Abstract
PI3K is activated in some cancers by direct mutation, but it is activated more commonly in cancer by mutation of upstream acting receptor tyrosine kinases (TKs). At present, there is no systematic method to determine which TK signaling cascades activate PI3K in certain cancers, despite the likely utility of such information to help guide selection of tyrosine kinase inhibitor (TKI) drug strategies for personalized therapy. Here we present a quantitative tandem mass spectrometry (LC/MS/MS) approach that identifies upstream activators of PI3K both in vitro and in vivo. Using non-small cell lung carcinoma (NSCLC) to illustrate this approach, we demonstrate a correct identification of the mechanism of PI3K activation in several models, thereby identifying the most appropriate TKI to down-regulate PI3K signaling. This approach also determined the molecular mechanisms and adaptors required for PI3K activation following inhibition of the mTOR kinase TORC1. We further validated the approach in breast cancer cells with mutational activation of PIK3CA, where tandem mass spectrometry detected and quantitatively measured the abundance of a helical domain mutant (E545K) of PIK3CA connected to PI3K activation. Overall, our findings establish a mass spectrometric approach to identify functional interactions that govern PI3K regulation in cancer cells. Using this technique to define the pathways which activate PI3K signaling in a given tumor could help inform clinical decision making by helping guide personalized therapeutic strategies for different patients.
Keywords: PI3K, mass spectrometry, targeted therapy, cancer, LC/MS/MS, personalized medicine
Introduction
The phosphatiditylinositide-3-kinase (PI3K) signaling pathway is central to growth and survival of many cancers. PI3K is a lipid kinase that converts phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] to phosphatidylinositol 3,4,5-triphosphate [PtdIns(3,4,5)P3] leading to membrane recruitment and activation of specific proteins with pleckstrin homology (PH) domains, including AKT. Although there are several classes of PI3K, class IA PI3Ks are the most intimately linked to cancer cell growth and survival (1). These enzymes are heterodimers, consisting of a regulatory subunit, p85, and a catalytic subunit, p110 (2). Recent cancer genome sequencing efforts have revealed that PI3K signaling can be directly activated by genetic mutations. Most commonly, these mutations are in PIK3CA, the gene encoding the p110α subunit, and in PTEN, the gene encoding the phosphatase that degrades PIP3, the lipid product of PI3K (3). Moreover, several studies have shown that PI3K signaling is critical for tumorigenesis and tumor maintenance, even in cancers that lack PIK3CA or PTEN mutations (4–9). In many of these cancers, class IA PI3K is activated upon direct binding to receptor tyrosine kinases (RTKs) and/or adaptor proteins. The p85 regulatory subunit binds to tyrosine phosphorylated proteins via two SH2 domains. The engagement of the p85 SH2 domains with tyrosine phosphorylated receptors and adaptors recruits PI3K to the membrane where its lipid substrate resides (10–12). Currently, there is no validated method to determine how PI3K is activated in different cancers, although this information would provide insights into potential therapeutic strategies.
Recent work has shown that when receptor tyrosine kinase (RTK) inhibitors are effective against a particular cancer, inhibition of the RTK invariably leads to down regulation of PI3K signaling (13). Thus, when a cancer is “oncogene addicted” to an RTK, PI3K is under the sole regulation of that RTK, and the corresponding tyrosine kinase inhibitor leads to suppression of PI3K signaling. Furthermore, cancers develop resistance to kinase inhibitors when secondary events restore PI3K signaling in the presence of the tyrosine kinase inhibitor (13). Thus, detailed understanding of the regulation of PI3K signaling is important for determining both sensitivity and resistance to targeted therapies.
Over the past few years, we and others have utilized immunoprecipitations (IPs) of PI3K to identify the associated phosphotyrosine proteins, and thus the pathways directly activating PI3K (14–18). Currently, this is primarily accomplished through biochemical approaches using multiple p85 IPs assessed by western blot analyses, and is only effective when sensitive and specific antibodies are available. To date, these approaches have been time consuming with low yield. In recent years, there has been a growing trend of using IP combined with mass spectrometry (MS) (19–21). In this study, we evaluated the efficacy of tandem MS using a targeted approach to quantify and assess the association of adaptors and RTKs with PI3K in several different cancer models and paradigms. We also test the mechanistic role of activating PIK3CA mutations in cells that harbor these mutations. These results demonstrate that MS can identify the mechanisms of PI3K activation in cancers, and can successfully point to the appropriate RTK inhibitor(s) that will lead to PI3K suppression.
Materials and Methods
Cell lines and reagents
The EGFR mutant NSCLC cell line HCC827 (del E746_A750) has been extensively characterized (15, 16). HCC827 cells were maintained in RPMI 1640 (Cellgro; Mediatech Inc., Herndon, CA) supplemented with 5% FBS. The EGFR-wild type (WT) squamous cancer cell line A431 was made resistant to 1µM gefitinib by Jeff Engelman (MGH, Boston, MA) as described previously (17) and named A431Gefitinib Resistant (A431GR). A431GR cells were maintained in RPMI 1640 supplemented with 10% FBS and 1µM gefitinib. EBC-1 cells containing MET amplification (15) were obtained from Jeff Settleman (MGH, Boston, MA), H3122 containing an echinoderm microtubule-associated protein-like 4–anaplastic lymphoma kinase (EML4-ALK) gene translocation (22) were obtained from Pasi Jänne (DFCI, Boston, MA) and H1703 NSCLC cells (23) were obtained from ATCC and all were grown in RPMI-1640 media with 10% FBS. ER positive MCF7, and HER2-amplified SKBR3 and BT474 breast cancer cell lines were grown in DMEM media with10% FBS. Three KRAS-mutant NSCLC cells lines (A549, H460 and H23) were cultured in RPMI 1640 media supplemented with 10% FBS. All growth media was supplemented with 100 units/mL penicillin, 100 units/mL streptomycin, and 2 mM glutamine. Growth factors (EGF, recombinant HGF and IGF) were purchased from R&D Systems and used at 50ng/mL. PHA-665752 was purchased from Tocris. The stock solutions were prepared in DMSO and stored at −20°C. Imatinib, gefitinib and NVP-AEW541 were obtained from American Custom Chemical and used at 1µM. TAE-684 was purchased from Selleck and was used at 100nM. Rapamycin was purchased from Sigma and used at 50nM. Cells were treated with tyrosine kinase inhibitors in 10% FBS for 6 hrs except for rapamycin which was used for 16 hrs.
Immunoprecipitation and western blotting
Cells were lysed in a 1% NP-40 containing lysis buffer. Lysates were centrifuged at 16,000 × g for 5 min at 4°C. The supernatant was used for subsequent procedures. Co-immunoprecipitations were performed by incubating 10 mg of the cell lysate with the p85α rabbit polyclonal antibody (Millipore) and protein A sepharose beads (GE Healthcare) overnight at 4°C. Beads were precipitated, washed with lysis buffer, and boiled in sample buffer containing beta mercaptoethanol. Western blot analyses were conducted after separation by SDS/PAGE and transfer to nitrocellulose or PVDF membranes. Antibodies against ERBB3 and AKT were purchased from Santa Cruz Biotechnology and antibodies against GAB1, GAB2, IRS1 and pTyr were purchased from Cell Signaling Technologies. All were used per manufacturer’s directions. Antibody binding was detected using enhanced chemiluminescence (PerkinElmer).Western blot images were captured using GeneSnap image acquisition software.
