Abstract
Coordinated global efforts to prevent and control malaria have been a tour-de-force for public health, but success appears to have reached a plateau in many parts of the world. While this is a multifaceted problem, policy strategies have largely ignored genetic variations in humans as a factor that influences both selection and dosing of antimalarial drugs. This includes attempts to decrease toxicity, increase effectiveness and reduce the development of drug resistance, thereby lowering health care costs. We review the potential hurdles to developing and implementing pharmacogenetic-guided policies at a national or regional scale for the treatment of uncomplicated falciparum malaria. We also consider current knowledge on some component drugs of artemisinin combination therapies and ways to increase our understanding of host genetics, with the goal of guiding policy decisions for drug selection.
ملخص
لقد حققت الجهود العالمية المنسقة نجاحاً باهراً في الوقاية من الملاريا ومكافحتها، ولكن هذا النجاح يبدو أنه قد وصل إلى مرحلة الثبات في كثير من أجزاء العالم. ومع أن هذه المشكلة تعود لأسباب متعددة، إلا أن سياسات الاستراتيجيات أهملت بدرجة كبيرة التباين الوراثي بين الأشخاص كعامل مؤثر في اختيار وجرعة الأدوية المضادة للملاريا. وهذا يشمل محاولات خفض السمية، وزيادة الفعالية، والحد من حدوث مقاومة الطفيلي للأدوية، ومن ثم خفض تكاليف الرعاية الصحية. وقد راجع الباحثون العقبات المحتملة أمام إعداد وتنفيذ سياسات تسترشد بعلم الجينوميات الدوائية على المجالين الوطني والإقليمي لمعالجة الملاريا المنجلية غير المصحوبة بمضاعفات. كما اهتم الباحثون بالمعلومات الراهنة حول بعض مكونات الأدوية في توليفة الأرتيميزين العلاجية، وطرق زيادة فهمنا لجينات الشخص المضيف، بغرض إرشاد صانعي السياسات نحو اختيار الدواء.
Resumen
Los esfuerzos coordinados que se están realizando en todo el mundo para prevenir y controlar la malaria han sido una auténtica hazaña para la salud pública, pero parece que el éxito conseguido se ha estabilizado en muchas partes del mundo. Debido a que se trata de un problema multifacético, muchas de las estrategias de las políticas han ignorado las variaciones genéticas en humanos como un factor que influye tanto en la selección como en las dosis de fármacos antimaláricos. Esto incluye los intentos de reducir la toxicidad, de aumentar la eficacia y de disminuir el desarrollo de resistencia a los fármacos. De esta manera, también se reducirían los costes de la asistencia sanitaria. Revisamos los posibles obstáculos para el desarrollo e implementación de las políticas orientadas a la farmacogenética a nivel nacional o regional para el tratamiento de la malaria por Plasmodium falciparum sin complicaciones. Con el objetivo de conseguir orientación para las decisiones sobre la selección de fármacos, también tenemos en cuenta los conocimientos disponibles sobre algunas terapias de combinación de fármacos componentes de artemisinina y las maneras de aumentar nuestros conocimientos sobre genética del huésped.
Résumé
Les efforts mondiaux coordonnés de prévention et de lutte contre le paludisme ont été un véritable tour de force pour la santé publique, mais le succès semble avoir atteint un plateau dans de nombreuses régions du monde. Bien que ce soit un problème à multiples facettes, les stratégies politiques ont largement ignoré les variations génétiques chez l'homme en tant que facteur influençant à la fois la sélection et le dosage des médicaments antipaludiques. Cela inclut des tentatives pour diminuer la toxicité, augmenter l'efficacité et réduire le développement de la résistance aux médicaments, réduisant ainsi les coûts des soins de santé. Nous passons en revue les obstacles potentiels au développement et à la mise en œuvre des politiques guidées par la pharmacogénétique à l'échelle nationale ou régionale pour le traitement du paludisme non compliqué à falciparum. Nous examinons également les connaissances actuelles sur certains médicaments qui composent les combinaisons thérapeutiques à base d'artémisinine et les moyens d'accroître notre compréhension de la génétique de l'hôte, dans l'objectif d'orienter les décisions politiques de sélection des médicaments.
Резюме
Скоординированные глобальные усилия по профилактике малярии и борьбе с этой болезнью стали важным достижением в сфере общественного здравоохранения. Однако во многих странах мира поступательное движение в этой области, по-видимому, затормозилось. Эта проблема имеет много аспектов, однако в политических стратегиях в значительной степени игнорировались генетические вариации людей как фактор, влияющий на выбор и дозирование противомалярийных препаратов. Учет этого фактора включает в себя попытки снизить токсичность, повысить эффективность и ограничить развитие лекарственной устойчивости, уменьшив тем самым расходы на медико-санитарную помощь. Мы исследуем потенциальные препятствия на пути разработки и внедрения политических мер, опирающихся на фармакогенетику, в общенациональном и региональном масштабе для лечения несложных случаев заболевания тропической малярией. Мы также рассматриваем накопленные в настоящее время знания о некоторых многокомпонентных лекарственных препаратах, применяемых в рамках артемизин-комбинированной терапии, а также способы углубления нашего понимания генетики хозяина с целью руководства политическими решениями в области подбора лекарств.
