Abstract
During polyadenylation, the multi-functional protein nucleophosmin (NPM1) is deposited onto all cellular mRNAs analysed to date. Premature termination of poly(A) tail synthesis in the presence of cordycepin abrogates deposition of the protein onto the mRNA, indicating natural termination of poly(A) addition is required for NPM1 binding. NPM1 appears to be a bona fide member of the complex involved in 3′ end processing as it is associated with the AAUAAA-binding CPSF factor and can be co-immunoprecipitated with other polyadenylation factors. Furthermore, reduction in the levels of NPM1 results in hyperadenylation of mRNAs, consistent with alterations in poly(A) tail chain termination. Finally, knockdown of NPM1 results in retention of poly(A)+ RNAs in the cell nucleus, indicating that NPM1 influences mRNA export. Collectively, these data suggest that NPM1 has an important role in poly(A) tail length determination and may help network 3′ end processing with other aspects of nuclear mRNA maturation.
Keywords: mRNA export, mRNA processing, nucleophosmin, polyadenylation
Introduction
Polyadenylation is vital to mRNA 3′ end formation in eukaryotes and is now clearly considered to be more than a default process along the pathway of mRNA maturation. Over 50% of genes contain alternative poly(A) signals (Tian et al, 2005), the choice of which is dynamically regulated by the cell based on its growth, developmental stage and cancerous phenotype (Sandberg et al, 2008; Ji et al, 2009; Mayr and Bartel, 2009). Furthermore, the polyadenylation process is closely networked with many aspects of gene expression. Therefore, a more detailed description of the polyadenylation process and mechanistic aspects of its connections to other aspects of RNA biogenesis will be important for a complete understanding of cellular gene expression.
Polyadenylation and transcription are co-regulated at a variety of levels. Polyadenylation can influence transcription termination (West and Proudfoot, 2009) as well as the efficiency of transcription re-initiation at upstream promoters (Mapendano et al, 2010). In addition to polyadenylation factors interacting with the C-terminal domain of RNA Pol II (Lunde et al, 2010), polyadenylation efficiency can be influenced by positioning of 3′ end processing signals relative to the promoter (Guo et al, 2011) and factors involved in transcription initiation can influence 3′ end processing and termination (Wang et al, 2010a).
Polyadenylation is also networked with a variety of nuclear mRNA processing events. The presence of a 5′ cap influences 3′ end processing efficiency (Flaherty et al, 1997). Polyadenylation influences splicing efficiency, particularly of the last exon (Cooke and Alwine, 1996) and the U1 spliceosomal snRNP has a role in regulating poly(A) site usage (Wilusz and Beemon, 2006; Hall-Pogar et al, 2007; Abad et al, 2008; Kaida et al, 2010). Polyadenylation factors have been shown to function in mRNA release from the nucleus (Ruepp et al, 2009), perhaps by recruiting export factors (Johnson et al, 2009) and export factors in turn have been demonstrated to influence polyadenylation (Qu et al, 2009). In yeast, coordination between 3′ end formation and export appears to occur at least in part through the THO/Sub2p protein complex (Rougemaille et al, 2008). Thus, the polyadenylation event may have a key coordinating or quality control role in nuclear mRNA production and maturation.
Given the extensive networking of polyadenylation with other nuclear processes, it is perhaps not surprising that the machinery associated with 3′ end processing in mammals may comprise as many as 85 proteins (Shi et al, 2009). Among these are several protein complexes and individual factors involved in promoting the enzymatic events of pre-mRNA cleavage and polyadenylation of pre-mRNAs (Millevoi and Vagner, 2010). CPSF recognizes the upstream core AAUAAA element and in conjunction with CstF (which recognizes the U/GU-rich downstream core element) serves as a platform for the assembly of enzymatic components onto pre-mRNA substrates. CFI and CFII are also required for the generation of the 3′ end of the transcript which is subsequently polyadenylated by poly(A) polymerase (PAP). Polyadenylation itself occurs in two stages, an initial distributive addition of ∼25 adenosines followed by the processive incorporation of more adenosines to form the mature tail (Bienroth et al, 1993). The size of the poly(A) tail in mammalian cells is directly influenced by nuclear poly(A) binding protein PABPN1. PABPN1 allows the AAUAAA-associated CPSF factor to stimulate PAP activity during the elongation phase and termination is thought to occur when the tail is too long for this interaction to effectively be maintained (Kühn et al, 2009). While the three factors PABPN1, CPSF and PAP will generate a tail of defined length in reconstituted reactions (Kühn et al, 2009), it is not clear whether additional factors may influence the termination event in living cells. Therefore, we set out to explore the potential function of nucleophosmin (NPM1), a protein we have previously reported to mark the 3′ UTR of mRNAs that have undergone successful polyadenylation (Palaniswamy et al, 2006).
