SUMMARY
Zebrafish (Danio rerio) can serve as a model system to study heritable skin diseases. The skin is rapidly developed during the first 5–6 days of embryonic growth, accompanied by expression of skin-specific genes. Transmission electron microscopy (TEM) of wild-type zebrafish at day 5 reveals a two-cell-layer epidermis separated from the underlying collagenous stroma by a basement membrane with fully developed hemidesmosomes. Scanning electron microscopy (SEM) reveals an ordered surface contour of keratinocytes with discrete microridges. To gain insight into epidermal morphogenesis, we have employed morpholino-mediated knockdown of the abca12 and snap29 genes, which are crucial for secretion of lipids and intracellular trafficking of lamellar granules, respectively. Morpholinos, when placed on exon-intron junctions, were >90% effective in preventing the corresponding gene expression when injected into one- to four-cell-stage embryos. By day 3, TEM of abca12 morphants showed accumulation of lipid-containing electron-dense lamellar granules, whereas snap29 morphants showed the presence of apparently empty vesicles in the epidermis. Evaluation of epidermal morphogenesis by SEM revealed similar perturbations in both cases in the microridge architecture and the development of spicule-like protrusions on the surface of keratinocytes. These morphological findings are akin to epidermal changes in harlequin ichthyosis and CEDNIK syndrome, autosomal recessive keratinization disorders due to mutations in the ABCA12 and SNAP29 genes, respectively. The results indicate that interference of independent pathways involving lipid transport in the epidermis can result in phenotypically similar perturbations in epidermal morphogenesis, and that these fish mutants can serve as a model to study the pathomechanisms of these keratinization disorders.
INTRODUCTION
Clinical and genetic heterogeneity of ichthyosis
Ichthyosis comprises a group of both acquired and heritable keratinization disorders characterized by hyperkeratotic and scaly skin (Brown and Irvine, 2008). Although the phenotypic spectrum of ichthyosiform dermatoses is extremely broad, with either limited or extensive involvement of the skin, among the inherited forms, three clinically and genetically distinct subtypes have been identified: ichthyosis vulgaris, X-linked ichthyosis and lamellar ichthyosis (LI) (McGrath and Uitto, 2008; Brown and Irvine, 2008; Brown and McLean, 2008; Elias et al., 2004). LI in itself is a heterogeneous group of autosomal recessive disorders with large plaque-like brown scales over most of the body, associated with ectropion and alopecia.
Harlequin ichthyosis (HI) is a rare, extremely severe form of ichthyosis, most closely associated with the LI group of these disorders (Akiyama, 2006a). Neonates are born encased in a thick skin that not only restricts their movement, but also distorts their facial features, averting their lips and eyelids. Although newborns with HI frequently die within the first few days of life, a few of these affected individuals do survive, and their skin eventually resembles severe non-bullous congenital ichthyosiform erythroderma or LI.
HI is an autosomal recessive disorder caused by mutations in the ATP-binding cassette, sub-family A, member 12 (ABCA12) gene, which encodes a lipid transporter protein localized to lamellar granules in epidermal keratinocytes (Sakai et al., 2007). Mutations in the ABCA12 gene result in congested lipid secretion and impaired barrier function of the stratum corneum (Kelsell et al., 2005). Thus, ABCA12 is crucial to the development of the skin-lipid barrier in the stratum corneum.
An Abca12−/− mouse model has been vital in confirming the role of this transporter molecule in the skin abnormalities seen in HI, i.e. hyperkeratosis, impaired barrier function, abnormal lamellar bodies and the retention of lipid droplets in the epidermis (Yanagi et al., 2008; Smyth et al., 2008; Sundberg et al., 1997). The role of Abca12 in transporting lipids was confirmed by culturing keratinocytes from Abca12−/− mice and observing impaired lipid efflux leading to intracellular accumulation of lipids, specifically ceramides (Akiyama et al., 2005). However, the drawback of the mouse model is the long gestation period and small number of offspring per litter.
In addition to nonsyndromic variants, ichthyosis can be associated with clinical manifestations in a number of organ systems besides the skin. An example of syndromic ichthyoses is the CEDNIK syndrome, a rare autosomal recessive disorder with cerebral dysgenesis, neuropathy, ichthyosis and keratoderma. This syndrome has been shown to be associated with mutations in the SNAP29 gene, which encodes soluble n-ethylmaleimide sensitive factor (NSF) attachment protein (SNAP)29, a member of the SNAP receptor (SNARE) family of proteins (Sprecher et al., 2005; Fuchs-Telem et al., 2011). SNARE proteins are required for vesicle trafficking and they mediate the fusion between the vesicles and their target membranes. SNAP29 deficiency has been suggested to result in impaired maturation and secretion of lamellar granules, particularly interfering with the transport of lipids to stratum corneum; however, no animal model for the CEDNIK syndrome exists.
In an attempt to create an alternative, and perhaps more expedient, model system to study ichthyosis, we have performed work on zebrafish (Danio rerio), which has nearly the same complement of genes as mammals. Some of the benefits to working with zebrafish include their rapid development and the ease with which one can manipulate their gene expression by morpholino-based antisense oligonucleotides (Kari et al., 2007; Li et al., 2011). Zebrafish develop rapidly, with all major organs, including the skin, having developed by 5–6 days post-fertilization (dpf). They also produce a large number of embryos per laying, approximately 50–100 per female. In this study, we performed experiments to show that abca12 and snap29 gene knockdown in zebrafish causes epidermal changes that are similar, attesting to the concept that diverse pathogenetic pathways, as a result of mutations in different genes, can result in phenotypes in the spectrum of ichthyotic diseases. Thus, zebrafish provide a novel and expedient model system to study this group of devastating, currently intractable, diseases.
RESULTS
Identification of an ABCA12-related gene in the zebrafish genome
Search of the online gene database (NCBI) identified one human ABCA12-related sequence, abca12, which mapped to zebrafish chromosome 9. This zebrafish abca12 gene had an open reading frame, and all splice sites appeared intact, which allowed deduction of the intron-exon organization. The abca12 gene consists of 53 exons, with sizes ranging from 55 to 2415 bp (Fig. 1A). The predicted primary sequence of the corresponding protein consists of 3634 amino acids, whereas the corresponding human primary sequence comprises 2595 amino acids. The overall conservation at the protein level was 49.3% and, consequently, the zebrafish abca12 gene can be considered to be the human ABCA12 homolog.
