness to iE-DAP and MDP seen in the present study. Hence, it is not the effects of the individual NLR ligand, but the combined actions of different signals that are of biological importance.
During activation, eosinophils up-regulate CD11b to facilitate firm adhesion to the vascular membrane, and simultaneously shed CD62L (also known as l-selectin) to permit rolling along the endothelium. These events in turn favour homing to inflamed tissues, which in the context of allergic diseases correspond to the nose and lungs, by the actions of different chemokines such as IL-8 and eotaxin.44 In addition, activated eosinophils up-regulate CD69, leading to an increased ability to interact with surrounding cells.45 They also release an array of cytokines and chemokines, including IL-8 and RANTES that affect cellular trafficking.46 Taken together, our results indicate that eosinophils activated via NOD1 and NOD2 acquire an activated phenotype that increases the ability of the cells to migrate to sites of inflammation, and to direct the traffic of other leucocytes into inflamed areas.
The present paper shows that eosinophils can be directly activated by the NOD1 and NOD2 agonists and that the responses are augmented in the presence of IL-5 and GM-CSF. This suggests a role for the NLR system in eosinophil activation. It is well-known that the increased responsiveness of the airways seen during periods of symptomatic allergic rhinitis can be further enhanced if the patient simultaneously suffers from a bacterial or viral infection. It might be that eosinophils, accumulated in the airways during such periods, are activated by bacterial or viral components to release mediators that in turn aggravate the allergic inflammation and cause tissue damage. In this context it is tempting to speculate on a role for the NLR system in linking respiratory infections with allergic disease exacerbations.
Acknowledgments
This study was supported by the Swedish Medical Research Council, the Swedish Heart-Lung Foundation and Karolinska Institutet, Sweden.
Glossary
Abbreviations
- ANXV
annexin V
- CD62L
CD62 ligand
- EDN
eosinophil-derived neurotoxin
- GM-CSF
granulocyte–macrophage colony-stimulating factor
- iE-DAP
γ-d-glutamyl-meso diaminopimelic acid
- IFN
interferon
- IL-5
interleukin-5
- MDA-5
melanoma differentiation associated gene-5
- MDP
muramyldipeptide
- NF-κB
nuclear factor-κB
- NLR
NOD-like receptor
- NLRP
NLR-family, pyrin domain containing
- NOD
nucleotide-binding oligomerization domain
- PI
propidium iodide
- RIG-I
retinoic acid-inducible gene-I
- RLR
RIG-I-like receptor
- Th2
T helper type 2
- TLR
Toll-like receptor
Disclosures
The authors declare that there are no conflicts of interest.
References
- 1.Bousquet J, Khaltaev N, Cruz AA, et al. Allergic Rhinitis and its Impact on Asthma (ARIA) 2008 update (in collaboration with the World Health Organization, GA(2)LEN and AllerGen) Allergy. 2008;63(Suppl 86):8–160. doi: 10.1111/j.1398-9995.2007.01620.x. [DOI] [PubMed] [Google Scholar]
- 2.Trivedi SG, Lloyd CM. Eosinophils in the pathogenesis of allergic airways disease. Cell Mol Life Sci. 2007;64:1269–89. doi: 10.1007/s00018-007-6527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kariyawasam HH, Robinson DS. The role of eosinophils in airway tissue remodelling in asthma. Curr Opin Immunol. 2007;19:681–6. doi: 10.1016/j.coi.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 4.Hogan SP, Rosenberg HF, Moqbel R, et al. Eosinophils: biological properties and role in health and disease. Clin Exp Allergy. 2008;38:709–50. doi: 10.1111/j.1365-2222.2008.02958.x. [DOI] [PubMed] [Google Scholar]
- 5.Micillo E, Bianco A, D'Auria D, Mazzarella G, Abbate GF. Respiratory infections and asthma. Allergy. 2000;55(Suppl 61):42–5. doi: 10.1034/j.1398-9995.2000.00506.x. [DOI] [PubMed] [Google Scholar]
- 6.Newcomb DC, Peebles RS., Jr Bugs and asthma: a different disease? Proc Am Thorac Soc. 2009;6:266–71. doi: 10.1513/pats.200806-056RM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sykes A, Johnston SL. Etiology of asthma exacerbations. J Allergy Clin Immunol. 2008;122:685–8. doi: 10.1016/j.jaci.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 8.Nagase H, Okugawa S, Ota Y, et al. Expression and function of Toll-like receptors in eosinophils: activation by Toll-like receptor 7 ligand. J Immunol. 2003;171:3977–82. doi: 10.4049/jimmunol.171.8.3977. [DOI] [PubMed] [Google Scholar]