摘要
全球在疟疾预防控制工作中的协调努力已经成为公共卫生领域的一大力作,然而在世界许多地区,工作业绩似乎已经达到一定高度而难以继续推进。尽管这个问题具有多方面制约因素,但是政策策略在很大程度上忽略了人类遗传变异情况也是一个影响抗疟药选择和剂量的因素,这包括试图降低毒性、提高药效和减少耐药性发生从而降低医疗成本。我们审查了在国家或区域范围内开发并实施以药物基因组学为指导治疗无并发症恶性疟疾政策的潜在障碍。我们还考虑了目前对青蒿素联合疗法中某些组分药物的分析以及增加对宿主基因了解的方法,旨在引导药物选择的政策决定。
Introduction
Renewed efforts to control and eliminate malaria are making advances using the current generation of tools: long-lasting insecticide treated nets, artemisinin combination therapies (ACTs) and indoor residual spraying. Despite gains made by bednets and spraying, malaria control and elimination efforts will continue to rely heavily on chemotherapy and, in particular, ACTs.
Currently, the World Health Organization (WHO) endorses four ACT combinations for uncomplicated falciparum malaria in non-pregnant adults (Table 1).1 In 2001, WHO recommended the use of ACTs due to high efficacy rates and potential to reduce the development of resistance.2 By 2009, 88 countries, including every country in Africa, had officially adopted ACTs as first-line agents for uncomplicated malaria.2 Despite the official guidelines, only 14 countries report distributing enough ACTs to treat at least 50% of reported malaria cases in the public system, and only 5 countries reported distributing enough ACTs to treat all cases in 2008.3 Over the next 5–10 years, an increasing number of patients, including children and pregnant women, will be given ACT therapy.
Table 1. WHO-recommended antimalarial regimens1.
| Regimen | Artemisinin | Partner antimalarial drug |
|---|---|---|
| AL | artemether | Lumefantrine |
| AS+AQ | artesunate | Amodiaquine |
| AS+MQ | artesunate | Mefloquine |
| AS+SP | artesunate | Sulfadoxine-pyrimethamine |
Combination therapies
Combination therapy, already in use for tuberculosis and HIV, is now recommended for malaria treatment to stem the tide of resistance and protect the widely available, highly efficacious class of drugs, the artemisinins. These combinations are based on the unlikely probability of a parasite becoming resistant simultaneously to two drugs with unrelated modes of action.4 With this in mind, the artemisinins are particularly good partner drugs for other antimalarials as they eliminate parasites rapidly from the blood and thereby limit the number of parasites exposed to the longer acting partner drug.
An example of host genetics
The success of drug therapy is dependent upon many contributing factors, including adherence to prescribed therapy, incorrect or suboptimal dosing, general health status and interactions with other drugs. Another crucial aspect of the lack of effectiveness (often due to low drug levels) and tolerability (often due to high drug levels) of chemotherapy is the way individuals metabolize the drug. Although the widespread use of modern antimalarials began in the 1940s, there was limited data about the absorption–distribution–metabolism–excretion parameters of antimalarials in humans until about 20 years ago.
The use of primaquine in the 1950s highlights the importance of pharmacogenetics for antimalarials, as severe reactions to the drug led to the discovery of glucose-6-phosphate dehydrogenase (G6PD) deficiency in 1956.5,6 The genetic variants among people with G6PD still confound our ability to develop effective antimalarials, illustrated by the withdrawal of the drug Lapdap® (chlorproguanil/dapsone) in 2008.7 This deficiency remains a cause for concern, as primaquine is used widely for terminal prophylaxis and to decrease transmission intensity by limiting reproduction of the malaria parasites.6
Certainly, scientists have investigated the contribution of parasite genetics, in particular relating to drug resistance. Several mutations in parasite drug target genes have been identified and associated with in vivo resistance to artemisinin partner drugs mefloquine, lumefantrine, amodiaquine and chlorproguanil.8–16 In addition to parasite genetics, host genetics can valuably inform national drug policy decisions for first-line antimalarials.
Selection of ACTs
New pharmacology data available for modern antimalarials has helped to identify the major metabolic pathways and to identify human genetic variations that affect the ability to metabolize these agents (Table 2). Beyond the case of G6PD, there remain numerous questions about the role that pharmacogenetics can play to predict adverse drug reactions to antimalarials. Clearly, point-of-care testing for genetic variants, such as those of the CYP2C8 gene, is far from a reality in the developing world. However, it is possible that pharmacogenetics could help inform drug formulary decisions on a regional or national scale.
Table 2. Human genetic variants important for antimalarial metabolism17.