Scrambled siRNA transfection and E545K expression
IRS and scrambled siRNA was purchased from Dharmacon (Lafayette CO) and transfection was performed using Qiagen (Valencia CA) HiPerFect Transfection Reagent. HA-tagged wild-type or E545K p110α coding sequences were excised from the JP1520 retroviral vector and cloned into the LZRS-Neo retrovirus (Gary Nolan laboratory, Stanford University). BT474 and SKBR3 cells were infected with viral supernatants produced by Phoenix cells and transfected with the LZRS-p110a retroviral construct. After transfection, cells were selected for 10–14 days in G418.
Xenograft studies
Nude mice (nu/nu; 6–8weeks old; Charles River Laboratories) were used for in vivo studies and were cared for in accordance with the standards of the Institutional Animal Care and Use Committee. A suspension of 5 × 106 H3122 lung cancer cells in 0.2 mL PBS were inoculated s.c. into the lower right quadrant of the flank of each mouse. When tumors reached ~ 400 mm3, tumors were treated with TAE-684 administered at 25 mg/kg/d or vehicle alone via orogastric gavage as described previously (22). After 2 days of treatment, tumors were excised and ~130 mg was homogenized and lysed. 12 mg of lysate was used for p85 IP. Tumors were measured twice weekly and mice were monitored daily for body weight and general condition. The experiment was terminated when the mean tumor volume of either the treated or control groups reached 2000 mm3.
Mass spectrometry
Targeted Ion MS/MS (TIMM)
Immunoprecipitations for the p85 complex were separated by SDS-PAGE until the 52 kDa marker was observed (short gel run, ~1/6 distance of mini gel lane). Gel sections were excised above the 55 kDa band (IgG heavy chain) to avoid antibody contamination and peptide signal suppression. Gel sections were reduced with DTT, alkylated with iodoacetamide and digested overnight with TPCK modified trypsin (Promega Corp.). Peptides were extracted, concentrated to 10µL using a SpeedVac and analyzed by positive ion mode reversed-phase liquid chromatography tandem mass spectrometry (LC/MS/MS) using a hybrid LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific). Peptides were delivered and separated using an EASY-nLC nanoflow HPLC (Proxeon Biosystems) at 300 nL/min using self-packed 15cm length × 75µm id C18 fritted microcapillary columns. Solvent gradient conditions were 50 min from 3% B buffer to 38% B (B buffer: 100% acetonitrile; A buffer: 0.1% formic acid/99.9% water). Peptide precursor m/z ratios representing p85 binding proteins (Table 1) were targeted in the ion trap portion of the LTQ-Orbitrap XL for MS/MS via CID using Xcalibur software (Thermo Fisher Scientific) across the entire chromatogram. For TIMM experiments, approximately 5–9 sequencing events were acquired per peptide sequence for average TIC calculations, depending upon sample abundance. TIMM cycle time for 16 IT MS/MS scans including 1 FT MS scan was ~ 2.4 sec using a MS2 max inject time of 100 msec. MS/MS spectra were analyzed using Sequest in Proteomics Browser Software (PBS) (W.S. Lane, Harvard University) by searching the reversed and concatenated Swiss-Prot protein database (version 57.5: 470,369 entries) with a parent ion tolerance of 50 ppm and fragment ion tolerance of 0.80 Da. Carbamidomethylation of cysteine (+57.0293 Da) was specified in Sequest as a fixed modification and oxidation of methionine (+15.9949) as a variable modification. Targeted peptide sequences were initially accepted if they matched the targeted protein from the forward database and met the following PBS scoring thresholds for 2+ ions: Xcorr ≥ 1.9, Sf ≥ 0.4, P ≥ 5, Δmass < 10 ppm. After passing the scoring thresholds, all MS/MS were then manually inspected to be sure that b- and y- fragment ions aligned with the assigned sequence. False discovery rates (FDR) for peptide identifications were calculated to be < 0.5%.
Table 1.
List of peptide sequences, MS charge states and peptide ion mass/charge (m/z) ratios for the PI3K adaptor and RTK proteins used for targeted ion MS/MS (TIMM) experiments. Note that peptide sequences containing methionine sulfoxide (Msx) were fully oxidized.
| Targeted protein | Peptide sequence | M/z ratios [M+2H]2+ |
|---|---|---|
| p85α | TWNVGSSNR | 510.747 |
| p85β | AALQALGVAEGGER | 671.360 |
| IRS1 | HTQRPGEPEEGAR | 732.353 |
| AAWQESTGVEMR | 711.328 | |
| IRS2 | PVSVAGSPLSPGPVR | 710.401 |
| SNTPESIAETPPAR | 735.365 | |
| GAB1 | LTGPDVLEYYK | 706.851 |
| APSASVDSSLYNLPR | 788.902 | |
| GAB2 | SSPAELSSSSQHLLR | 799.910 |
| SAESMsxSDGVGSFLPGK | 792.864 | |
| ERBB3 | ESGPGIAPGPEPHGLTNK | 879.444 |
| GESIEPLDPSEK | 650.817 | |
| VLGSGVFGTVHK | 600.840 | |
| LAEVPDLLEK | 563.821 | |
| PDGFR | VVEGTAYGLSR | 576.306 |
| ATSELDLEMsxEALK | 725.361 |
Relative quantification via average TIC
The total ion current (TIC) from each identified MS/MS spectrum was recorded in PBS. Validated data files were imported as .txt into in-house developed NakedQuant (v1.1, Beth Israel Deaconess Medical Center, Boston, MA) software for MS/MS based quantification (24). NakedQuant was programmed in MatLab and designed to perform as a platform for quantifying proteins across biological conditions according to average TIC, spectral counts and sum TIC. It contains a protein grouping algorithm and normalization calculations based on median TIC signal or a single protein (bait) and calculates ratio changes between selected samples. Here, the level of p85α/β was normalized across biological conditions. Relative quantities of each protein were calculated by averaging the TIC values from all targeted peptide MS/MS spectra per protein and compared across sample conditions. After NakedQuant performed quantitative calculations, data were exported to Excel for plotting. Note that Scaffold 3.1 software (Proteome Software) can also be used for MS/MS TIC calculations. Biological and/or technical replicates were performed for all targeted experiments, including xenografts where three mouse tumors were used and HCC827 where biological triplicates were performed. CV values were calculated and used in error bars on plots.
Results
To determine how Class IA PI3K is activated in various cancer models, we performed label-free quantitative mass spectrometry on p85 IPs from cell lysates using antibodies that recognize both the p85α and p85β regulatory subunits. The complex of p85-associated proteins was purified via SDS-PAGE, excised above 55 kDa to avoid antibody contamination, digested with trypsin and analyzed by LC/MS/MS. The known activators of PI3K that bind directly to p85 all have MWs greater than 55kDa. Initially, we assessed these PI3K complexes using a non-targeted or “shotgun” data-dependent LC/MS/MS label-free method termed spectral TIC or “average TIC” that averages the MS/MS total ion current (TIC) values across all identified peptides per protein as we previously described (24–28). However, the shotgun results suffered from high levels of non-specific protein associations, a common difficulty for antibody based IP-MS experiments (29, 30). This led to unreliable detection of the critical p85 binding proteins that we were attempting to quantify likely due to MS signal suppression of adaptor peptides.