- 9.Wong CK, Cheung PF, Ip WK, Lam CW. Intracellular signaling mechanisms regulating toll-like receptor-mediated activation of eosinophils. Am J Respir Cell Mol Biol. 2007;37:85–96. doi: 10.1165/rcmb.2006-0457OC. [DOI] [PubMed] [Google Scholar]
- 10.Cheung PF, Wong CK, Ip WK, Lam CW. FAK-mediated activation of ERK for eosinophil migration: a novel mechanism for infection-induced allergic inflammation. Int Immunol. 2008;20:353–63. doi: 10.1093/intimm/dxm146. [DOI] [PubMed] [Google Scholar]
- 11.Mansson A, Cardell LO. Role of atopic status in Toll-like receptor (TLR)7- and TLR9-mediated activation of human eosinophils. J Leukoc Biol. 2009;85:719–27. doi: 10.1189/jlb.0808494. [DOI] [PubMed] [Google Scholar]
- 12.Mansson A, Fransson M, Adner M, Benson M, Uddman R, Bjornsson S, Cardell LO. TLR3 in human eosinophils: functional effects and decreased expression during allergic rhinitis. Int Arch Allergy Immunol. 2010;151:118–28. doi: 10.1159/000236001. [DOI] [PubMed] [Google Scholar]
- 13.Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat Immunol. 2006;7:1250–7. doi: 10.1038/ni1412. [DOI] [PubMed] [Google Scholar]
- 14.Ting JP, Lovering RC, Alnemri ES, et al. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–7. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaparakis M, Philpott DJ, Ferrero RL. Mammalian NLR proteins; discriminating foe from friend. Immunol Cell Biol. 2007;85:495–502. doi: 10.1038/sj.icb.7100105. [DOI] [PubMed] [Google Scholar]
- 16.Le Bourhis L, Benko S, Girardin SE. Nod1 and Nod2 in innate immunity and human inflammatory disorders. Biochem Soc Trans. 2007;6:1479–84. doi: 10.1042/BST0351479. [DOI] [PubMed] [Google Scholar]
- 17.Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol. 2009;21:317–37. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kool M, Petrilli V, De Smedt T, et al. Cutting edge: alum adjuvant stimulates inflammatory dendritic cells through activation of the NALP3 inflammasome. J Immunol. 2008;181:3755–9. doi: 10.4049/jimmunol.181.6.3755. [DOI] [PubMed] [Google Scholar]
- 19.Eisenbarth SC, Colegio OR, O'Connor W, Sutterwala FS, Flavell RA. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008;453:1122–6. doi: 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eder W, Klimecki W, Yu L, et al. Association between exposure to farming, allergies and genetic variation in CARD4/NOD1. Allergy. 2006;61:1117–24. doi: 10.1111/j.1398-9995.2006.01128.x. [DOI] [PubMed] [Google Scholar]
- 21.Hysi P, Kabesch M, Moffatt MF, et al. NOD1 variation, immunoglobulin E and asthma. Hum Mol Genet. 2005;14:935–41. doi: 10.1093/hmg/ddi087. [DOI] [PubMed] [Google Scholar]
- 22.Weidinger S, Klopp N, Rummler L, et al. Association of NOD1 polymorphisms with atopic eczema and related phenotypes. J Allergy Clin Immunol. 2005;116:177–84. doi: 10.1016/j.jaci.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 23.Bogefors J, Rydberg C, Uddman R, et al. Nod1, Nod2 and Nalp3 receptors, new potential targets in treatment of allergic rhinitis? Allergy. 2010;65:1222–6. doi: 10.1111/j.1398-9995.2009.02315.x. [DOI] [PubMed] [Google Scholar]
- 24.Creagh EM, O'Neill LA. TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol. 2006;27:352–7. doi: 10.1016/j.it.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 26.Mansson A, Bachar O, Adner M, Cardell LO. Nasal CpG oligodeoxynucleotide administration induces a local inflammatory response in nonallergic individuals. Allergy. 2009;64:1292–300. doi: 10.1111/j.1398-9995.2009.02012.x. [DOI] [PubMed] [Google Scholar]
- 27.Ekman AK, Cardell LO. The expression and function of Nod-like receptors in neutrophils. Immunology. 2009;130:55–63. doi: 10.1111/j.1365-2567.2009.03212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petterson T, Jendholm J, Mansson A, Bjartell A, Riesbeck K, Cardell LO. Effects of NOD-like receptors in human B lymphocytes and crosstalk between NOD1/NOD2 and Toll-like receptors. J Leukoc Biol. 2011;89:177–87. doi: 10.1189/jlb.0210061. [DOI] [PubMed] [Google Scholar]