NPM1, which is also referred to as numatrin, B23 and NO38, is a conserved multi-functional nuclear/nucleolar chaperone protein that has been implicated in both growth promotion and tumour suppression (Lindström, 2011). NPM1 is overexpressed in most tumours (Pianta et al, 2010), is a common target for oncogenic translocations (Falini et al, 2007) and approximately one third of adult acute myeloid leukaemias display aberrant cytoplasmic localization of NPM1 due to frameshift mutations in exon 12 (Meani and Alcalay, 2009). NPM1 knockout mice die early in embryogenesis due to multiple defects in organogenesis (Grisendi et al, 2005). The protein contains oligomerization, chaperone, nucleic acid binding and a variety of subcellular localization domains that are involved in nucleo-cytoplasmic shuttling (Okuwaki, 2008). The protein can be extensively modified post-translationally by phosphorylation, acetylation, ubiquitinylation and SUMOylation (Lindström, 2011) which likely regulates its various functions. NPM1 has been implicated in numerous cellular processes, including ribosome biogenesis (Maggi et al, 2008), chromatin remodelling (Hisaoka et al, 2010), DNA repair (Lin et al, 2010), ARF/p53 regulation (Li and Hann, 2009) and SUMO regulation (Yun et al, 2008).
We have previously identified NPM1 as a factor that is specifically deposited on the 3′ UTR of viral mRNAs following successful 3′ end cleavage and polyadenylation (Palaniswamy et al, 2006) and propose that NPM1 may function as a mark to identify mRNAs that have undergone polyadenylation. Given the extensive networking of polyadenylation with other nuclear RNA synthesis and processing events, we wished to pursue the role of NPM1 deposition on polyadenylated nuclear mRNA. We specifically focused on the influence of the type of poly(A) signal on NPM1 deposition, the effect of NPM1 on the polyadenylation process itself and whether NPM1 has a role in the coordination of other events in mRNA biogenesis with polyadenylation.
In this study, polyadenylation in HeLa cell nuclear extracts and crosslinking/co-immunoprecipitation analyses in living cells suggest that NPM1 is associated with many if not all poly(A)+ mRNAs. NPM1 associates with the CPSF polyadenylation factor in cells. Interestingly, the deposition of NPM1 on the 3′ UTR of polyadenylated mRNAs requires proper termination of polyadenylation. Reduction in the level of NPM1 in cells or nuclear extracts results in hyperadenylation. These data suggest that NPM1 may have a role in influencing the termination of poly(A) tail synthesis. Finally, in addition to causing mRNA hyperadenylation, NPM1 knockdown in living cells also results in nuclear accumulation of poly(A)+ mRNA, suggesting that NPM1 deposition may influence mRNA export. In summary, these data identify a novel function for the NPM1 oncoprotein and provide further insight into factors involved in the networking of polyadenylation with other aspects of gene expression.
Results
Polyadenylation mediated by a variety of cellular polyadenylation signals causes deposition of NPM1 on transcripts
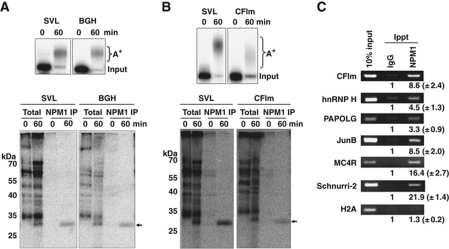
Our previous studies used three independent viral polyadenylation signals to demonstrate the deposition of NPM1 on the 3′ UTR of RNA substrates undergoing 3′ end processing in HeLa nuclear extracts (Palaniswamy et al, 2006). NPM1 deposition required the presence of at least 10 bases upstream of the AAUAAA element. Moreover, the mere presence of a long poly(A) tail alone was not sufficient to induce NPM1 deposition as RNA substrates with a preformed ∼200 nt poly(A) tail failed to crosslink to the protein. While crosslinking studies in living cells showed that NPM1 can also be found on poly(A)+ RNAs in the nucleus (Palaniswamy et al, 2006), it remained important to directly demonstrate that cellular poly(A) signals also mediate the polyadenylation-dependent deposition of NPM1. We first assessed the proteins deposited as a result of in vitro 3′ end processing on RNA substrates bearing the well-characterized bovine growth hormone (BGH) poly(A) signal (Goodwin and Rottman, 1992). As seen in Figure 1A (top panel), pre-cleaved RNAs containing the BGH poly(A) signal were polyadenylated with an efficiency similar to the SV40 late (SVL) viral poly(A) signal. Furthermore, NPM1 specifically associated with the body of both RNA substrates (labelled at internal U residues) following polyadenylation (Figure 1A, lower panel). Similar data were obtained for the cellular CFIm (CPSF6) poly(A) signal (Figure 1B). Thus, we conclude that cellular poly(A) signals, like previously tested viral poly(A) signals, mediate efficient association of NPM1 with the body of the RNA in conjunction with the 3′ end polyadenylation event in nuclear extracts.