Targeted MS/MS of PI3K interactions
Since most of the major PI3K-activating proteins are well-known, we targeted specific tryptic peptide precursor ions from the known PI3K activating proteins for MS/MS fragmentation via collisionally induced dissociation (CID) over their chromatographic elution, a label-free quantitative experiment referred to as “Targeted Ion MS/MS” (TIMM). Fig. S1 shows the schematic of the TIMM approach using hybrid linear ion trap/orbitrap mass spectrometry technology. Using a single LC/MS/MS run from a tryptic digestion of a gel purified p85 complex, we targeted and quantified only 16 peptides representing p85α, p85β and six proteins (IRS1, IRS2, GAB1, GAB2, PDGFR and ERBB3) known to bind directly to p85α and p85β due to their multiple pYXXM motifs binding to SH2 domains on p85. The peptides were chosen because they represent the most consistent and abundant peptide signals from tryptic digestions and shotgun LC/MS/MS performed in our laboratory and are from regions where phosphorylation or other post-translational modifications were not significantly detected. The individual peptides were isolated, fragmented, identified via database searching, and quantified by averaging the TIC values across all identified MS/MS spectra per protein. Table 1 lists the peptides used for the TIMM experiment. Of note, four ERBB3 peptides were used while other adaptor proteins were represented by only one or two peptides. More ERBB3 peptides were used because it was more difficult to identify this adaptor and the quantitative information was more robust with multiple peptides. Adding peptides to other proteins did not significantly improve sensitivity. Since the number of total peptides for targeting was low, chromatographic scheduling, a frequently used feature for multiple reaction monitoring (MRM) experiments, was not required. Non-scheduled targeted runs allow for potential shifts in chromatographic elution that could otherwise result in missed information and allows for the technology transfer across different chromatographic platforms. It is important to note that average TIC values are dependent upon MS ionization and fragmentation efficiency and can be reliably assessed relative to other samples in a quantitative manner because TIMM data do not represent absolute concentrations of tryptic peptides and their protein precursors in the absence of isotope labeled standards. However, by normalizing the TIC values for each adaptor protein to the values for p85 from the same immunoprecipitate, it is possible to compare relative amounts of p85-associated proteins in different cells or compare changes in these ratios in response to growth factors or drugs.
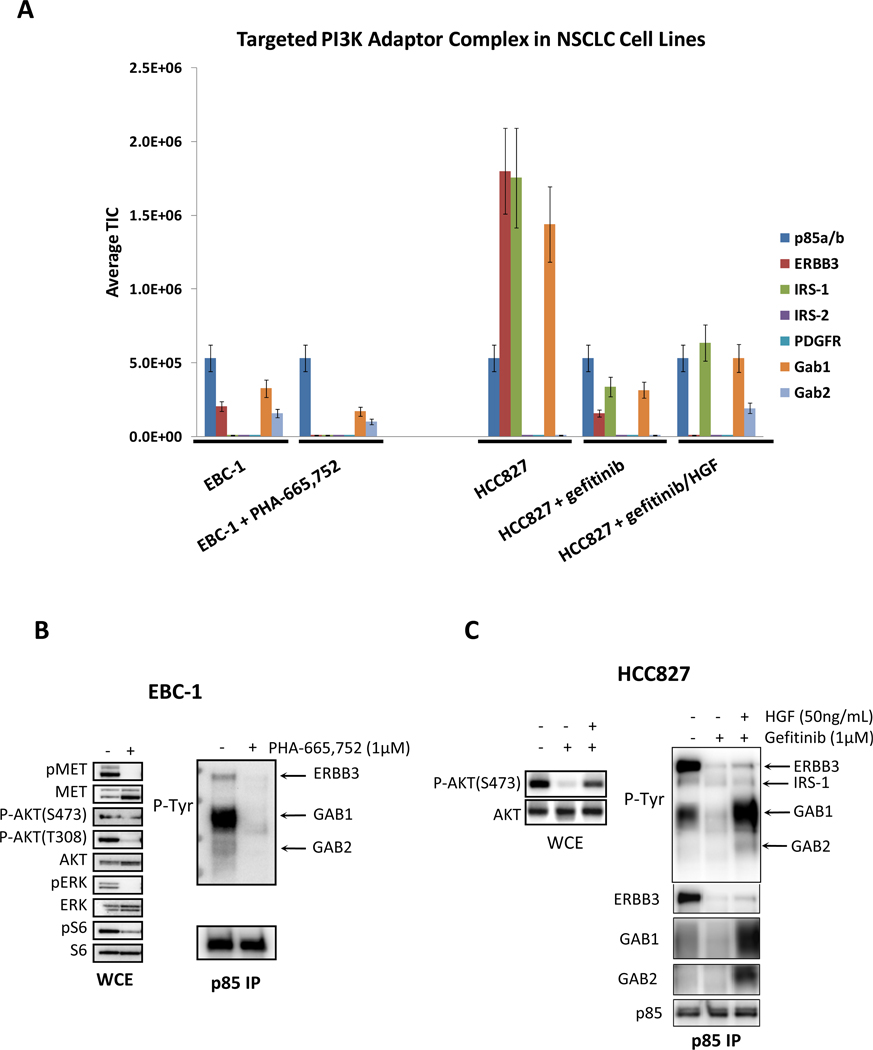
This targeted approach was examined initially in the non-small cell lung carcinoma (NSCLC) EBC-1 and HCC827 cell lines (Fig. 1A). EBC-1 cells have amplification of the MET (HGF receptor) tyrosine kinase, and treatment with a MET tyrosine kinase inhibitor (TKI) leads to suppression of PI3K signaling (15). EBC-1 cells were assessed in the absence and presence of the MET inhibitor PHA-665,752. As shown by western blot analyses, treatment with the MET inhibitor led to a loss of AKT phosphorylation and a marked reduction in the binding of several phosphotyrosine proteins to PI3K that correspond to ERBB3, GAB1 and GAB2 (Fig. 1B). These results were recapitulated independently by the TIMM approach (Fig. 1A). Please note that the quantitative changes observed by western and TIMM analyses were slightly different for HCC827 cells. This is partly due to the fact that the TIMM and western experimenters were prepared from different cell preparations.
Figure 1.
(A) The quantitative output of the TIMM approach in EBC-1 and HCC827cells subjected to the indicated conditions. Cells were treated with the indicated drugs and ligands for 6 hours before lysis. The relative signal level of each detected adaptor (normalized for p85 levels) is shown. (B) EBC-1 cells were treated in the absence or presence of the MET inhibitor, PHA-665,752 (1µM) for 6 hours and then subjected to lysis. (C) HCC827 cells were treated in the absence or presence of gefitinib (1µM) and HGF (50 ng/mL) for 6 hours. Left) Whole cell extracts were probed with the indicated antibodies. Right) Extracts were subjected to a p85 IP, and the IP was probed with an anti-PTyr and anti-p85 antibodies.
An EGFR mutant NSCLC cell line, HCC827 is highly sensitive to EGFR tyrosine kinase inhibitors. Treatment with an EGFR inhibitor leads to profound loss of PI3K-AKT signaling and decreased association between PI3K and phosphotyrosine adaptor proteins (14–17). Therefore, in this cell line, we compared the p85 complexes in the absence and presence of the EGFR kinase inhibitor, gefitinib. In EGFR mutant NSCLCs, we recently determined that HGF causes resistance to EGFR inhibitors by rescuing PI3K signaling via a mechanism independent of ERBB3 via GAB1 (16). As shown in Fig. 1A, the TIMM method shows that treatment of HCC827 cells with gefitinib decreases the binding of p85 to ERBB3, IRS1, GAB1 and GAB2, and that HGF partially restores binding to the phosphotyrosine proteins except for ERBB3. These results are in agreement with the western blot studies (Fig. 1C).