| Gene | Phenotype | Phenotype frequency in African populations (%) | Phenotype frequency in Asian populations (%) | Phenotype frequency in white populations (%) | First-line antimalarial |
|---|---|---|---|---|---|
| Drug metabolizing enzymes | |||||
| CYP2A6 | Poor metabolizer | 2 | 4–12 | 1 | artesunate |
| CYP2C8 | Poor metabolizer | 1.5–4 | < 0–1 | 2 | amodiaquine |
| CYP3A5 | Poor metabolizer with residual CYP3A5 activity | 12–40 | 60–75 | 85–95 | artemether, lumefantrine, mefloquine |
| Poor metabolizer with no CYP3A5 activity | 10–22 | 0 | 0 | artemether, lumefantrine, mefloquine | |
| UGT1A9 | Poor metabolizer | < 0–1 | unknown | ≤ 1 | DHA |
| UGT2B7 | Poor metabolizer | 4–10 | 6–7 | 20–25 | DHA |
| Drug transporters | |||||
| ABCB1 | Reduced function | 21 | 69 | 46 | mefloquine |
| Higher concentration of drug substrate | < 1–16 | 40–45 | 46–56 | mefloquine |
DHA, dihydroartemisinin
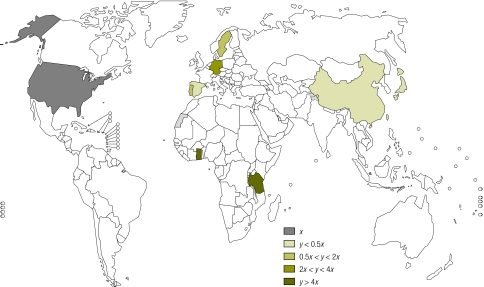
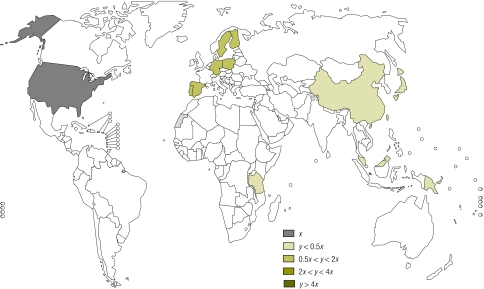
Currently, national malaria control programmes select the most appropriate drugs for use in the respective country based upon costs, in vitro Plasmodium falciparum resistance patterns and the risk of adverse drug reactions. Very little attention has been paid to host genetics and their potential to affect major formulary decisions by decreasing toxicity, increasing efficacy and improving the time in the therapeutic range to combat resistance. This is especially important when ministries of health, national medicines review authorities or national malaria control programmes must choose between seemingly equal drugs without the aid of comprehensive local pharmacogenetic data (Fig. 1 and Fig. 2). Importantly, knowledge of genetic differences in populations could reduce costs by decreasing adverse events and increasing efficacy of drugs.
Fig. 1.
World map showing frequencies of the CYP2C8*2 allele in different populations
Source: Pharmacogenetics for Every Nation Initiative.
x = allele frequency in reference population (US caucasians), y = allele frequency in country with data
Fig. 2.
World map showing frequencies of the CYP2C8*3 allele in different populations
Source: Pharmacogenetics for Every Nation Initiative.
x = allele frequency in reference population (US caucasians), y = allele frequency in country with data
This paper looks at drugs commonly used as part of ACTs to treat uncomplicated falciparum malaria in adults and provides specific examples of how genetics may affect drug efficacy and toxicity at a population level. It does not serve as an exhaustive review of all relationships between host genetics and antimalarial drugs. Several high-quality comprehensive reviews have been done elsewhere.17,18 Table 3 lists the drugs and their potential relationships with various genes. This paper discusses ways to improve knowledge of genetic data in malaria-endemic regions and explores the issues that need to be addressed to make pharmacogenetic-guided drug policy a reality.
Table 3. Potential drug–gene relationships for first-line treatments of uncomplicated falciparum malaria in non-pregnant adults.
| Drug | Gene |
|---|---|
| Artesunate | CYP2B6 |
| UGT1A9 | |
| UGT2B7 | |
| Artemether | CYP3A4/5 |
| CYP2C8 | |
| UGT1A9 | |
| UGT2B7 | |
| Amodiaquine | CYP2C8 |
| ABCB1 | |
| Mefloquine | ABCB1 |
| Sulfadoxine/pyrimethamine | No data |
| Lumefantrine | No data |
Genetics to guide drug policy
The case of amodiaquine
The antimalarial amodiaquine is generally well tolerated, but its use in primarily Caucasian populations in the 1980s and 1990s was associated with high rates of agranulocytosis (1 in 2100 with a case fatality rate of 1 in 31 000) and hepatotoxicity (1 in 15 600).19–21 WHO removed amodiaquine from the Essential Medicines List due to the discovery of these toxicities, but it was added back on the list in the late 1990s as resistance to alternative drugs began to appear. Recent evidence points to frequent mild adverse events caused by amodiaquine. A trial in 15 subjects in Africa evaluating the pharmacokinetics of amodiaquine and the combination of amodiaquine and artesunate revealed the common adverse events of hepatotoxicity and leukopenia after just two doses given three weeks apart.22
People with mutant genotypes of CYP2C8, CYP1A1 and CYP1B1 have been found to have immunogenic adverse reactions to amodiaquine.23–28 Wide variations in the CYP2C8 genotype mean that individuals experience broad differences in the drug’s efficacy and toxicity. Evidence suggests that decreased function of the CYP2C8 enzyme impairs metabolism of the drug and can form a toxic metabolite that causes hepatotoxicity and agranulocytosis. Two more common CYP2C8 variants, CYP2C8*2 and *3, decrease the metabolizing ability of CYP2C8 as evidenced by reduced clearance of the anticancer drug paclitaxel and arachidonic acid, respectively.29,30 In fact, CYP2C8*3 displays no detectable metabolizing ability in vitro.30