Figure 1.
Nucleophosmin is deposited on cellular poly(A)+ mRNAs in both HeLa cells and an in vitro polyadenylation system. (A, B) Radiolabelled RNAs containing polyadenylation signals from the SV40 late (SVL), bovine growth hormone (BGH) or the 68-kDa component of cleavage factor Im (CFIm) were incubated with HeLa nuclear extract in an in vitro polyadenylation assay for the times indicated. RNA products were analysed on a 5% acrylamide gel containing urea (top panel). A+ indicates the migration of the polyadenylated product RNAs, while ‘input’ indicates the size of the RNA substrate. In the lower panel, in parallel experiments in the cell-free polyadenylation system radioactive RNAs were crosslinked by UV light to closely associated proteins, treated with RNase and total crosslinked proteins were analysed by SDS–PAGE (total lanes) or immunoprecipitated using an anti-NPM1 antisera before electrophoresis (NPM1 IP lanes). The arrow on the right indicates the migration of the NPM1 crosslinked product while molecular weight markers are indicated on the left. (C) HeLa cells were treated with formaldehyde to stabilize mRNP complexes and lysed. Lysates were immunoprecipitated using equal amounts of either IgG control sera or NPM1-specific antibodies. Immunoprecipitated RNA was extracted and analysed by RT–PCR using the primers listed on the left of the panels and products were resolved on 2% agarose gels. The numbers below each band in the immunoprecipitated lanes represent the relative fold enrichment of PCR product obtained compared with the control IgG lanes. Results shown are the mean of three experiments with the standard deviations indicated.
We next assessed whether individual cellular mRNAs could be found associated with NPM1 in living cells. Cells were treated with formaldehyde to stabilize protein–RNA complexes. Extracts were subjected to immunoprecipitation using NPM1-specific or control antisera and co-precipitated mRNAs were assessed using RT–PCR analysis. As seen in Figure 1C, CF1m mRNA, along with five other polyadenylated mRNAs, was specifically co-precipitated using NPM1 antiserum. This subset of mRNAs was chosen to represent mRNAs with conventional AAUAAA-driven polyadenylation signals (CFIm and hnRNP H), signals containing a core upstream UGUAN motif (PAPOLG; Venkataraman et al, 2005), an upstream A-rich core motif (JunB and MC4R; Nunes et al, 2010) or a putative ‘poly(A) limiting element’ that restricts the overall size of the tail (Schnurri-2; Gu et al, 1999) in Jurkat cells. In HeLa cells, however, the poly(A) tail of Schnurri-2 mRNAs was longer (up to ∼150 adenosines) (data not shown). Thus, the regulation of poly(A) tail size of the Schnurri-2 mRNA appears to be cell and/or tissue specific. As a control, histone H2A mRNAs, which generally lack a poly(A) tail, failed to significantly interact with NPM1. Collectively, these data suggest that NPM1 is deposited on poly(A)+ cellular mRNAs independent of the type of upstream polyadenylation signal they possess. While the significance of the differences in the overall efficiency of NPM1 co-precipitation among individual polyadenylated mRNAs remains to be elucidated, the mode of recognition used to initiate 3′ end processing does not appear to have a role in deposition of NPM1.