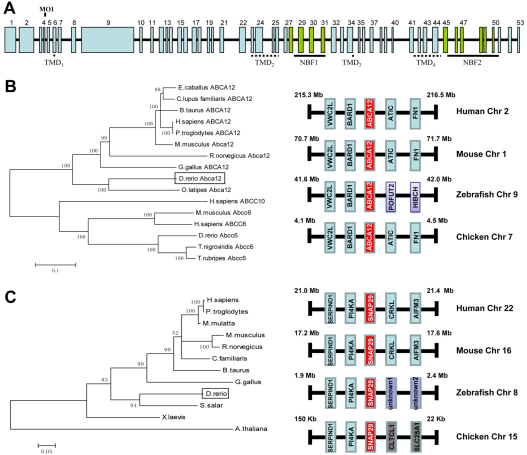
Fig. 1.
Schematic representation of the zebrafish abca12 gene, and the phylogenetic trees of the protein sequences of Abca12 and Snap29, together with syntenic analysis of the corresponding genes. (A) The abca12 gene consists of 53 exons, which are numbered on the top, and the coding segments for transmembrane domains (TMDs) and nucleotide binding folds (NBFs; green) are underlined. Note the location corresponding to the morpholino (MO1) at the exon-4–intron-4 junction. (B) The phylogenetic relationship between zebrafish Abca12 and the other members of the ABC family of transporters estimated by the neighbor-joining method (left panel). The syntenic analysis of the abca12 and flanking genes in human, mouse, zebrafish and chicken chromosomes is shown on right. (C) Cladogram and syntenic analysis of snap29. The unknown genes 1 and 2 in zebrafish chromosome 8 have been designated as si:dkey-178e17.1 and si:dkeyp-117b11.1, respectively.
Alignment of human and zebrafish protein sequences revealed that zebrafish Abca12 has an extended 486 amino acid N-terminal sequence, as well as a number of insertions in the N-terminal half of the protein. However, alignment of zebrafish and human sequences identified conservation of domains that are characteristic of the ABC transporter proteins. Specifically, the zebrafish sequence, similar to the human sequence, was predicted to consist of four transmembrane domains (TMD1-4) and to contain two nucleotide binding fold domains (NBF1 and NBF2) (Tusnády et al., 2006) (Fig. 1A). The NBFs displayed characteristic sequences for Walker A and B motifs, as well as a highly conserved ABC signature sequence. Comparison of the deduced amino acid sequence within the NBF1 domain of zebrafish Abca12 showed 74% identity to the corresponding NBF1 domain in the human protein, whereas the NBF2 domain had 68% identity to human NBF2.
Evolutionary conservation of zebrafish abca12
Differences between the zebrafish abca12 gene and homologous genes in other species were examined by phylogenetic analysis of the corresponding protein sequence by cladistic measurement (Fig. 1B). The cladogram suggested that the zebrafish gene is distant from most of the other ABCA12-related genes in a number of species, and, therefore, presumably diverged early. However, inclusion of other members of the ABC transporter family, such as ABCC10 and ABCC6, in different species, serving as an outgroup, indicated that the zebrafish Abca12 protein sequence is closer to human ABCA12 than it is to the sequences in the outgroup. To confirm that the zebrafish abca12 is the correct ortholog of human ABCA12, syntenic analysis of Abca12 in different species was performed (Fig. 1B). These analyses revealed that ABCA12 and its flanking genes, VWC2L and BARD1, were located on the same chromosome in the same gene order in human, mouse, zebrafish and chicken genomes (Fig. 1B).
Expression of the zebrafish abca12 gene during early embryonic development
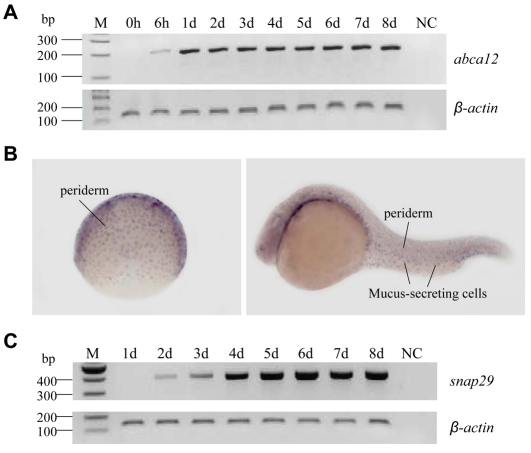
The temporal expression profile of abca12 was examined in embryos collected during the first 8 days of development, and the corresponding mRNA levels were determined by reverse transcriptase (RT)-PCR. An undetectable level of expression was noted in embryos at the time of fertilization [0 hours post-fertilization (hpf)], but detectable levels of mRNA transcripts were noted at 6 hpf, with a significant further increase by 1 dpf. During the subsequent days (2–8 dpf), the expression levels remained relatively constant in comparison with the control gene, β-actin (Fig. 2A).
Fig. 2.
abca12 and snap29 gene expression in normal zebrafish. (A–C) Zebrafish embryos were collected at 0 and 6 hpf and 1–8 dpf, and total RNA was isolated and cDNA prepared. The abca12 (A) and snap29 (C) mRNA expression levels were measured by RT-PCR and standardized against the mRNA expression level of the β-actin gene. (B) Whole-mount in situ hybridization of embryos at different stages of early development for abca12 expression; gastrula period (left panel), 24 hpf (right panel).
Whole-mount in situ hybridization of abca12 in zebrafish
To determine the spatial expression of abca12 during different stages of zebrafish development, whole-mount in situ hybridization was performed using probes specific for the abca12 gene (Fig. 2B). An antisense probe for abca12 gave specific expression patterns. During the gastrula period, expression of abca12 was observed in cells of the enveloping layer (EVL; Fig. 2B). Expression of the abca12 gene in this tissue, which is named periderm after the end of gastrulation, is observed until the end of embryonic development. After 24 hpf, expression of abca12 was also observed, although at lower levels, in the olfactory vesicle as well as in mucus-secreting cells (Fig. 2B). At the end of embryonic development, expression was observed mainly in olfactory vesicle, pharynx and mucus-secreting cells. A sense probe was used as a control and did not give a specific expression pattern.