In a study by Parikh et al., 275 subjects with malaria from Burkina Faso were evaluated for amodiaquine metabolism.30 The subjects with the CYP2C8*2 genotype (more common African variant) experienced more abdominal pain than subjects with the wild-type (i.e. the normal, non-mutated gene) (52% versus 30%; P < 0.01).30 Other adverse effects did not differ.30 However, as in many pharmacogenetic studies, this study used a rather small sample to evaluate rare toxicities and did not test for liver function change as a marker for hepatotoxicity or pharmacokinetic changes to test for drug concentrations or toxic intermediates. Studies in Burkina Faso and Ghana have not revealed a relationship between drug efficacy and CYP2C8 genotype.30,31
While the percentage of the population with an inactivating CYP2C8 genetic variant is not high in most malaria-endemic regions, the overall incidence of risk is significant due to the burden of disease. For example, Cavaco et al. estimated that the CYP2C8*2 and CYP2C8*3 genetic variants occur in 2.1% of the population in Zanzibar, United Republic of Tanzania: this translates into more than 30 000 patients per year at risk for severe drug reactions to amodiaquine (of a malaria patient population of ~1 000 000).32
Ghana example
In Ghana, with a total population of more than 25 million, 5 270 108 cases of malaria were documented in 2007. The majority of these cases were likely treated with artesunate plus amodiaquine, the first-line treatment of choice on the Ghanaian Essential Medicines List.33 Based on a 1.5% population incidence of low-metabolizing CYP2C8, almost 80 000 (79 052) of these malaria cases would have occurred in low metabolizers (Fig. 1 and Fig. 2). Collecting data on adverse drug reactions and their cost is the best way to evaluate amodiaquine-related adverse effects. Without pharmacovigilance and cost data to model the medical costs associated with potential adverse drug reactions among these low metabolizers, giving this drug combination to 79 000 low metabolizers each year wastes an estimated 0.57 United States dollars per treatment course (average cost per treatment ranges from US$ 0.57–0.88), a total of approximately US$ 45 000 a year.34
The known literature on CYP2C8 allele (gene) frequencies reveals a lack of data in malaria-endemic Africa but, in those African countries with data, there is a higher frequency than in American Caucasians (Fig. 1, Fig. 2 and Table 2). With the Ghana example showing a loss of more than US$ 45 000, the national control programme or national medicines review authority could choose an alternative from the three first-line antimalarial combinations to treat malaria.
Artemisinins
The artemisinins as a group differ in the routes they take in the human body to convert to the major active metabolite, dihydroartemisinin (DHA). Artesunate undergoes rapid and extensive conversion via CYP2A6 and, to a lesser extent, CYP2B6 and CYP1A1 and CYP1A2.35 Almost 40 gene variants for CYP2A6 have been identified; at least 13 of these show decreased metabolizing function and five show no activity in vivo.36 The major antimalarial activity of artesunate is performed by DHA.37 People with poorly functioning CYP2A6 will have higher concentrations of artesunate and lower concentrations of DHA. This could reduce the drug’s antimalarial activity, kill fewer parasites and increase the potential for artesunate resistance.
Malaria endemicity is high in many areas of sub-Saharan Africa, such as Ghana. In contrast, in the Sabah region of Malaysia (which has the highest endemicity for malaria in this region) the burden of disease is considerably less than in Ghana. Almost two thirds of a Ghanaian population exhibited the wild-type genotype of CYP2A6 with more than 80% allele frequency of CYP2A6*1A.38 In contrast, a Malaysian population had an observed frequency for wild-type genotype of only 8% and an allele frequency for CYP2A6*1A of approximately 32%.39 With wild-type responsible for the majority of Ghanaian CYP2A6 activity, artesunate conversion to DHA and antimalarial activity are expected to be normal. In the Malaysian population with a low wild-type genotype frequency, CYP2A6 activity is decreased, the conversion to DHA decreased, and the antimalarial activity of artesunate may be compromised. Studies in other Asian populations also reveal a high frequency of non-wild-type alleles and an especially high frequency of the alleles that translate to no CYP2A6 activity (11.5–20.1% for Chinese, Japanese, Korean and Thai people)38
Resistance to artemisinin-based therapies is emerging in Thailand and approaches 10% in some areas.40 While not established, a potential contributor to artemisinin resistance in Thailand may be related to CYP2A6 activity (at least 14% frequency of the CYP2A6 alleles with no activity) and the inability to convert artesunate to DHA.41
What next?
Our understanding of the host factors contributing to the effectiveness of antimalarials is still in its infancy. As a community we need to determine ways to enhance our ability to study the pharmacokinetics and pharmacogenetics of antimalarials to help decrease the burden imposed by drug toxicity and to help protect their longevity. We can do this in three main ways: (i) integrating pharmacogenetic studies with the pharmacokinetic studies of antimalarials; (ii) increasing the generation of human genetic data and (iii) enhancing the infrastructure for pharmacogenetic research of antimalarial use in endemic countries.