NPM1 is deposited on mRNAs as a result of the natural process of termination of polyadenylation
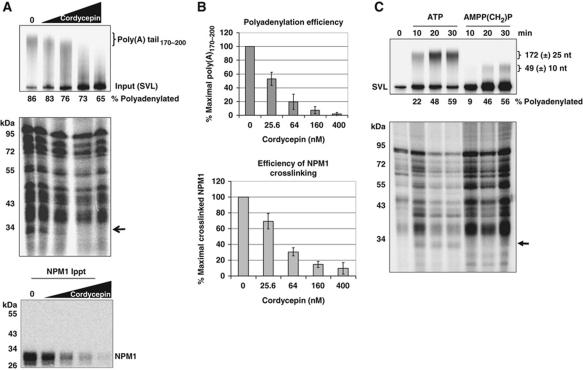
Our previous results clearly indicated that NPM1 deposition on mRNAs was associated with the process of poly(A) tail addition rather than 3′ end cleavage of pre-mRNA substrates (Palaniswamy et al, 2006). Furthermore, RNAs containing preformed poly(A) tails of ∼200 bases failed to associate with NPM1, suggesting that NPM1 deposition was intimately linked with the polyadenylation event itself and was not a simple consequence of the presence of a poly(A) tail on an RNA. In order to begin to gain more mechanistic insight into NPM1 deposition onto mRNAs, we addressed which step of the polyadenylation process (initiation, elongation or termination of poly(A) synthesis) was associated with NPM1 deposition. We approached this experimentally via the addition of increasing amounts of cordycepin triphosphate into polyadenylation reactions mediated by HeLa nuclear extracts. Cordycepin will act as a chain terminator when incorporated into the growing adenylate tract by PAP, allowing us to determine the effect of artificially terminating poly(A) synthesis on NPM1 deposition. As seen in Figure 2A, when cordycepin is added in sufficient concentrations to terminate polyadenylation prematurely, NPM1 was not deposited on the pre-mRNA substrate. This suggests that NPM1 deposition does not occur during the initiation or elongation steps of poly(A) tail synthesis. However, when cordycepin is titered into reactions to cause a block in poly(A) tail elongation and a concomitant decrease in the amount of natural termination of poly(A) tail synthesis, a direct association between NPM1 deposition and natural poly(A) tract termination can be observed (Figure 2B). Thus, we conclude that deposition of NPM1 on RNAs undergoing polyadenylation is closely associated with the termination step of poly(A) tail synthesis. Two additional observations support this conclusion. First, RNAs with preformed poly(A) tails of 60 to ∼200 bases in length fail to interact with NPM1 (Palaniswamy et al, 2006). Thus, the length of the poly(A) tail per se does not appear to be important for NPM1 interactions. Second, if an ATP analogue with a non-cleavable β-γ bond (AMPP(CH2)P) is used instead of ATP in in vitro polyadenylation assays, the poly(A) tails generated in the reaction are much shorter (49±10 bases), consistent perhaps with failure of the poly(A) tail addition to effectively switch to its processive mode (Wahle, 1995; Figure 2C, top panel). As seen in Figure 2C (lower panel), NPM1 deposition also fails to occur under these conditions of AMPP(CH2)P-induced inappropriate termination of poly(A) synthesis. Quantification of the total amount of products containing poly(A) tails of any length indicated that changes in overall polyadenylation efficiency when using ATP analogues cannot account for the decrease in NPM1 binding that is observed (Figure 2A and C). Therefore, collectively these data suggest that NPM1 deposition is associated with the natural termination step of poly(A) tail synthesis.
Figure 2.
Deposition of NPM1 on RNAs is associated with natural termination of poly(A) synthesis. (A) RNAs containing the SVL polyadenylation signal were incubated in cell-free polyadenylation reactions using HeLa nuclear extract in the presence of the indicated amount of cordycepin. RNA products of the reaction were analysed on a 5% denaturing acrylamide gel (top); total proteins that were UV crosslinked to the radioactive body of the RNA were analysed by 10% SDS–PAGE (middle gel); and crosslinked proteins immunoprecipitated with NPM1-specific antisera were analysed by 10% SDS–PAGE (bottom gel). The arrow in the middle gel indicates the position of NPM1. The percentage of RNA that received a poly(A) tail of any length is indicated under the top gel. (B) Quantification of the results shown in (A). Error bars represent the standard deviation of three experiments. (C) RNAs containing the SVL polyadenylation signal were incubated in cell-free polyadenylation reactions using HeLa nuclear extract in the presence of either ATP or AMPP(CH2)P. RNA products of the reaction were analysed on a 5% denaturing acrylamide gel (top gel); total proteins that were UV crosslinked to the radioactive body of the RNA were analysed by 10% SDS–PAGE (bottom gel). The arrow indicates the position of crosslinked NPM1. Poly(A) tail sizes are expressed as the mean size along with the size range (±) of 80% of the poly(A) synthesized in the reaction. The percentage of RNA that received a poly(A) tail of any length is indicated under the top gel.