Morpholino knockdown of abca12 expression results in an altered skin phenotype
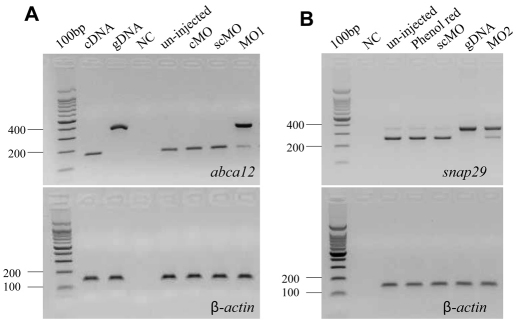
Morpholino antisense oligonucleotide (MO1) directed against a splice donor site in abca12 was injected into one- to four-cell-stage embryos, and amplification of total RNA was performed by primers corresponding to exons 4 and 5. Using these primers, PCR amplification of abca12 cDNA resulted in a 189 bp product, whereas amplification of genomic DNA generated a 356 bp product (Fig. 3A). RT-PCR of total RNA extracted from zebrafish 3 days after injection with MO1 revealed that essentially all (>90%) of the pre-mRNA remained unprocessed, attesting to the efficiency of the morpholino knockdown (Fig. 3A). Injection of control morpholinos, either a global standard control MO (scMO) or 5-bp mismatched control (cMO), had no effect on pre-mRNA processing (Fig. 3A).
Fig. 3.
Knockdown of abca12 and snap29 expression by morpholinos. (A) Knockdown of abca12. (B) Knockdown of snap29. MO1 and MO2 morpholinos (right lanes, the upper panel), which target the splice donor site at the exon-4–intron-4 border of the corresponding genes, prevents pre-mRNA splicing. The consequences of MO1 on abca12 pre-mRNA splicing and MO2 on snap29 mRNA splicing were determined by RT-PCR. The results showed the retention of intron 4 in the majority of mRNA transcripts (>90%) as compared with the normally transcribed control. The mRNA levels were normalized by the level of β-actin mRNA (lower panels). cDNA and gDNA represent amplification of the corresponding complementary DNA and genomic DNA, respectively. Injections with the 5-bp mismatched control morpholino for abca12 (cMO) or global standard control morpholino (scMO) did not alter pre-mRNA processing, similar to the uninjected controls or those injected with phenol red.
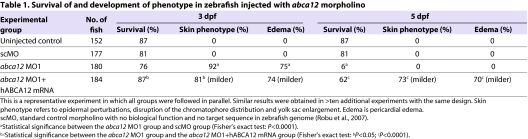
The effect of the injection of morpholinos into one- to four-cell embryos was first examined by determining the survival of the embryos. Of the 180 embryos injected with abca12 MO1, 76% survived at 3 dpf, a number that did not statistically differ from the survival of embryos injected with standard control morpholino (81%) (Table 1). At 5 dpf, the survival of embryos injected with MO1 was only 6%, a statistically significant reduction from the survival noted with scMO and uninjected controls (81% and 87%, respectively; P<0.0001) (Table 1).
Table 1.
Survival of and development of phenotype in zebrafish injected with abca12 morpholino
Examination of the morphology of zebrafish larvae injected with MO1 (n=180) revealed profound changes during development. Although no differences were noted between the morphant and control larvae at 1 dpf, by 3 dpf the morphants had developed noticeable changes in the distribution of pigment along their trunk and tail, in addition to pericardial edema (Fig. 4A). Upon careful examination at 3 dpf, 92% of larvae displayed yolk sac enlargement and severe disruption of their chromatophore distribution, with 75% exhibiting concomitant pericardial edema (Table 1).
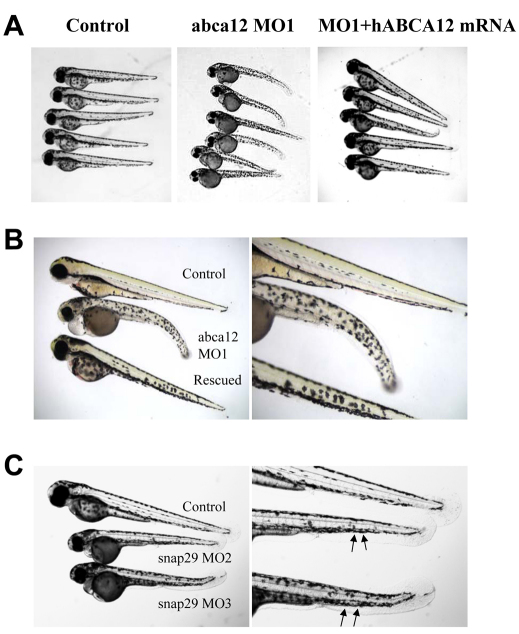
Fig. 4.
Zebrafish phenotypes and their mRNA rescue at 3 dpf. (A) Phenotypic appearance of zebrafish larvae after injection with an abca12 MO1 morpholino (middle panel) compared with control larvae (left panel), and partial rescue with human ABCA12 mRNA (right panel). (B) Higher magnification of the larvae shown in A. (C) Phenotype of larvae at 3 dpf injected with snap29 morpholinos MO2 or MO3. The irregular contour of the epidermis is noted by arrows.
Altered epidermal morphology in the morphant larvae
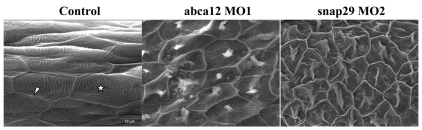
To examine the consequences of the morpholino-mediated knockdown of abca12 expression in the skin of zebrafish, we first used scanning electron microscopy (SEM) to examine the surface contour and cellular morphology of the epidermis. In 3-dpf controls (n=21), well-demarcated keratinocytes with distinct borders and characteristic microridges were observed (Fig. 5). Examination of the skin surface of the morphant larvae (n=4) revealed perturbations in the architecture of the microridges, with spicules protruding from the center of each keratinocyte. Thus, the development of the top layer of skin during the first 3 days of zebrafish development was perturbed in the absence of Abca12 activity.
Fig. 5.
SEM of the skin surface. The skin of the tail of the control larvae injected with the global standard control morpholino at 3 dpf shows the presence of keratinocytes with well-demarcated cell-cell borders (arrowhead) containing microridges (star; left panel). The morphant larvae injected with MO1 morpholino for abca12 (middle) or snap29 (MO2; right panel) revealed perturbed microridge formation with spicules in the center of the keratinocytes.
Alterations in the epidermis at 3 dpf were further examined by transmission electron microscopy (TEM) both in control and morphant larvae (n=4 in each group). At this developmental stage, normal epidermis consists of two unicellular layers, the superficial layer and the basal layer. The contour of the outer surface of the superficial layer is studded with spicules that correspond to the microridges noted previously on SEM (Fig. 6A). The epidermis rests on a basement membrane, which separates the epidermis from the underlying developing dermis.