Integrating research
Much of the data concerning the pharmacogenetics of antimalarials is derived from small and under-powered studies conducted in a retrospective manner. In particular, we are lacking significant data for some ACT partner drugs, such as sulfadoxine-pyrimethamine and lumefantrine. A resurgent interest in the pharmacology and pharmacokinetics of antimalarials in the malaria drug resistance community has spurred the development of another potential new resource for gathering pharmacogenetic data. As part of the Worldwide Antimalarial Resistance Network, a module has been developed to improve the quality of antimalarial pharmacology data generated, to use the new information to define drug resistance and to inform optimal dosing schedules. The goal is to facilitate global cooperation between groups active in the antimalarial pharmacology field (available at: http://www.wwarn.org). Pharmacology researchers, especially pharmacokinetic researchers, of antimalarials should obtain informed consent from study participants to allow use of their samples for collaborative pharmacogenetic analysis, either prospectively or in future retrospective analyses.
Generating data
Another potential resource for increasing our understanding of the pharmacogenetics of antimalarials are national control programmes, and the researchers and funding agencies associated with these programmes. During in vivo monitoring and efficacy studies carried out by these groups, samples obtained as dried blood spots are often collected and stored. The amount of DNA collected in these samples would not allow for assessment of a large number of genetic variations but it could allow for targeted testing of a small number that are vetted in strong pharmacogenetic studies. The key would be to encourage these groups to obtain consent from patients for retrospective human DNA testing. For example, considering the four first-line treatments, blood spots could provide a way to use pharmacogenetics to prioritize pharmacotherapy based on genetic differences within a defined population and to evaluate CYP2C8 genetics. Since amodiaquine requires CYP2C8 for metabolism to an active agent, national control programmes or the national medicines review authorities could use genotype frequencies in that population to determine the priority of a regimen containing amodiaquine.
Enhanced infrastructure
Ethnic variability is known for its association with antimalarial metabolism, but genetic data are missing for much of the world’s population. An increasing number of resources and efforts has been put into place to help with gathering information about the genetic make-up of different ethnic groups. In malaria-endemic areas, there are no pharmacogenetics organizations, such as the National Institute of Genomic Medicine (INMEGEN) in Mexico, to evaluate population genetic differences. However, the ideas explored in the white paper, Harnessing genomic technologies towards improving health in Africa, detail the systematic process for improving the research infrastructure in Africa.42
There is a strong need for genetic research in Africa. The overarching needs are for infrastructure, education and training, disease-related research and a comprehensive and continued focus on ethical, legal and social issues. To address these needs the Human Health and Heredity in Africa Initiative aims to integrate pharmacogenetics in the selection of national formularies in Africa.
In the meantime, to address the gap in knowledge and need for resources to evaluate genetic differences in developing countries, the Pharmacogenetics for Every Nation Initiative (PGENI) plans to sample 500 people from every ethnic group that represents 10% or more of the population in 104 developing countries.25,43 To avoid any conflicts of interest, PGENI declines funding from any drug manufacturers. PGENI aims to start the development of basic genetic knowledge in traditionally underserved environments. With the generation of medication prioritization algorithms for diseases of global importance, PGENI produces country-specific, genetically-enhanced treatment algorithms for diseases such as malaria. Using known genetic information and the WHO recommended treatment for uncomplicated malaria, a prioritization algorithm includes entry points where pharmacogenetics may augment decision-making, such as for CYP2C8 and CYP2A6 polymorphism frequencies when evaluating the use of amodiaquine and artesunate, respectively. When considering local parasite resistance and drug costs, the pharmacogenetically-enhanced, medical decision-making algorithm becomes a practical mechanism for weighing the options for formulary inclusion or exclusion by national medicines review authorities or national control programmes.
Countries such as the United Kingdom of Great Britain and Northern Ireland and the United States of America have made tremendous investments in research towards genomic medicine. However, in parts of the world where national health expenditures are as low as US$ 30 a day, the investment in genomic research has been extremely low.44 Importantly however, some malaria-endemic countries, such as India, South Africa and Thailand, have begun to invest in large-scale studies of human genomic variation.45,46 In addition, smaller projects, such as the DNA data bank in the Gambia and a biobank and pharmacogenetics database in Harare, Zimbabwe, are emerging and may be able to provide valuable data to inform national control programmes and medicines review authorities.47,48
Make it practical
While research is necessary to develop an objective strategy, individualized antimalarial therapy based on genetics will not be practical in most endemic countries for decades. However, that should not prevent the application of pharmacogenetic knowledge in public policy decisions. Just as regional sampling of a small number of malaria (or HIV or tuberculosis) isolates is used for sensitivity predictions for the infecting agents, we can use regional data from the population as part of the selection of therapy. The hope is that international cooperation through agencies such as PGENI, the Public Population Project in Genomics and the Genome-based Research and Population Health International Network, will help to build infrastructure in endemic countries and provide the training necessary to support a sustained skilled human resource. Enhancing the research infrastructure in developing countries should improve the capability for future genetic research and support continued efforts to address the gaps in knowledge of host genetics. By encouraging collaboration with the malaria control programmes and parasitologists, as well as with the national medicines review authorities, the stakeholders in genomic research will grow substantially and sustainably. Any use of human samples will require an informed consent that allows future use in research, including genetic research. As the pharmacogenetic markers with significant impact on drug response and/or toxicity are vetted, using the malaria control programme samples in malaria-endemic countries provides a good first step to expanding the potential to impact global drug use through improved efficacy and decreased drug toxicity. This should help to populate a map of pharmacogenetic data to guide national formulary decision-making by using existing samples, data and processes.