- 29.Mansson A, Bogefors J, Cervin A, Uddman R, Cardell LO. NOD-like receptors in the human upper airways: a potential role in nasal polyposis. Allergy. 2011;66:621–8. doi: 10.1111/j.1398-9995.2010.02527.x. [DOI] [PubMed] [Google Scholar]
- 30.Petterson T, Mansson A, Riesbeck K, Cardell LO. Nucleotide-binding and oligomerization domain-like receptors and retinoic acid inducible gene-like receptors in human tonsillar T lymphocytes. Immunology. 2011;133:84–93. doi: 10.1111/j.1365-2567.2011.03414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tada H, Aiba S, Shibata K, Ohteki T, Takada H. Synergistic effect of Nod1 and Nod2 agonists with toll-like receptor agonists on human dendritic cells to generate interleukin-12 and T helper type 1 cells. Infect Immun. 2005;73:7967–76. doi: 10.1128/IAI.73.12.7967-7976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang S, Tamai R, Akashi S, Takeuchi O, Akira S, Sugawara S, Takada H. Synergistic effect of muramyldipeptide with lipopolysaccharide or lipoteichoic acid to induce inflammatory cytokines in human monocytic cells in culture. Infect Immun. 2001;69:2045–53. doi: 10.1128/IAI.69.4.2045-2053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uehara A, Fujimoto Y, Fukase K, Takada H. Various human epithelial cells express functional Toll-like receptors, NOD1 and NOD2 to produce anti-microbial peptides, but not proinflammatory cytokines. Mol Immunol. 2007;44:3100–11. doi: 10.1016/j.molimm.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 34.Aimanianda V, Haensler J, Lacroix-Desmazes S, Kaveri SV, Bayry J. Novel cellular and molecular mechanisms of induction of immune responses by aluminum adjuvants. Trends Pharmacol Sci. 2009;30:287–95. doi: 10.1016/j.tips.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 35.Willingham SB, Allen IC, Bergstralh DT, et al. NLRP3 (NALP3, Cryopyrin) Facilitates In Vivo Caspase-1 Activation, Necrosis, and HMGB1 Release via Inflammasome-Dependent and -Independent Pathways. J Immunol. 2009;183:2008–15. doi: 10.4049/jimmunol.0900138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harenberg A, Guillaume F, Ryan EJ, Burdin N, Spada F. Gene profiling analysis of ALVAC infected human monocyte derived dendritic cells. Vaccine. 2008;26:5004–13. doi: 10.1016/j.vaccine.2008.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guarda G, Zenger M, Yazdi AS, et al. Differential expression of NLRP3 among hematopoietic cells. J Immunol. 2011;186:2529–34. doi: 10.4049/jimmunol.1002720. [DOI] [PubMed] [Google Scholar]
- 38.Ting JP, Duncan JA, Lei Y. How the noninflammasome NLRs function in the innate immune system. Science. 2010;327:286–90. doi: 10.1126/science.1184004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem. 2001;276:4812–18. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto-Furusho JK, Barnich N, Xavier R, Hisamatsu T, Podolsky DK. Centaurin beta1 down-regulates nucleotide-binding oligomerization domains 1- and 2-dependent NF-kappaB activation. J Biol Chem. 2006;281:36060–70. doi: 10.1074/jbc.M602383200. [DOI] [PubMed] [Google Scholar]
- 41.Yamaguchi T, Kimura H, Kurabayashi M, Kozawa K, Kato M. Interferon-γ enhances human eosinophil effector functions induced by granulocyte–macrophage colony-stimulating factor or interleukin-5. Immunol Lett. 2008;118:88–95. doi: 10.1016/j.imlet.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 42.Rosenberg HF, Phipps S, Foster PS. Eosinophil trafficking in allergy and asthma. J Allergy Clin Immunol. 2007;119:1303–10. doi: 10.1016/j.jaci.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 43.Codarri L, Gyulveszi G, Tosevski V, et al. RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol. 2011;12:560–7. doi: 10.1038/ni.2027. [DOI] [PubMed] [Google Scholar]
- 44.Broide D, Sriramarao P. Eosinophil trafficking to sites of allergic inflammation. Immunol Rev. 2001;179:163–72. doi: 10.1034/j.1600-065x.2001.790116.x. [DOI] [PubMed] [Google Scholar]
- 45.Dallaire MJ, Ferland C, Page N, Lavigne S, Davoine F, Laviolette M. Endothelial cells modulate eosinophil surface markers and mediator release. Eur Respir J. 2003;21:918–24. doi: 10.1183/09031936.03.00102002. [DOI] [PubMed] [Google Scholar]
- 46.Rothenberg ME, Hogan SP. The eosinophil. Annu Rev Immunol. 2006;24:147–74. doi: 10.1146/annurev.immunol.24.021605.090720. [DOI] [PubMed] [Google Scholar]