Fig. 6.
TEM of 3-dpf larvae injected with abca12 or snap29 morpholinos. (A,E) Control morpholino (scMO); (B) abca12 morpholino (MO1). Boxes surrounding electron-dense subcellular structures in B were examined at higher magnification and are shown in C and D. (F) Injection with snap29 morpholino (MO2). (A–F) Arrows point to microridges; open arrowheads indicate basement membrane; solid black arrowheads point to the areas of accumulation of putative lipids within the electron-dense granules in C and D; solid white arrowheads in F point to apparently empty vesicles. e, epidermis; d, developing dermis.
The epidermis of the morphant larvae similarly consisted of two cell layers resting on a basement membrane (Fig. 6B). However, in contrast to the control larvae, both layers of the morphant epidermis contained an abundance of electron-dense granules, approximately 440 nm in average diameter. Closer examination of these aggregates at higher magnification suggested the presence of lipid-like vesicles within the larger electron-dense granules (Fig. 6C,D). It should be noted that, although somewhat similar aggregates of electron-dense material were noted in the epidermis of the control specimens, they were localized only to the area of the superficial layer just below the microridges.
Co-injection of human ABCA12 mRNA rescues the morpholino-mediated phenotype
To test the specificity of the phenotypic changes associated with MO1 injection, a rescue experiment with co-injection of in vitro transcribed human ABCA12 mRNA was performed. The injection of MO1 alone caused characteristic phenotypic changes, whereas co-injection of human mRNA together with MO1 partially rescued the phenotype (Fig. 4A,B). Specifically, at 5 dpf, the survival of the co-injected larvae was 62%, which is statistically different from the 6% in those injected with MO1 alone (P<0.0001) (Table 1). Also, 27% of co-injected larvae (n=184) had a phenotype that was indistinguishable from the controls. In the remaining 73% of co-injected larvae, the degree of yolk sac enlargement and chromatophore disorganization was noticeably less than in the larvae injected with MO1 alone. Of this 73%, 70% also manifested pericardial edema. The rescue experiment, in addition to injection of control morpholinos, attested to the specificity of the phenotype documented in the morphant larvae. These experiments also confirmed that the zebrafish abca12 gene is the functional homolog of human ABCA12.
Epidermal perturbations in zebrafish injected with snap29 morpholino
Because knockdown of abca12 expression was speculated to interfere with lipid secretion by lamellar granules, resulting in a characteristic epidermal phenotype, we proceeded to test this postulate by interfering with the lipid transport by another, independent mechanism: knockdown of the expression of the snap29 gene. The corresponding protein, Snap29, has been suggested to mediate lipid transport within the epidermis and the deficiency of SNAP29 expression results in syndromic ichthyosis in patients with CEDNIK syndrome (Rapaport et al., 2010; Sprecher et al., 2005).
Surveying the zebrafish genome database revealed the presence of one SNAP29-related gene, snap29, on chromosome 8. This gene product had 52% identity with the human gene product at the protein level, and cladogram and syntenic analyses suggested that zebrafish snap29 is the ortholog of human SNAP29 (Fig. 1C). The expression of the gene was readily detectable at 2 dpf by RT-PCR and the expression level increased during 3–8 dpf (Fig. 2C). In situ hybridization of larvae showed weak, ubiquitous expression with accentuation of the labeling in the central nervous system marginal zone (not shown).
Injection of a morpholino (MO2) placed on the exon-4–intron-4 junction of the snap29 gene into one- to four-cell-stage embryos inhibited the processing of pre-mRNA by >90% (Fig. 3B). A second, non-overlapping morpholino (MO3), placed on the intron-4–exon-5 border of the snap29 gene similarly resulted in >90% inhibition of the splicing of intron 4 (data not shown). Examination of the morphant larvae at 3 dpf (n=165 for MO2, and n=203 for MO3) revealed a phenotype consisting of pigmentary changes, somewhat analogous with those noted with the abca12 morpholino, in 80% of larvae, and the contour of the epidermis in the tail section was irregular (Fig. 4C). SEM of 20 morphant larvae revealed perturbations in the morphology of the epidermis that were very similar to those noted as a result of abca12 knockdown (Fig. 5). Examination of the epidermis of the snap29 morphant larvae (n=4) by TEM at 3 dpf revealed an increase in vesicles, which appeared empty under the same fixation conditions that revealed accumulation of lipid-like material in abca12 morphant fish (Fig. 6E,F). Thus, interference by morpholino knockdown of the expression of two independent genes, abca12 and snap29, that are involved in lipid transport in the epidermis can lead to similar phenotypic alterations in the epidermal morphology.
DISCUSSION
ABCA12 mutations underlie HI
The molecular basis of HI is linked to mutations in the ABCA12 gene (Akiyama et al., 2005; Kelsell et al., 2005). Initial approaches utilizing single-nucleotide polymorphism-based chip technology and homozygosity mapping of families with individuals affected with HI placed the candidate gene locus on chromosomal region 2q35, and microsatellite markers narrowed the interval to consist of six genes (Kelsell et al., 2005). Several previous observations pointed to ABCA12 as a candidate gene within the critical region. First, a characteristic ultrastructural feature of the epidermis in HI is an abnormality in the localization of epidermal lipids, together with abnormal ultrastructural epidermal lamellar granules (Akiyama, 2006b). ABCA12 has been suggested to encode a transmembrane transporter protein, which, from sequence homology with several other ABC family members, was thought to be involved in the transport of lipids (Kaminski et al., 2006). Second, the ABCA12 gene was previously shown to harbor missense mutations in a milder form of ichthyosis, lamellar ichthyosis type 2 (LI2), with some resemblance to the phenotype in patients with HI who survive beyond the immediate postnatal period (Lefèvre et al., 2003). Currently, a total of 53 distinct mutations have been identified in the ABCA12 gene (Akiyama, 2010).