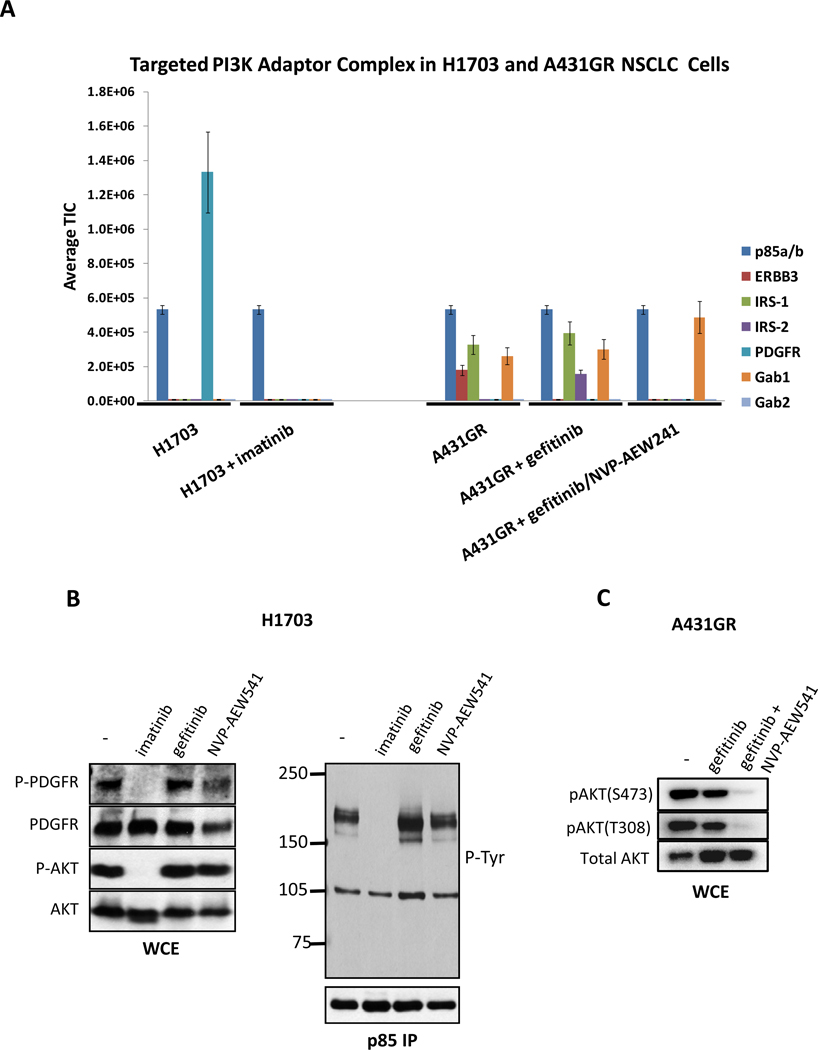
The encouraging results from the EBC-1 and HCC827 cells prompted us to test this targeted MS based methodology in other NSCLC models. We examined H1703 cells, and interestingly, found that PDGFR was the only adaptor bound to PI3K (Fig. 2A). This interaction was obliterated by the PDGFR inhibitor, imatinib. Using western blots, we confirmed that the association between PI3K and a 160 kDa phosphotyrosine protein (consistent with PDGFR) was disrupted only by imatinib and not by gefitinib or the by IGF-IR/InsR inhibitor, NVP-AEW541 (Fig. 2B). Accordingly, only imatinib led to loss of AKT phosphorylation (Fig. 2B). These results are in agreement with a recent report demonstrating that this cell line has high activation of PDGFR and is sensitive to imatinib in vitro (23). In addition, we assessed a cell line that was made resistant to gefitinib, A431GR. We previously showed that these cells are resistant to EGFR inhibitors through activation of IGF1-R that leads to persistent PI3K signaling despite EGFR inhibition (17). When these cells were analyzed by TIMM, we observed that treatment with gefitinib led to loss of ERBB3 binding to p85, but combined treatment with both an EGFR and IGF1-R/InsR inhibitor (NVP-AEW541) abrogated the interaction between p85 and both IRS proteins and ERBB3, consistent with our previously published western blot analyses (17). In agreement with the TIMM data, concomitant EGFR and IGF1-R/InsR inhibition was required to suppress AKT phosphorylation in A431GR cells (Fig. 2C).
Figure 2.
(A) H1703 and A431 GR (gefitinib resistant) cells were treated with the indicated inhibitors and p85 adaptors were quantified using TIMM as in Figure 1. (B) H1703 cells were treated with imatinib (1µM), gefitinib (1µM), or NVP-AEW541 (1µM) for the 6 hours. Left) Whole cell extracts were probed with the indicated antibodies. Right) Extracts were subjected to a p85 IP, and the IP was probed with an anti-PTyr and anti-p85 antibodies. (C) A431 GR cells were treated with gefitinib (1µM) or gefitinib + NVP-AEW541 (1µM) for six hours. Extracts were probed with the indicated antibodies.
Mouse xenograft model
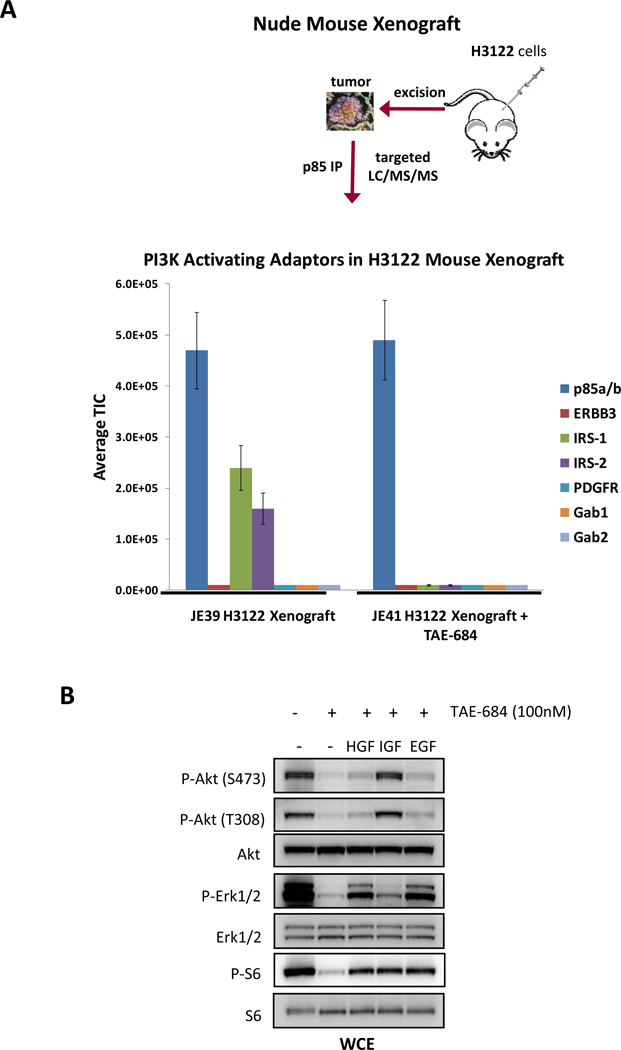
The encouraging results using cell lines led us to ask whether we could utilize this methodology to determine PI3K activators in vivo. For this study, immunodeficient mice were injected with H3122 NSCLC cells and tumors were allowed to develop (~400 mm3). H3122 tumors harbor an EML4-ALK translocation and are sensitive to ALK inhibitors in vivo and in vitro (22). Mice bearing tumors were treated with an ALK inhibitor, TAE-684, or vehicle (DMSO) control for 2 days. Tumors (~130 mg) were excised and used to prepare ~ 12 mg of protein lysate for p85 IP and TIMM analysis. Fig. 3A shows that the primary adaptors for PI3K activation are IRS1 and IRS2 in H3122 mouse xenografts. Interestingly, treatment with TAE-684 eliminates these interactions, suggesting that ALK signals to PI3K via IRS2 and IRS1. This is consistent with previous reports suggesting that NPM-ALK signals to PI3K via IRS proteins (31, 32). The finding that IRS proteins were utilized to activate PI3K suggested that these cells might activate PI3K in response to IGF1. Indeed, treatment of H3122 cells with IGF1, but not EGF or HGF, rescued PI3K-AKT signaling with TAE-684 treatment (Fig. 3B). Of note, EGF and HGF rescued ERK signaling demonstrating that these cells do respond to those ligands, but that they are unable to rescue PI3K-AKT signaling.