Conclusion
To accomplish the considerable goal of developing pharmacogenetically-informed drug policies, there is a need for strong collaboration between parasitologists studying parasite resistance, public health officials from control programmes and researchers identifying genetic causes of differences in drug toxicity and efficacy. The African Medicines Regulatory Harmonization Initiative will provide a regional regulatory process and may allow improvements not only at the national but also at the regional level.49 These collaborations, in conjunction with others investigating new drug entities, may help to affect the delivery of care to real patients in malaria-endemic regions. The work to create collaborations and to develop systems that allow for sampling large populations to establish a map of genetic differences will not be easy, but it is certainly worthwhile to include pharmacogenetic information when considering the data and defining the limited number of drugs that will be available to a population.
Acknowledgements
Jonathan J Juliano was supported by award number KL2RR025746 from the National Center for Research Resources, National Institutes of Health, USA. This work is part of the Pharmacogenetics for Every Nation Initiative. Mary W Roederer and Howard McLeod complete research with PGENI.
Competing interests:
None declared.
References
- 1.Guidelines for the treatment of malaria. Geneva: World Health Organization; 2006. Available from: http://whqlibdoc.who.int/publications/2006/9241546948_eng_full.pdf [accessed 23 August 2011].
- 2.Lin JT, Juliano JJ, Wongsrichanalai C. Drug-resistant malaria: the era of ACT. Curr Infect Dis Rep. 2010;12:165–73. doi: 10.1007/s11908-010-0099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World malaria report 2009 Geneva: World Health Organization; 2009. Available from: http://whqlibdoc.who.int/publications/2009/9789241563901_eng.pdf [accessed 23 August 2011].
- 4.Yeung S, Pongtavornpinyo W, Hastings IM, Mills AJ, White NJ. Antimalarial drug resistance, artemisinin-based combination therapy, and the contribution of modeling to elucidating policy choices. Am J Trop Med Hyg. 2004;71:179–86. [PubMed] [Google Scholar]
- 5.Carson PE, Flanagan CL, Ickes CE, Alving AS. Enzymatic deficiency in primaquine-sensitive erythrocytes. Science. 1956;124:484–5. doi: 10.1126/science.124.3220.484-a. [DOI] [PubMed] [Google Scholar]
- 6.Luzzatto L. The rise and fall of the antimalarial Lapdap: a lesson in pharmacogenetics. Lancet. 2010;376:739–41. doi: 10.1016/S0140-6736(10)60396-0. [DOI] [PubMed] [Google Scholar]
- 7.Update on GSK’s malaria treatments: Dacart and Lapdap. [media release]. 29 February 2008. Brentford: GlaxoSmithKline;2008. Available from: http://www.gsk.com/media/pressreleases/2008/2008_pressrelease_0014.htm. [accessed 23 August 2011].
- 8.Basco LK, Ringwald P. Analysis of the key pfcrt point mutation and in vitro and in vivo response to chloroquine in Yaounde, Cameroon. J Infect Dis. 2001;183:1828–31. doi: 10.1086/320726. [DOI] [PubMed] [Google Scholar]
- 9.Berens N, Schwoebel B, Jordan S, Vanisaveth V, Phetsouvanh R, Christophel EM, et al. Plasmodium falciparum: correlation of in vivo resistance to chloroquine and antifolates with genetic polymorphisms in isolates from the south of Lao PDR. Trop Med Int Health. 2003;8:775–82. doi: 10.1046/j.1365-3156.2003.01099.x. [DOI] [PubMed] [Google Scholar]
- 10.Happi TC, Thomas SM, Gbotosho GO, Falade CO, Akinboye DO, Gerena L, et al. Point mutations in the pfcrt and pfmdr-1 genes of Plasmodium falciparum and clinical response to chloroquine, among malaria patients from Nigeria. Ann Trop Med Parasitol. 2003;97:439–51. doi: 10.1179/000349803235002489. [DOI] [PubMed] [Google Scholar]
- 11.Maguire JD, Susanti AI, Kristin, Sismadi P, Fryauff DJ, Baird JK. The T76 mutation in the pfcrt gene of Plasmodium falciparum and clinical chloroquine resistance phenotypes in Papua, Indonesia. Ann Trop Med Parasitol. 2001;95:559–72. doi: 10.1080/00034980120092516. [DOI] [PubMed] [Google Scholar]
- 12.Nelson AL, Purfield A, McDaniel P, Uthaimongkol N, Buathong N, Sriwichai S, et al. pfmdr1 genotyping and in vivo mefloquine resistance on the Thai-Myanmar border. Am J Trop Med Hyg. 2005;72:586–92. [PubMed] [Google Scholar]
- 13.Ochong EO, van den Broek IV, Keus K, Nzila A. Short report: association between chloroquine and amodiaquine resistance and allelic variation in the Plasmodium falciparum multiple drug resistance 1 gene and the chloroquine resistance transporter gene in isolates from the upper Nile in southern Sudan. Am J Trop Med Hyg. 2003;69:184–7. [PubMed] [Google Scholar]
- 14.Omar SA, Adagu IS, Warhurst DC. Can pretreatment screening for dhps and dhfr point mutations in Plasmodium falciparum infections be used to predict sulfadoxine-pyrimethamine treatment failure? Trans R Soc Trop Med Hyg. 2001;95:315–9. doi: 10.1016/S0035-9203(01)90250-0. [DOI] [PubMed] [Google Scholar]