Expression of ABCA12 has been localized to lamellar granules. In normal epidermal keratinocytes there is an upregulation of ABCA12 expression in association with physiological keratinization of the human epidermis (Sakai et al., 2007). Mutations in the ABCA12 gene result in congested lipid retention in the skin of individuals with HI. It has been suggested that ABCA12 transports ceramides, the major lipid of the stratum corneum of the epidermis. Finally, lamellar-granule-mediated lipid secretion was resumed in the cultured keratinocytes of patients with HI upon transfer of the wild-type ABCA12 gene (Akiyama et al., 2005). Thus, it is clear that mutations in the ABCA12 transporter gene underlie HI.
abca12 and zebrafish skin development
In this study, we have demonstrated that zebrafish abca12 is the ortholog of human ABCA12. There is a high degree of conservation of the Walker A and B motifs in addition to the retention of the four transmembrane domains containing one, five, one and five transmembrane segments, respectively. Zebrafish NBF1 and NBF2 domains in the Abca12 protein have 74% and 68% similarity with human NBF1 and NBF2 domains, respectively, at the amino acid level.
Whole-mount in situ hybridization in developing zebrafish embryos revealed that abca12 was expressed in the EVL and the periderm. The EVL first appears at the 64-cell stage of development (∼2 hpf) and is the outermost monolayer of cells surrounding the embryo. The EVL eventually gives rise to the periderm, which is thought to ultimately be replaced by the superficial stratum of the epidermis (Kimmel et al., 1995; Le Guellec et al., 2004). Although the physiology of zebrafish skin is still largely unexplored, the fact that abca12 is expressed in the skin suggests its importance in normal skin development. This hypothesis is further strengthened by the results from our morpholino experiments. Injecting a morpholino that inhibited pre-mRNA splicing by >90% produced alterations in chromatophore distribution and the abnormal retention of lipids in both layers of the epidermis. Not only does this suggest that abca12 is responsible for lipid transport in zebrafish, but the abnormal accumulation of lipids throughout the epidermis is a frequent finding in individuals with HI. Finally, the rescue of this phenotype by co-injection of human ABCA12 mRNA shows that the phenotype is the result of abca12 knockdown and not due to an off-target effect. In this context, it should be emphasized that the EVL-derived skin in zebrafish is embryologically different from the mammalian skin. Specifically, zebrafish epidermis does not undergo terminal differentiation, which in human skin culminates in the development of stratum corneum with barrier function. Emphasizing this difference is the fact that survey of the current zebrafish genome database (Ensembl, Zebrafish Zv9; http://www.ensembl.org/Danio_rerio/Info/Index) does not reveal the presence of filaggrin, involucrin and trichohyalin genes, which are crucial for development of the stratum corneum in human epidermis (Li et al., 2011).
The role of lipids in epidermal development is further emphasized by our findings that knockdown of the expression of an independent gene, snap29, results in a similar epidermal phenotype as noted in abca12 mutant larvae. Snap29 has been postulated to mediate lipid transport in the epidermis by facilitating membrane fusion of lamellar granules (Sprecher et al., 2005). Thus, interference of lipid trafficking by knockdown of two independent genes results in phenocopies of the epidermal perturbations in zebrafish, mimicking epidermal alterations in different forms of ichthyosis. It should be noted that, similar to the CEDNIK syndrome, ichthyosis has been reported in association with mental retardation, enteropathy, deafness, peripheral neuropathy and keratodermia, dubbed as the MEDNIK syndrome (Montpetit et al., 2008). This constellation was shown to be associated with a homozygous splice-site mutation in the AP1S1 gene, encoding a subunit of the adaptor protein complex that regulates clathrin-coated vesicle assembly, protein cargo sorting, and vesicular trafficking between organelles in eukaryotic cells (Montpetit et al., 2008). The pathogenic effect of this mutation was validated by knockdown of ap1s1 expression in zebrafish by a morpholino, resulting in perturbation in skin formation, reduced pigmentation and motility deficits. These findings, together with our observations in snap29 mutant larvae, attest to the importance of vesicular trafficking in epidermal morphogenesis.
As indicated by morphological observations of the developing epidermis in zebrafish in comparison with human skin, there are clear differences. For example, the embryological origin of the EVL (periderm) in zebrafish is distinct from the basal layer in embryonic skin. In spite of this difference, there is an increasing body of evidence suggesting that the underlying molecular differentiation pathways are conserved between mammals and the zebrafish epidermis, based on molecular homologies (Sabel et al., 2009; Slanchev et al., 2009). Our work highlighting the early abca12 expression in the EVL seems to support the conclusions that EVL forms the external layer of the embryonic and larval dermis and represents the initial differentiation of a true epidermis (Fukazawa et al., 2010).
Collectively, our results highlight the role of lipid transport and vesicular trafficking in epidermal development, and the results further suggest that zebrafish can serve as a model system to study different variants of ichthyosis, such as HI and the CEDNIK syndrome. Besides increasing our understanding of the disease mechanisms involved in ichthyotic syndromes, this model system is potentially useful for testing novel treatment modalities, for example by performing a small molecule library screen for compounds that are able to suppress the phenotype.
METHODS
Maintenance of zebrafish
Adult wild-type zebrafish were maintained under standard conditions at 28.5°C. Zebrafish embryos and larvae were also maintained at 28.5°C in a special embryo medium. All animals were housed in the zebrafish facility at Thomas Jefferson University and were cared for and used in accordance with University Institutional Animal Care and Use Committee guidelines and permission.
Phylogenetic and syntenic analyses
The genomic sequences of zebrafish were extracted from the Ensembl database. The zebrafish protein sequences were aligned with the corresponding proteins in different species by using ClustalW software (http://www.ebi.ac.uk/clustalw/).
The accession numbers for the abca12 gene products in different species are: E. caballus (ENSECAP00000007797), C. lupus familiaris (XP_536058), B. taurus (XP_001788086), H. sapiens (NP_775099), P. troglodytes (XP_516070), M. musculus (XP_001002308), R. norvegicus (XP_237242), G. gallus (XP_421867), D. rerio (XP_686632) and O. latipes (ENSORLP00000020129). The accession numbers for ABCC10 in H. sapiens is NP_258261. The accession numbers for Abcc6 in different species are: M. musculus (NP_061265), H. sapiens (NP_001162), D. rerio (ENSDARP00000065432), T. nigroviridis (ENSTNIP00000015029) and T. rubripes (ENSTRUP00000029065).
The accession numbers for the snap29 gene products in different species are: H. sapiens (NP_004773.1), P. troglodytes (XP_514997.2), M. mulatta (XP_001086227.1), M. musculus (NP_075837.3), R. norvegicus (NP_446262.3), C. familiaris (XP_543568.2), B. taurus (NP_001069427.1), G. gallus (NP_001025823.1), D. rerio (XP_700124.3), S. salar (NP_001134759.1), X. laevis (NP_001080076.1) and A. thaliana (NP_196405.1).