Figure 3.
(A) H3122 xenografts harboring the EML4-ALK translocation were treated with control vehicle or the ALK inhibitor, TAE-684, for 2 days; the tumors were excised and lysates were prepared. The TIMM results for the control and treated animals are shown. (B) H3122 cells were treated in the presence or absence of TAE-684 (100 nM) for 6 hours in the presence or absence of the indicated ligands [EGF (50 ng/mL), IGF1 (50 ng/mL), and HGF (50 ng/mL)]. Extracts were probed with the indicated antibodies.
Effect of rapamycin on the PI3K complex
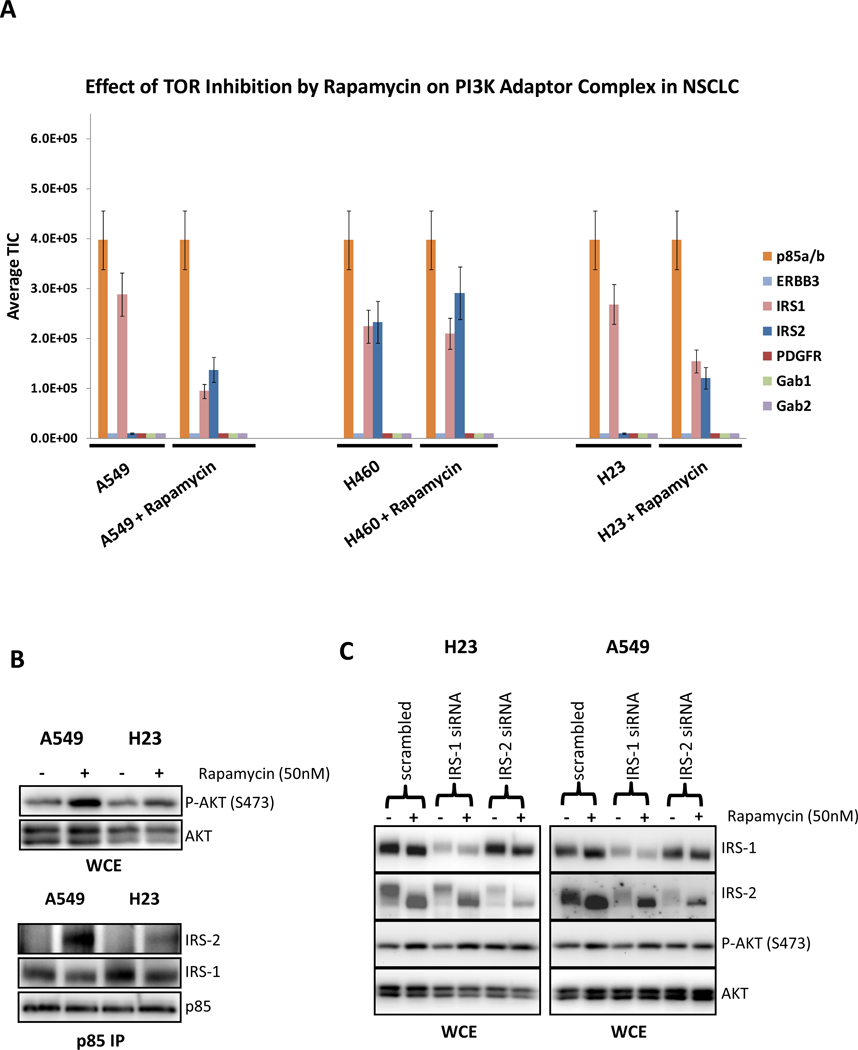
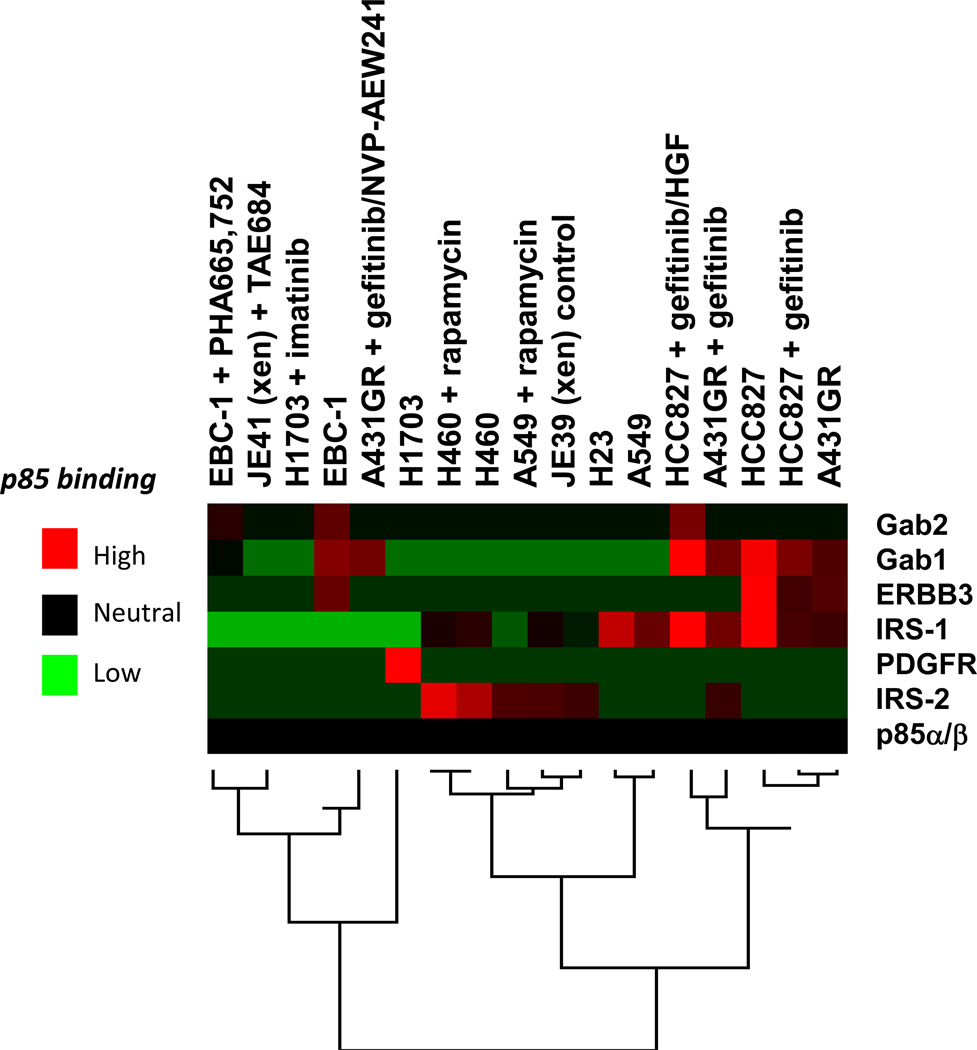
Previous studies have demonstrated that inhibition of TORC1 with rapamycin induces PI3K-AKT signaling by de-repressing a negative feedback. In some cancer types, this is mediated by de-repression of IRS1 and activation of IGF-IR signaling (33). Thus, we utilized the TIMM approach to determine the molecular mechanism of this feedback. Several NSCLC cell lines were serum starved then treated with vehicle or rapamycin for 16 hours, lysed, subjected to immunoprecipitation with anti-p85 antibodies and analyzed by mass spectrometry. As shown in Fig. 4A, in the KRAS mutant H23, A549, and H460 cells rapamycin induced an interaction of p85 with IRS2 but decreased the interaction with IRS1. As shown in Fig. 4B, western blot analysis of the A549 and H23 cells confirms that rapamycin enhanced the interaction between p85 and IRS2 but impaired the interaction between p85 and IRS1, thereby verifying the mass spectrometry data. Furthermore, knockdown of IRS2, but not IRS1, abrogated the capacity of rapamycin to induce AKT phosphorylation in these cells (Fig. 4C). The reason why p85 switches from IRS1 to IRS2 in response to rapamycin treatment of these KRAS transformed cell lines is not clear. Apparently, the IRS2-PI3K complex is more critical than the IRS1-PI3K complex for AKT activation. IRS protein band shifts are due to loss of previously characterized phosphoserine and phosphothreonine sites (34). Fig. 5 shows an unsupervised hierarchical clustering heat map for all of the p85 IP-TIMM experiments in this study. The data generally cluster according to treatment and cell line. The expansion of this heat map with data from other cell lines, xenograft models and, eventually, human cancer specimens may ultimately provide a reference that will predict mechanisms of PI3K activation in different cancer types. For example, the heat map clearly indicates that PI3K in H1703 cells is driven solely by PDGFR, a situation where imatinib was the effective treatment.