- 15.Pillai DR, Labbe AC, Vanisaveth V, Hongvangthong B, Pomphida S, Inkathone S, et al. Plasmodium falciparum malaria in Laos: chloroquine treatment outcome and predictive value of molecular markers. J Infect Dis. 2001;183:789–95. doi: 10.1086/318836. [DOI] [PubMed] [Google Scholar]
- 16.Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–47. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerb R, Fux R, Morike K, Kremsner PG, Gil JP, Gleiter CH, et al. Pharmacogenetics of antimalarial drugs: effect on metabolism and transport. Lancet Infect Dis. 2009;9:760–74. doi: 10.1016/S1473-3099(09)70320-2. [DOI] [PubMed] [Google Scholar]
- 18.Mehlotra RK, Henry-Halldin CN, Zimmerman PA. Application of pharmacogenomics to malaria: a holistic approach for successful chemotherapy. Pharmacogenomics. 2009;10:435–49. doi: 10.2217/14622416.10.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatton CS, Peto TE, Bunch C, Pasvol G, Russell SJ, Singer CR, et al. Frequency of severe neutropenia associated with amodiaquine prophylaxis against malaria. Lancet. 1986;327:411–4. doi: 10.1016/S0140-6736(86)92371-8. [DOI] [PubMed] [Google Scholar]
- 20.Phillips-Howard PA, West LJ. Serious adverse drug reactions to pyrimethamine-sulphadoxine, pyrimethamine-dapsone and to amodiaquine in Britain. J R Soc Med. 1990;83:82–5. doi: 10.1177/014107689008300208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raymond JM, Dumas F, Baldit C, Couzigou P, Beraud C, Amouretti M. Fatal acute hepatitis due to amodiaquine. J Clin Gastroenterol. 1989;11:602–3. doi: 10.1097/00004836-198910000-00034. [DOI] [PubMed] [Google Scholar]
- 22.Orrell C, Little F, Smith P, Folb P, Taylor W, Olliaro P, et al. Pharmacokinetics and tolerability of artesunate and amodiaquine alone and in combination in healthy volunteers. Eur J Clin Pharmacol. 2008;64:683–90. doi: 10.1007/s00228-007-0452-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerb R, Fux R, Morike K, Kremsner PG, Gil JP, Gleiter CH, et al. Pharmacogenetics of antimalarial drugs: effect on metabolism and transport. Lancet Infect Dis. 2009;9:760–74. doi: 10.1016/S1473-3099(09)70320-2. [DOI] [PubMed] [Google Scholar]
- 24.Li XQ, Bjorkman A, Andersson TB, Ridderstrom M, Masimirembwa CM. Amodiaquine clearance and its metabolism to N-desethylamodiaquine is mediated by CYP2C8: a new high affinity and turnover enzyme-specific probe substrate. J Pharmacol Exp Ther. 2002;300:399–407. doi: 10.1124/jpet.300.2.399. [DOI] [PubMed] [Google Scholar]
- 25.Roederer MW. Cytochrome P450 enzymes and genotype-guided drug therapy. Curr Opin Mol Ther. 2009;11:632–40. [PubMed] [Google Scholar]
- 26.Clarke JB, Maggs JL, Kitteringham NR, Park BK. Immunogenicity of amodiaquine in the rat. Int Arch Allergy Appl Immunol. 1990;91:335–42. doi: 10.1159/000235138. [DOI] [PubMed] [Google Scholar]
- 27.Harrison AC, Kitteringham NR, Clarke JB, Park BK. The mechanism of bioactivation and antigen formation of amodiaquine in the rat. Biochem Pharmacol. 1992;43:1421–30. doi: 10.1016/0006-2952(92)90198-R. [DOI] [PubMed] [Google Scholar]
- 28.Jewell H, Maggs JL, Harrison AC, O'Neill PM, Ruscoe JE, Park BK. Role of hepatic metabolism in the bioactivation and detoxication of amodiaquine. Xenobiotica. 1995;25:199–217. doi: 10.3109/00498259509061845. [DOI] [PubMed] [Google Scholar]
- 29.Dai D, Zeldin DC, Blaisdell JA, Chanas B, Coulter SJ, Ghanayem BI, et al. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenetics. 2001;11:597–607. doi: 10.1097/00008571-200110000-00006. [DOI] [PubMed] [Google Scholar]
- 30.Parikh S, Ouedraogo JB, Goldstein JA, Rosenthal PJ, Kroetz DL. Amodiaquine metabolism is impaired by common polymorphisms in CYP2C8: implications for malaria treatment in Africa. Clin Pharmacol Ther. 2007;82:197–203. doi: 10.1038/sj.clpt.6100122. [DOI] [PubMed] [Google Scholar]
- 31.Adjei GO, Kristensen K, Goka BQ, Hoegberg LC, Alifrangis M, Rodrigues OP, et al. Effect of concomitant artesunate administration and cytochrome P4502C8 polymorphisms on the pharmacokinetics of amodiaquine in Ghanaian children with uncomplicated malaria. Antimicrob Agents Chemother. 2008;52:4400–6. doi: 10.1128/AAC.00673-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cavaco I, Strömberg-Nörklit J, Kaneko A, Msellem MI, Dahoma M, Ribeiro VL, et al. CYP2C8 polymorphism frequencies among malaria patients in Zanzibar. Eur J Clin Pharmacol. 2005;61:15–8. doi: 10.1007/s00228-004-0871-8. [DOI] [PubMed] [Google Scholar]
- 33.The health sector in Ghana: facts and figures, 2008. Accra: Ghana Health Service; 2008. Available from: http://www.ghanahealthservice.org/includes/upload/publications/Facts%20and%20Figures%202008.pdf [accessed 23 August 2011].