Phylogenetic analyses were conducted in the Molecular Evolution Genetics Analysis software (MEGA) version 4.0 (Tamura et al., 2007). The cladogram was constructed using the Neighbor-Joining method (Saitou and Nei, 1987). The Kimura two-parameter method was used to compute the evolutionary distances (Zuckerkandl and Pauling, 1965). The statistical reliance of NJ tree branches was evaluated using 1000 bootstrap samples.
For syntenic analysis, the orientation and chromosomal positions of abca12 and snap29 and their adjacent genes were determined manually from the gene orientations in the current Ensembl database. The zebrafish (Zv9), human (GRCh37/hg19), mouse (NCB137/mm9) and chicken (WUGSC2.1/galGal3) genome assembly versions were used for this analysis.
In situ hybridization
Whole-mount in situ hybridization was performed as described previously (Thisse and Thisse, 2008). Collected zebrafish embryos were fixed in 4% paraformaldehyde before hybridization. Digoxigenin (DIG)-labeled antisense and sense probes were synthesized. After hybridization, detection was performed with an anti-DIG antibody coupled to alkaline phosphatase.
Morpholinos and microinjection
Morpholino oligonucleotides were obtained from Gene Tools, LLC (Corvalis, OR). The morpholino oligomer sequences were written from 5′ to 3′, to correspond to the following genomic sequences (brackets surround the morpholino target sequence, exon sequences are capitalized, intron sequences are in lowercase, and nucleotide substitutions are bolded). For abca12 knockdown: splice donor site morpholino (MO1), tgggaaataaatgtaatttacctgt, targets the exon-4–intron-4 junction, AAATGAAATAACTGA[ACAGgta-aattacatttatttccca]acggtc; 5-base pair mismatched control morpholino for abca12 (cMO): tggcaaaaaaatctaatttacgtct. For snap29 knockdown: splice donor site morpholino (MO2), ctgctcttgtgtttctcacccaggt, targets the exon-4–intron-4 junction, GACAGAA[ACCTGGgtgagaaacacaagacag]cttctctcata; a second snap29 splice junction morpholino (MO3) targets the intron 4-exon 5 border, ctcatctggaggacacaaacacaca, agtgtgtgtg[tgtgtg-tttgtgtcctccagAT GAG]ATGTCTCTGGGTC. Global standard control morpholino (scMO), cctcttacctcagttacaatttata, has no target sequence in the zebrafish genome and is, therefore, inactive.
Embryos at the one- to four-cell stage were injected with an abca12 morpholino (MO1, 25.6 ng) or snap29 morpholinos (MO2, 2.6 ng and MO3, 5.2 ng) using glass microelectrodes fitted to a gas pressure injector (PL1-100, Harvard Apparatus). Electrodes were pulled (P-97, Flaming/Brown) and filled with morpholino and phenol red (final concentration 0.025%) to visualize the injected embryos. The embryos were then followed for viability, morphology and mRNA expression levels.
Total RNA isolation and cDNA synthesis
Zebrafish embryos were collected at 0 as well as 6 hpf and 1–8 dpf. They were disintegrated by pipetting through a 21 gauge needle and total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA). To remove contaminating genomic DNA, RNase-free DNase I digestion (Fisher Scientific, St Louis, MO) was performed. 1 μg of total RNA was reverse transcribed using the Superscript III First-Strand cDNA synthesis kit (Invitrogen) according to the manufacturer’s protocol. Controls were performed by omitting the reverse transcriptase enzyme. All cDNA samples were stored at −20°C for future use.
PCR amplification of cDNA
abca12 cDNA was amplified by PCR using a forward primer on exon 4 (5′-ATCTGGGACAACTTGGGCAACT-3′), and a reverse primer on exon 5 (5′-TCATCTGGTCAGCAGTTCCAGAGA-3′). The snap29 cDNA was amplified using a forward primer on exon 4 (5′-TTCTGCTGCTCTTGATAACGGCT-3′), and a reverse primer on exon 5 (5′-TTTAAGGCTTTTGAGCTGCCGGTT-3′). Primers for the zebrafish β-actin gene (fwd: 5′-ATCTGGCACCACACCTTCTACAATG; rev: 5′-GGGGTG-TTGAAGGTCTCAAACATGAT) were used as a positive control. PCR was performed using Taq polymerase and Q buffer (Qiagen, Valencia, CA), according to the manufacturer’s instructions. The PCR conditions were as follows: an initial denaturation at 94°C for 5 minutes, followed by 35 cycles of 94°C for 1 minute; 58°C for 1 minute; 72°C for 1 minute; and finally 72°C for 10 minutes. The intensity of the bands was quantified using ImageQuant version 5.0 software (Molecular Dynamics, Sunnyvale, CA).
mRNA rescue experiments
Capped full-length human mRNA corresponding to ABCA12 was transcribed from an expression vector pCMV-Tag4B using the T3 mMessage mMachine kit (Ambion, Austin, TX). The morpholino was injected into one- to four-cell-stage embryos either alone or in combination with mRNA (2.3 ng) and followed for viability and morphology.
Scanning electron microscopy
Samples were fixed in neutral buffered formalin at room temperature for 2 hours, followed by a rinse with phosphate buffered saline and an ethanol dehydration series of exchanges by completely replacing each successively higher ethanol solution with the next higher (20, 30, 50, 75, 95 and 100%). Samples were then incubated for 15 minutes in a 1.5 ml micro test tube containing 1,1,2-Trichloro-1,2,2-trifluoroethane before covering the open micro test tube with parafilm, punching holes in it with a 30G needle, and situating it under a fume hood where it was dried by turbulent air flow. Samples were then mounted onto stubs with carbon paint and coated in 50 nm of gold using a sputter coater. Specimens were imaged in a JEOL-T330A scanning electron microscope (JEOL, Tokyo, Japan) at 15 kV.
Transmission electron microscopy
Samples were collected and fixed overnight at 4°C in 2.5% gluteraldehyde, 2% paraformaldehyde and 0.1 M sodium cacodylate. Samples were then washed in 0.1 M sodium cacodylate before undergoing secondary fixation in 2% osmium tetroxide, 1.5% potassium ferricyanide and 0.1 M sodium. Samples were again washed with 0.1 M sodium cacodylate followed by deionized water before undergoing en block staining with 2% uranyl acetate. Samples were washed again with deionized water, then dehydrated in a graded ethanol series and embedded in EMbed-812 (EMS, Hatfield, PA). Ultrathin sections (60 nm) were cut and analyzed using a JEOL JEM-1010 transmission electron microscope fitted with a Hamamatsu digital camera (Hamamatsu Photonics, Hamamatsu City, Japan) and AMT Advantage image capture software (AMT, Danvers, MA).