Figure 4.
(A) The TIMM results from the p85 adaptor complexes in KRAS mutated A549, H460 and H23 NSCLC cells in the absence or presence of rapamycin (50 nM) for 16 hours. Results were quantified as in Figure 2. (B) Extracts from the A549 and H23 treated as in (A) were probed with the indicated antibodies. (C) H23 and A549 cells were transiently transfect with scramble, IRS1, or IRS2 siRNA. Cells were treated in the absence or presence of rapamycin (50 nM) for 16 hours. Cells were lysed and probed with the indicated antibodies.
Figure 5.
Unsupervised hierarchical clustering heat map for the targeted p85 IP mass spectrometry results performed in Figures 1–4. TIC was normalized to p85α/p85β across all experiments.
Quantifying the E545K PIK3CA mutation
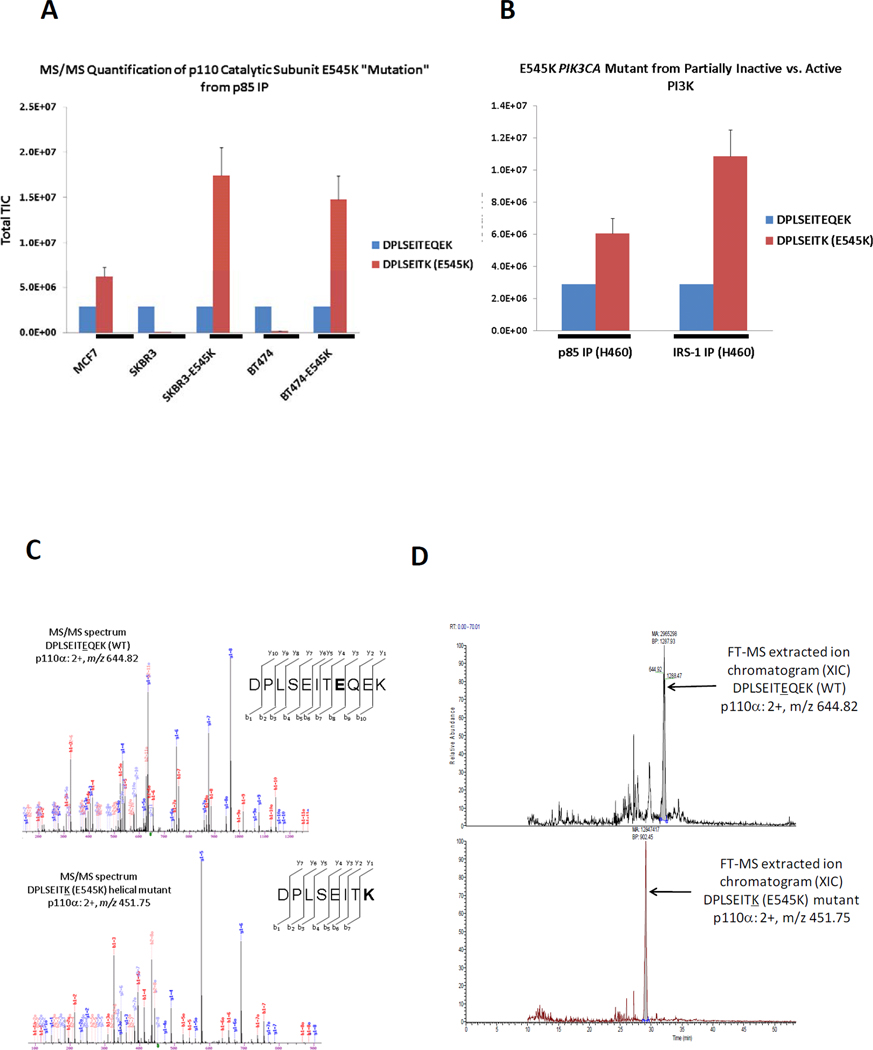
In addition to determining the mechanism of PI3K activation, we tested whether the same p85 IPs could quantify the amount of mutant p110 in those cancers harboring a somatic mutation. PIK3CA, the gene encoding p110α, is mutated in some cancers. These mutations are usually located in one of the two “hotspots” in the gene (35–37). One exists in the helical domain (E545K) and the other in the kinase domain (H1047R). Both have been linked to aberrant PI3K-AKT activation (36, 38, 39). We obtained several cancer cell lines known to possess the E545K mutation including breast carcinoma MCF7 and NSCLC H460. As controls, we included two p110α WT breast cancer cell lines, SKBR3 and BT474 that were infected with retroviruses encoding E545K p110α mutant. The total level of the WT E545 p110α peptide DPLSEITEQEK (2+, m/z 644.82) was normalized across all samples. Since only a single peptide was quantified across conditions, sum TIC was used rather than average TIC. The WT and mutated [DPLSEITK (2+, m/z 451.75)] tryptic peptides are different lengths and contain amino acid differences, so it is possible that the ionization efficiency of each peptide varies. However, a difference in the TIC value ratio of the E545 mutant to the WT p110α between cell types or in different immunoprecipitates from the same cell provides useful information. As shown in Fig. 6A, the TIMM method correctly identified the presence of the E545K mutation in MCF7 cells (40) and the lack of this mutant protein in p85 immunoprecipitates from WT SKBR3 and BT474 cells (41). The TIMM method also detected the E545K mutant in SKBR3 and BT474 cells that were engineered to express this protein, and the relative TIC values indicate that the ratio of mutant to WT p110α is considerably higher (nearly 10-fold) in the engineered cells, although this increase may be inflated since the total level of p110 is higher in the over expressed cells.
Figure 6.