- 34.Drug prices for artesunate/amodiaquine. International Drug Price Indicator Guide. Cambridge: Management Sciences for Health; 2009. Available from: http://erc.msh.org [accessed 23 August 2011].
- 35.Li XQ, Björkman A, Andersson TB, Gustafsson LL, Masimirembwa CM. Identification of human cytochrome P(450)s that metabolise anti-parasitic drugs and predictions of in vivo drug hepatic clearance from in vitro data. Eur J Clin Pharmacol. 2003;59:429–42. doi: 10.1007/s00228-003-0636-9. [DOI] [PubMed] [Google Scholar]
- 36.Di YM, Chow VD, Yang LP, Zhou SF. Structure, function, regulation and polymorphism of human cytochrome P450 2A6. Curr Drug Metab. 2009;10:754–80. doi: 10.2174/138920009789895507. [DOI] [PubMed] [Google Scholar]
- 37.Ilett KF, Ethell BT, Maggs JL, Davis TM, Batty KT, Burchell B, et al. Glucuronidation of dihydroartemisinin in vivo and by human liver microsomes and expressed UDP-glucuronosyltransferases. Drug Metab Dispos. 2002;30:1005–12. doi: 10.1124/dmd.30.9.1005. [DOI] [PubMed] [Google Scholar]
- 38.Gyamfi MA, Fujieda M, Kiyotani K, Yamazaki H, Kamataki T. High prevalence of cytochrome P450 2A6*1A alleles in a black African population of Ghana. Eur J Clin Pharmacol. 2005;60:855–7. doi: 10.1007/s00228-004-0854-9. [DOI] [PubMed] [Google Scholar]
- 39.Yusof W, Gan SH. High prevalence of CYP2A6*4 and CYP2A6*9 alleles detected among a Malaysian population. Clin Chim Acta. 2009;403:105–9. doi: 10.1016/j.cca.2009.01.032. [DOI] [PubMed] [Google Scholar]
- 40.White NJ. Qinghaosu (artemisinin): the price of success. Science. 2008;320:330–4. doi: 10.1126/science.1155165. [DOI] [PubMed] [Google Scholar]
- 41.Noedl H, Socheat D, Satimai W. Artemisinin-resistant malaria in Asia. N Engl J Med. 2009;361:540–1. doi: 10.1056/NEJMc0900231. [DOI] [PubMed] [Google Scholar]
- 42.Harnessing genomic technologies towards improving health in Africa. Bethesda: Human Heredity and Health in Africa Initiative; 2011. Available from: http://www.h3africa.org/whitepaper.cfm [accessed 23 August 2011].
- 43.Marsh S, Van Booven DJ, McLeod HL. Global pharmacogenetics: giving the genome to the masses. Pharmacogenomics. 2006;7:625–31. doi: 10.2217/14622416.7.4.625. [DOI] [PubMed] [Google Scholar]
- 44.Hardy BJ, Seguin B, Goodsaid F, Jimenez-Sanchez G, Singer PA, Daar AS. The next steps for genomic medicine: challenges and opportunities for the developing world. Nat Rev Genet. 2008;9(Suppl 1):S23–7. doi: 10.1038/nrg2444. [DOI] [PubMed] [Google Scholar]
- 45.Hardy BJ, Seguin B, Singer PA, Mukerji M, Brahmachari SK, Daar AS. From diversity to delivery: the case of the Indian Genome Variation initiative. Nat Rev Genet. 2008;9(Suppl 1):S9–14. doi: 10.1038/nrg2440. [DOI] [PubMed] [Google Scholar]
- 46.Hardy BJ, Seguin B, Ramesar R, Singer PA, Daar AS. South Africa: from species cradle to genomic applications. Nat Rev Genet. 2008;9(Suppl 1):S19–23. doi: 10.1038/nrg2441. [DOI] [PubMed] [Google Scholar]
- 47.Sirugo G, Schim van der Loeff M, Sam O, Nyan O, Pinder M, Hill AV, et al. A national DNA bank in The Gambia, West Africa, and genomic research in developing countries. Nat Genet. 2004;36:785–6. doi: 10.1038/ng0804-785. [DOI] [PubMed] [Google Scholar]
- 48.Matimba A, Oluka MN, Ebeshi BU, Sayi J, Bolaji OO, Guantai AN, et al. Establishment of a biobank and pharmacogenetics database of African populations. Eur J Hum Genet. 2008;16:780–3. doi: 10.1038/ejhg.2008.49. [DOI] [PubMed] [Google Scholar]
- 49.Ndomondo-Sigonda M, Ambali A. The african medicines regulatory harmonization initiative: rationale and benefits. Clin Pharmacol Ther. 2011;89:176–8. doi: 10.1038/clpt.2010.299. [DOI] [PubMed] [Google Scholar]