Statistical analysis
Risk differences and 95% confidence intervals were calculated between experimental groups with regards to survival, skin phenotype and edema in Table 1, for 3 dpf and 5 dpf separately. Fisher’s exact test was used to determine the difference between proportions because of the presence of cells with zero observations. Adjustments for multiple comparisons were performed using False Discovery Rate, and it is these adjusted P-values that are reported. Analyses were conducted using SAS 9.2 (SAS Institute, Cary, NC).
TRANSLATIONAL IMPACT.
Clinical issue
Ichthyosis comprises a group of cutaneous disorders characterized by dry, scaly skin and a broad spectrum of other phenotypic manifestations. One of the most severe forms of ichthyosis is known as harlequin ichthyosis (HI); neonates affected with HI are born encased in a thick skin that restricts their movement and frequently die shortly after birth. Some forms of ichthyosis are syndromic; for example, CEDNIK syndrome is so-named because it consists of cerebral dysgenesis, neuropathy, ichthyosis and keratoderma. Details of the pathomechanisms of HI have recently been revealed through molecular genetics, which showed that patients with this disorder carry mutations in the ABCA12 gene. Examination of Abca12−/− mice suggested that this gene encodes a transmembrane transporter present in the epidermis that is postulated to transport lipids (specifically ceramides) and that is required for formation of the stratum corneum on the surface of the skin. Although the mouse model is useful in that it recapitulates features of human HI, drawbacks include the long gestational period and the small number of offspring produced per litter. CEDNIK syndrome is caused by mutations in the SNAP29 gene, which is required for normal vesicle trafficking and lipid transport in the epidermis. There is no animal model for this syndrome.
Results
To create alternative, more expedient model systems to investigate pathological mechanisms of both HI and CEDNIK syndrome, the authors of this study knocked down the homologs of ABCA12 and SNAP29 in zebrafish embryos (Danio rerio). Morpholino antisense oligonucleotides targeted to exon-intron splice junctions were used to inhibit the splicing of abca12 or snap29 pre-mRNA. Inhibition of processing of either one of these mRNAs was accompanied by changes in the distribution of pigment along the trunk and tail of the fish as early as 2 days post-fertilization (dpf). Examination of epidermal morphology by scanning electron microscopy revealed perturbations in the surface contour of the keratinocytes, with loss of characteristic microridges and development of pathological spicules protruding from the center of each keratinocyte. These epidermal changes were accompanied by premature demise of the fish by 5 dpf. Transmission electron microscopy revealed an abundance of electron-dense granules in both morphants: lipid-like vesicles were seen in abca12 knockdown fish, whereas the epidermis of snap29 knockdown animals showed the presence of apparently empty vesicles.
Implications and future directions
This study demonstrates that inhibition of abca12 or snap29 gene splicing in zebrafish leads to epidermal perturbations that are similar to those seen in human patients with various forms of ichthyosis. In addition, it suggests that interfering with two independent pathways involved in lipid transport can result in phenotypically similar perturbations in epidermal morphogenesis. These systems can serve as models to study ichthyosis, and provide a means to develop pharmacological approaches towards treatment of this currently intractable group of diseases. Finally, in a broader sense, this study attests to the feasibility of using zebrafish as a model system to study heritable skin diseases.
Acknowledgments
The authors thank the following colleagues for advice and assistance: Wolfgang Driever, Institute for Biology I, University of Freiburg; Raymond Meade, Biomedical Imaging Core Facility, University of Pennsylvania; Gerald Harrison, Department of Biochemistry, School of Dental Medicine, University of Pennsylvania; Jean-Yves Sire, Equipe “Evolution et Développement du Squelette”, Université Paris; Eijiro Adachi, Department of Matrix Biology and Tissue Regeneration, Graduate School of Medical Sciences, Kitasato University; Ulrich Rodeck, Gabor Kari, April Aguillard, Adele Donahue and Andrzej Fertala, Department of Dermatology and Cutaneous Biology, Thomas Jefferson University; Terry Hyslop and Jocelyn Andrel, Department of Pharmacology and Experimental Therapeutics, Thomas Jefferson University. Carol Kelly assisted in manuscript preparation. This study was supported by the NIH/NIAMS grant R01AR055225 to J.U.; by the University of Virginia to C.T. and B.T. Q.L. is a recipient of a Research Career Development Award from the Dermatology Foundation.
Footnotes
COMPETING INTERESTS
The authors declare that they do not have any competing or financial interests.
AUTHOR CONTRIBUTIONS
Q.L., M.F., C.T. and B.T. performed the experiments; M.A. provided reagents; H.S. and E.S. interpreted the data and edited the manuscript; S.-Y.H. contributed to the data analysis; J.U. developed the concept, interpreted the data and prepared the manuscript.