(A) Quantitative analyses of E545K peptide targeted by mass spectrometry from the MCF7 cells and from SKBR3 and BT-474 cells engineered to express the WT (WT) p110α or the E545K mutant. (B) H460 cells were lysed and immunoprecipitated with an anti-p85 antibody or an anti-IRS1 antibody. The TIMM data show that E545K is enriched more than two-fold in the IRS1 complex. (C) The MS/MS fragmentation spectra via CID identifying the p110α WT and E545K somatic mutant peptide quantified from H460 cells. (D) The extracted ion chromatograms for the WT and E545K mutant p110α peptides. These represent the typical TIMM peak elution profile for targeted tryptic peptides using hybrid linear ion trap-orbitrap technology via LC/MS/MS.
In addition, we took advantage of the quantitative nature of this assessment to determine if the E545K mutant is preferentially recruited to phosphotyrosine adaptors. We utilized the H460 NSCLC cells that harbor the E545K p110α catalytic site mutation and use IRS proteins to associate with p85 for activation of PI3K/AKT (Fig. 4A). We compared the ratio of the mutant PI3K in p85 IPs (holoenzyme bound to adaptor and free) versus IRS1 IPs (PI3K bound to adaptor in active state). We observed that the relative amount of E545K is greater than 2-fold higher in the IRS1 IPs (fully active form) (Fig. 6B). This result is consistent with structural analyses of E545K suggesting that the mutation abrogates an intermolecular interaction with an SH2 domain in p85 (11, 12). Therefore, in the PI3K holoenzymes containing E545K, the untethered SH2 domain could be more available to bind to phosphotyrosine adaptors. The MS/MS fragmentation spectra and the extracted ion chromatograms for the WT and mutant peptides are shown in Fig. 6C and 6D, respectively. We also attempted to target the H1047R p110α mutation from BT474 engineered breast cancer cells; however, this mutation resulted in a tryptic peptide (QMNDAR) of weak signal in MS and its MW was too small for reliable MS/MS identification. Alternative proteolytic enzymes and/or MRM may be better suited for quantifying H1047R by MS.
Discussion
The central role of PI3K in the sensitivity and resistance to targeted therapies has increased the need for understanding the molecular regulation of this enzyme in cancer cells. In this study, we utilized a targeted MS approach. This approach effectively identifies the activators of PI3K and holds promise for understanding how various cancers regulate PI3K in a dynamic manner.
Initially, we utilized a non-targeted tandem MS (shotgun) approach to determine the adaptor proteins bound to p85 in cancer cells. Although this method effectively identified several PI3K-activating proteins, it was not sufficiently sensitive or quantitative to specifically determine the phosphotyrosine proteins activating PI3K across NSCLC cell lines. Thus, we switched to a targeted approach to quantitatively measure the association of PI3K with key adaptors and RTKs that are the most common activators of PI3K. While there are several methods for performing quantitative analyses of protein interactions by MS, we utilized hybrid ion trap-orbitrap technology whereby we targeted peptide ions for isolation and fragmentation in the ion trap component. The trap fills with product ions of each peptide precursor ion across the chromatogram and generates sufficient and quantitative signal when the peptide of interest elutes from the column. We have used a similar technique to quantify phosphopeptide signals from various signaling proteins (42–44). Alternatively, one can use stable isotope labeling approaches such as SILAC (45) or chemical tagging approaches such as iTRAQ (46) in addition to label-free quantitative methodologies including spectral counting (47) and MRM (48). SILAC is a metabolic labeling approach that is useful in cell culture but is not readily adapted to in vivo tissue sources while iTRAQ requires clean-up and a chemical labeling step that can be affected by the sample matrix (49). In this study, we observed that hybrid linear ion trap–orbitrap mass spectrometers can be used for successful label-free quantification in targeted mode if the total number of peptides is kept low since the cycle time is slower than that for triple quadrupole mass spectrometers in MRM mode.
Currently, there are several methods for assessing the status of PI3K signaling pathway in cancers including genetic analyses and assessment of the abundance of downstream signaling events such as phospho-AKT. However, there have not been any validated methods for determining how PI3K is activated in cancers, especially those without PIK3CA or PTEN mutations. This study validates a simple mass spectrometry method to make such determinations. Although we determined how PI3K was activated in a variety of cancer paradigms in this study, there are clearly other applications. For example, MEK inhibitors (8) and other cellular stresses such as radiation (50) can lead to activation of PI3K signaling, and this method could be used to determine the molecular mechanisms of activation. Indeed, this approach could identify potential therapeutic targets that would prevent PI3K activation in response to these stresses. In addition, one can include hot spot mutations in oncogenes such as PIK3CA in the targeted approach to correlate mutation status with adaptor activation. We also evaluated other RTKs for PI3K activation such as EGFR and MET receptor but did not find evidence of direct p85 binding since these receptors lack multiple pYXXM motifs, although they associated with known adaptors at low levels in some p85 IPs.
Herein, we demonstrated that mass spectrometry in combination with immuno-purification of signaling complexes can be used to identify oncogenic pathways driving tumor growth using milligram quantities of tumor tissue. As these MS results are coupled with comprehensive genetic analyses of cancers, the mechanisms of PI3K activation may be predicted by the genetic abnormalities harbored by a particular cancer, thus leading to personalized therapeutic strategies that block PI3K activation.
Supplementary Material
Acknowledgements
The work was supported in part by NIH 5P01CA120964-04 and NIH DF/HCC Cancer Center Support Grant 5P30CA006516-46 (J.M.A.), NIH K08 CA120060 (J.A.E.), R01CA137008 (J.A.E., P.A.J.), R01CA140594 (J.A.E), NIH DF/HCC Gastrointestinal Cancers SPORE P50 CA127003 (J.A.E. and L.C.C.), NCI Lung SPORE P50CA090578 (J.A.E., P.A.J.), NIH R01-GM41890 (L.C.C), NIH R01CA135257-01 (P.A.J. and J.A.E) and NIH R01CA136851 (to P.A.J.). NIH Breast Cancer SPORE P50CA98131, NIH Vanderbilt-Ingram Cancer Center Support Grant P30CA68485, ACS Clinical Research Professorship Grant CRP-07-234 (to C.L.A.), and Stand Up to Cancer/AACR Dream Team Translational Cancer Research Grant, Grant No. SU2C-AACR-DT0209 (to C.L.A. and L.C.C.).