NPM1 interacts with factors directly implicated in the determination of poly(A) tail length
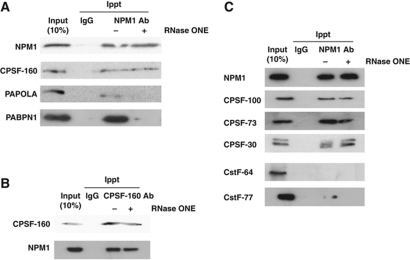
Previous studies indicated that three factors have a major role in determining the length of poly(A) synthesized on a given mRNA: CPSF (which interacts with the AAUAAA core upstream element of the poly(A) signal), PABPN1 (which interacts with CPSF and the growing adenylate tail) and PAP (which is the enzyme responsible for adding adenylate residues) (Wahle, 1995; Kühn et al, 2009; Millevoi and Vagner, 2010). Poly(A) tail length can also be affected post-synthesis through remodelling by deadenylases (Brown et al, 1996). In order to gain further insight into the mechanism of NPM1 deposition on polyadenylated mRNAs, we determined whether NPM1 associates with factors known to have an influence on poly(A) tail length. Co-immunoprecipitations were performed using antisera specific for NPM1 and the blots were probed for a variety of polyadenylation factors. As seen in Figure 3A, NPM1 specifically co-precipitated with CPSF-160. The interaction between NPM1 and the CPSF complex was direct as it was insensitive to degradation of RNA by RNase ONE. Immunoprecipitations using antibodies against CPSF-160 and probing for co-precipitated NPM1 confirmed the interaction (Figure 3B). Interestingly, the major nuclear PAP and PABPN1 were also co-immunoprecipitated with NPM1 antibody, but in an RNase-sensitive manner (Figure 3A). As a control, only trace amounts of the nuclear deadenylase PARN was immunoprecipitated with NPM1, but this weak interaction was indirect and likely due to RNA bridging as it was sensitive to ribonuclease (data not shown). As seen in Figure 3C, the CPSF-100, CPSF-73 and CPSF-30 components of the CPSF complex also co-immunoprecipitated with NPM1 in an RNase-resistant manner. The CstF-64 component of the CstF complex that interacts with the downstream element of the polyadenylation signal, failed to detectably interact with NPM1 (Figure 3B). The CstF-77 component of this complex interacted only weakly with NPM1 and in an RNase-sensitive manner (Figure 3B). Thus, we conclude that NPM1 can interact with the CPSF factor that binds the AAUAAA element in the 3′ UTR of the pre-mRNA. In addition, NPM1 interacts indirectly through RNA bridging with the enzyme that adds adenylate residues to the 3′ end of the pre-mRNA (PAP) as well as a key factor in poly(A) length determination (PABPN1). Thus, NPM1 may be part of the complex present on pre-mRNAs that are being actively polyadenylated. Collectively, these data suggest that NPM1 may directly influence factors that have a role in the natural process of termination of poly(A) tail addition.
Figure 3.
NPM1 is directly associated with the core polyadenylation factor CPSF. (A) HeLa cell lysates were immunoprecipitated with either control mouse IgG or NPM1 antibodies before (− lanes) or after (+ lanes) treatment with RNase ONE. Precipitated proteins were separated on a 10% SDS-acrylamide gel and analysed by western blotting using the antisera indicated on the left. Input lanes represent 10% of the total amount of protein used for the immunoprecipitation reactions. (B) Same as in (A), but antibodies against CPSF-160 were used for the immunoprecipitation. (C) Co-immunoprecipitation analysis was performed as described in (A) and blots were probed with the antibodies indicated on the left.