REFERENCES
- Akiyama M. (2006a). Harlequin ichthyosis and other autosomal recessive congenital ichthyoses: the underlying genetic defects and pathomechanisms. J. Dermatol. Sci. 42, 83–89 [DOI] [PubMed] [Google Scholar]
- Akiyama M. (2006b). Pathomechanisms of harlequin ichthyosis and ABCA transporters in human diseases. Arch. Dermatol. 142, 914–918 [DOI] [PubMed] [Google Scholar]
- Akiyama M. (2010). ABCA12 mutations in harlequin ichthyosis, congenital ichthyosiform erythroderma and lamellar ichthyosis. Hum. Mutat. 31, 1090–1096 [DOI] [PubMed] [Google Scholar]
- Akiyama M., Sugiyama-Nakagiri Y., Sakai K., McMillan J. R., Goto M., Arita K., Tsuji-Abe Y., Tabata N., Matsuoka K., Sasaki R. (2005). Mutations in lipid transporter ABCA12 in harlequin ichthyosis and functional recovery by corrective gene transfer. J. Clin. Invest. 115, 1777–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. J., Irvine A. D. (2008). Atopic eczema and the filaggrin story. Semin. Cutan. Med. Surg. 27, 128–137 [DOI] [PubMed] [Google Scholar]
- Brown S. J., McLean W. H. I. (2008). Eczema genetics: current state of knowledge and future goals. J. Invest. Dermatol. 129, 543–552 [DOI] [PubMed] [Google Scholar]
- Elias P. M., Crumrine D., Rassner U., Hachem J. P., Menon G. K., Man W., Choy M. H., Leypoldt L., Feingold K. R., Williams M. L. (2004). Basis for abnormal desquamation and permeability barrier dysfunction in RXLI. J. Invest. Dermatol. 122, 314–319 [DOI] [PubMed] [Google Scholar]
- Fuchs-Telem D., Stewart H., Rapaport D., Nousbeck J., Gat A., Gini M., Lugassy Y., Emmert S., Eckl K., Hennies H. C., et al. (2011). CEDNIK syndrome results from loss-of-function mutations in SNAP29. Br. J. Dermatol. 164, 610–616 [DOI] [PubMed] [Google Scholar]
- Fukazawa C., Santiago C., Park K. M., Deery W. J., Gomez de la Torre Canny S., Holterhoff C. K., Wagner D. A. (2010). poly/chuk/ikk1 is required for differentiation of the zebrafish embryonic epidermis. Dev. Biol. 346, 272–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski W. E., Piehler A., Wenzel J. J. (2006). ABC A-subfamily transporters: structure, function and disease. Biochim. Biophys. Acta 1762, 510–524 [DOI] [PubMed] [Google Scholar]
- Kari G., Rodeck U., Dicker A. P. (2007). Zebrafish: an emerging model system for human disease and drug discovery. Clin. Pharmacol. Ther. 82, 70–80 [DOI] [PubMed] [Google Scholar]
- Kelsell D. P., Norgett E. E., Unsworth H., Teh M. T., Cullup T., Mein C. A., Dopping-Hepenstal P. J., Dale B. A., Tadini G., Fleckman P. (2005). Mutations in ABCA12 underlie the severe congenital skin diseases harlequin ichthyosis. Am. J. Hum. Genet. 76, 794–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 [DOI] [PubMed] [Google Scholar]
- Le Guellec D., Morvan-Dubois G., Sire J. Y. (2004). Skin development in bony fish with particular emphasis on collagen deposition in the dermis of the zebrafish (Danio rerio). Int. J. Dev. Biol. 48, 217–232 [DOI] [PubMed] [Google Scholar]
- Lefèvre C., Audebert S., Jobard F., Bouadjar B., Lakhdar H., Boughdene-Stambouli O., Blanchet-Bardon C., Heilig R., Foglio M., Weissenbach J. (2003). Mutations in the transporter ABCA12 are associated with lamellar ichthyosis type 2. Hum. Mol. Genet. 12, 2369–2378 [DOI] [PubMed] [Google Scholar]
- Li Q., Frank M., Thisse C., Thisse B., Uitto J. (2011). Zebrafish: a model system to study heritable skin diseases. J. Invest. Dermatol. 131, 565–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J. A., Uitto J. (2008). The filaggrin story: novel insights into skin-barrier function and disease. Trends Mol. Med. 14, 20–27 [DOI] [PubMed] [Google Scholar]
- Montpetit A., Côté S., Brustein E., Drouin C. A., Lapointe L., Boudreau M., Meloche C., Drouin R., Hudson T. J., Drapeau P., et al. (2008). Disruption of AP1S1, causing a novel neurocutaneous syndrome, perturbs development of the skin and spinal cord. PloS Genet. 4, e1000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D., Lugassy Y., Sprecher E., Horowitz M. (2010). Loss of SNAP29 impairs endocytic recycling and cell motility. PloS ONE 5, e9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabel J. L., d’Alençon C., O’Brien E. K., Van Otterloo E., Lutz K., Cuykendall T. N., Schutte B. C., Houston D. W., Cornell R. A. (2009). Maternal interferon regulatory factor 6 is required for the differentiation of primary superficial epithelia in Danio and Xenopus embryos. Dev. Biol. 325, 249–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- Sakai K., Akiyama M., Sugiyama-Nakagiri Y., McMillan J. R., Sawamura D., Shimizu H. (2007). Localization of ABCA12 from golgi apparatus to lamellar granules in human upper epidermal keratinocytes. Exp. Dermatol. 16, 920–926 [DOI] [PubMed] [Google Scholar]
- Slanchev K., Carney T. J., Stemmler M. P., Koschorz B., Amsterdam A., Schwarz H., Hammerschmidt M. (2009). The epithelial cell adhesion molecule EpCAM is required for epithelial morphogenesis and integrity during zebrafish epiboly and skin development. PLoS Genet. 5, e1000563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth I., Hacking D. F., Hilton A. A., Mukhamedova N., Meikle P. J., Ellis S., Satterley K., Collinge J. E., de Graaf C. A., Bahlo M. (2008). A mouse model of harlequin ichthyosis delineates a key role for Abca12 in lipid homeostasis. PLoS Genet. 4, e1000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprecher E., Ishida-Yamamoto A., Mizrahi-Koren M., Rapaport D., Goldsher D., Indelman M., Topaz O., Chefetz I., Keren H., O’Brien T. J., et al. (2005). A mutation in SNAP29, coding for a SNARE protein involved in intracellular trafficking, causes a novel neurocutaneous syndrome characterized by cerebral dysgenesis, neuropathy, ichthyosis, and palmoplantar keratoderma. Am. J. Hum. Genet. 77, 242–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg J. P., Boggess D., Hogan M. E., Sundberg B. A., Rourk M. H., Harris B., Johnson K., Dunstan R. W., Davisson M. T. (1997). Harlequin ichthyosis (ichq): a juvenile lethal mouse mutation with ichthyosiform dermatitis. Am. J. Pathol. 151, 293–310 [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thisse C., Thisse B. (2008). High resolution in situ hybridization on whole-mount zebrafish embryo. Nat. Protoc. 3, 59–69 [DOI] [PubMed] [Google Scholar]
- Tusnády G. E., Sarkadi B., Simon I., Váradi A. (2006). Membrane topology of human ABC proteins. FEBS Lett. 580, 1017–1022 [DOI] [PubMed] [Google Scholar]
- Yanagi T., Akiyama M., Nishihara H., Sakai K., Nishie W., Tanaka S., Shimizu H. (2008). Harlequin ichthyosis model mouse reveals alveolar collapse and severe fetal skin barrier defects. Hum. Mol. Genet. 17, 3075–3083 [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L. (1965). Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins (ed. Bryson V., Vogel H. J.), pp. 97–166 New York: Academic Press [Google Scholar]