References
- 1.Engelman JA. Targeting PI3K signaling in cancer: opportunities, challenges and limitations. Nat. Rev. Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 2.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositols 3-kinases as regulators of growth and metabolism. Nat Rev. Genet. 2006;7:606–619. doi: 10.1038/nrg1879. Review. [DOI] [PubMed] [Google Scholar]
- 3.Samuels Y, Ericson K. Oncogenic PI3K and its role in cancer. Curr. Opin. Oncol. 2006;18:77–82. doi: 10.1097/01.cco.0000198021.99347.b9. [DOI] [PubMed] [Google Scholar]
- 4.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat. Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 6.Engelman JA, Mukohara T, Zejnullahu K, Lifshits E, Borrás AM, Gale CM, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J. Clin. Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sos ML, Fischer S, Ullrich R, Peifer M, Heuckmann JM, Koker M, et al. Identifying genotype-dependent efficacy of single and combined PI3K- and MAPK-pathway inhibition in cancer. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18351–18356. doi: 10.1073/pnas.0907325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faber AC, Li D, Song Y, Liang MC, Yeap BY, Bronson RT, et al. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19503–19508. doi: 10.1073/pnas.0905056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, et al. Downward J. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 10.Shekar SC, Wu H, Fu Z, Yip SC, Nagajyothi, Cahill SM, et al. Mechanism of constitutive phosphoinositide 3-kinase activation by oncogenic mutants of the p85 regulatory subunit. J. Biol. Chem. 2005;280:27850–27855. doi: 10.1074/jbc.M506005200. [DOI] [PubMed] [Google Scholar]
- 11.Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Y, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 12.Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, et al. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 13.Engelman JA, Jänne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin. Cancer Res. 2008;14:2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 14.Engelman JA, Jänne PA, Mermel C, Pearlberg J, Mukohara T, Fleet C, et al. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non small cell lung cancer cell lines. Proc. Natl. Acad. Sci. U.S.A. 2005;102:3788–3793. doi: 10.1073/pnas.0409773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 16.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guix M, Faber AC, Wang SE, Olivares MG, Song Y, Qu S, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J. Clin. Invest. 2008;118:2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Chaerkady R, Wu J, Hwang HJ, Papadopoulos N, Kopelovich L, et al. Mutant proteins as cancer-specific biomarkers. Proc. Natl. Acad. Sci. U S A. 2011;108:2444–2449. doi: 10.1073/pnas.1019203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang G, Fang B, Liu RZ, Lin H, Kinose F, Bai Y, et al. Mass spectrometry mapping of epidermal growth factor receptor phosphorylation related to oncogenic mutations and tyrosine kinase inhibitor sensitivity. J. Proteome Res. 2011;10:305–319. doi: 10.1021/pr1006203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.ten Have S, Boulon S, Ahmad Y, Lamond AI. Mass spectrometry-based immuno-precipitation proteomics - The user's guide. Proteomics. 2011;11:1153–1159. doi: 10.1002/pmic.201000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, Holmes AJ, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin. Cancer Res. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDermott U, Ames RY, Iafrate AJ, Maheswaran S, Stubbs H, Greninger P, et al. Ligand-dependent platelet-derived growth factor receptor (PDGFR)-alpha activation sensitizes rare lung cancer and sarcoma cells to PDGFR kinase inhibitors. Cancer Res. 2009;69:3937–3946. doi: 10.1158/0008-5472.CAN-08-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X, Friedman A, Nagpal S, Perrimon N, Asara JM. Use of a label-free quantitative platform based on MS/MS average TIC to calculate dynamics of protein complexes in insulin signaling. J. Biomol. Tech. 2009;20:272–277. [PMC free article] [PubMed] [Google Scholar]
- 25.Asara JM, Christofk HR, Freimark LM, Cantley LC. A label-free quantification method by MS/MS TIC compared to SILAC and spectral counting in a proteomics screen. Proteomics. 2008;8:994–999. doi: 10.1002/pmic.200700426. [DOI] [PubMed] [Google Scholar]
- 26.Jiang X, Chen S, Asara JM, Balk SP. Phosphoinositide 3-kinase pathway activation in phosphate and tensin homolog (PTEN) deficient prostate cancer cells is independent of receptor tyrosine kinases and mediated by the p110beta and p110delta catalytic subunits. J. Biol. Chem. 2010;285:14980–14989. doi: 10.1074/jbc.M109.085696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clements RT, Smejkal G, Sodha NR, Ivanov AR, Asara JM, Feng J, et al. Pilot proteomic profile of differentially regulated proteins in right atrial appendage before and after cardiac surgery using cardioplegia and cardiopulmonary bypass. Circulation. 2008;118:S24–S31. doi: 10.1161/CIRCULATIONAHA.107.792747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kesavan K, Ratliff J, Johnson EW, Dahlberg W, Asara JM, Misra P, et al. Annexin A2 is a molecular target for TM601, a peptide with tumor-targeting and anti-angiogenic effects. J Biol Chem. 2010;285:4366–4374. doi: 10.1074/jbc.M109.066092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L, Zhang H, Bruce JE. Optimizing the detergent concentration conditions for immunoprecipitation (IP) coupled with LC-MS/MS identification of interacting proteins. Analyst. 2009 Apr;134(4):755–762. doi: 10.1039/b813335b. [DOI] [PubMed] [Google Scholar]
- 30.Malovannaya A, Li Y, Bulynko Y, Jung SY, Wang Y, Lanz RB, et al. Streamlined analysis schema for high-throughput identification of endogenous protein complexes. Proc. Natl. Acad. Sci. U.S.A. 2010;107:2431–2436. doi: 10.1073/pnas.0912599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slupianek A, Nieborowska-Skorska M, Hoser G, Morrione A, Majewski M, Xue L, et al. Role of phosphatidylinositol 3-kinase-AKT pathway in nucleophosmin/anaplastic lymphoma kinase-mediated lymphomagenesis. Cancer Res. 2001;61:2194–2199. [PubMed] [Google Scholar]
- 32.Marzec M, Kasprzycka M, Liu X, El-Salem M, Halasa K, Raghunath PN, et al. Oncogenic tyrosine kinase NPM/ALK induces activation of the rapamycin-sensitive mTOR signaling pathway. Oncogene. 2007;26:5606–5614. doi: 10.1038/sj.onc.1210346. [DOI] [PubMed] [Google Scholar]
- 33.O'Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates AKT. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah OJ, Hunter T. Turnover of the Active Fraction of IRS1 Involves Raptor-mTOR and S6K1-Dependent Serine Phosphorylation in Cell Culture Models of Tuberous Sclerosis. Mol. Cell. Biol. 2006;26:6425–6434. doi: 10.1128/MCB.01254-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 36.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc. Natl. Acad. Sci. U.S.A. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikenoue T, Kanai F, Hikiba Y, Obata T, Tanaka Y, Imamura J, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65:4562–4567. doi: 10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- 38.Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle. 2004;3:1221–1224. doi: 10.4161/cc.3.10.1164. [DOI] [PubMed] [Google Scholar]
- 39.Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65 doi: 10.1158/0008-5472.CAN-05-2612. 10992-1000. [DOI] [PubMed] [Google Scholar]
- 40.Hollestelle A, Elstrodt F, Nagel JH, Kallemeijn WW, Schutte M. Phosphatidylinositol-3-OH kinase or RAS pathway mutations in human breast cancer cell lines. Mol. Cancer Res. 2007;5:195–201. doi: 10.1158/1541-7786.MCR-06-0263. [DOI] [PubMed] [Google Scholar]
- 41.Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 42.Dibble CC, Asara JM, Manning BD. Characterization of Rictor phosphorylation sites reveals direct regulation of mTOR complex 2 by S6K1. Mol. Cell Biol. 2009;29:5657–5670. doi: 10.1128/MCB.00735-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng B, Jeong JH, Asara JM, Yuan YY, Granter SR, Chin L, et al. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Mol. Cell. 2009;33:237–247. doi: 10.1016/j.molcel.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29:124–130. doi: 10.1016/s1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
- 46.Chen X, Sun L, Yu Y, Xue Y, Yang P. Amino acid-coded tagging approaches in quantitative proteomics. Expert Rev. Proteomics. 2007;4:25–37. doi: 10.1586/14789450.4.1.25. [DOI] [PubMed] [Google Scholar]
- 47.Lundgren DH, Hwang SI, Wu L, Han DK. Role of spectral counting in quantitative proteomics. Expert Rev. Proteomics. 2010;7:39–53. doi: 10.1586/epr.09.69. [DOI] [PubMed] [Google Scholar]
- 48.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quaglia M, Pritchard C, Hall Z, O'Connor G. Amine-reactive isobaric tagging reagents: requirements for absolute quantification of proteins and peptides. Anal. Biochem. 2008;379:164–169. doi: 10.1016/j.ab.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 50.Li HF, Kim JS, Waldman T. Radiation-induced Akt activation modulates radioresistance in human glioblastoma cells. Radiat. Oncol. 2009;4:43. doi: 10.1186/1748-717X-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.