Knockdown of NPM1 in cells results in hyperadenylation of mRNA
If NPM1 does have a role in the termination of poly(A) synthesis, then one would predict that poly(A) tail length might be affected in the absence of the protein. In order to address this, we assessed poly(A) tail length of mRNAs isolated from HeLa cells depleted of NPM1 using shRNAs. We established two NPM1 knockdown HeLa cell lines using the Mission® pLKO.1 shRNA expression system. As seen in Supplementary Figure S1A and B, NPM1 protein and RNA levels were knocked down >90% in these cells. Similar levels of NPM1 depletion could be achieved with other shRNAs that target independent regions of the mRNA (data not shown). An analysis of cell doubling rates indicated that while viable, HeLa cells that were knocked down for NPM1 did grow significantly slower than their wild-type counterparts (Supplementary Figure S1C). These data confirm previously published work indicating that NPM1 has a significant role in cellular growth and metabolism (Grisendi et al, 2005; Brady et al, 2009).
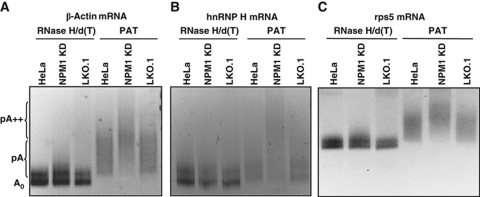
We performed linker ligation-mediated poly(A) tail (LLM-PAT) length assays (Garneau et al, 2008) for a series of individual mRNAs. As seen in Figure 4, the length of the poly(A) tail on the β-actin, hnRNP H and rps5 mRNAs was clearly increased in NPM1 knockdown cells compared with controls. The effect was specific for mRNAs as no evidence was obtained for an increase in polyadenylated subforms of rRNAs (data not shown). Therefore, we conclude that knockdown of NPM1 in HeLa cells results in hyperadenylation of mRNAs.
Figure 4.
Knockdown of NPM1 results in an increase in mRNA poly(A) tail length. Total RNA was isolated from untreated HeLa cells (HeLa lanes), HeLa cells containing a vector only control (LKO.1 lanes) or HeLa cells knocked down for NPM1 (NPM1 KD lanes) and analysed by linker ligation-mediated PCR-based poly(A) tail length assay. A set of samples was treated with oligo dT and RNAse H to remove the poly(A) tail before analysis (RNase H/d(T) lanes). The PAT lanes represent samples where mRNAs with intact poly(A) tails were analysed. Primers for β-actin mRNA were used in (A), hnRNP H mRNA in (B) and rps5 mRNA in (C). PCR products were analysed on a 5% acrylamide gel. The positions of the unadenylated RNA (A0), normally polyadenylated (pA) and hyperadenylated (pA++) are indicated on the left.
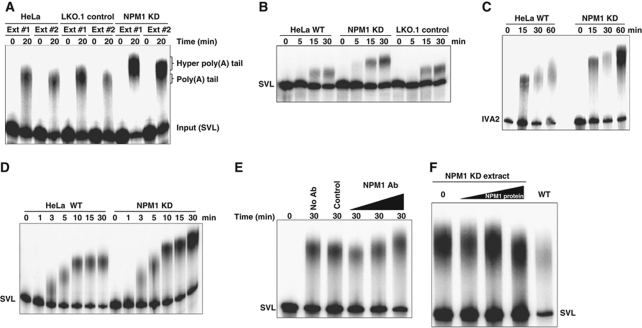
We hypothesized that the hyperadenylation of mRNAs observed in NPM1 knockdown cells is due to a direct disruption in regulation of poly(A) tail length during synthesis or remodelling. Alternatively, hyperadenylation could be an indirect effect of a block in nuclear export or as a consequence of aberrant mRNA quality control. In order to begin to differentiate between these mechanistic possibilities, we prepared nuclear extracts from wild-type, control and NPM1 knockdown HeLa cells. Pre-cleaved RNAs containing the SVL poly(A) signal (which get a poly(A) tail added directly to their 3′ end without the need for a prior cleavage reaction) were incubated in cell-free polyadenylation reactions using nuclear extracts and poly(A) tail length was assessed on acrylamide gels. As seen in Figure 5A, while extracts from untransfected HeLa cells and control cells stably transfected with an empty LKO.1 vector gave polyadenylated products of similar size, nuclear extracts from stably transfected NPM1 knockdown lines gave hyperadenylated tails ∼80 bases longer than those seen in control extracts. Importantly, independent extracts gave similar, highly reproducible results (Figure 5A). Similar hyperadenylation in NPM1 knockdown extracts was observed on SVL RNA substrates that were both cleaved and polyadenylated in the reaction (Figure 5B). A similar hyperadenylation of RNA substrates in NPM1 knockdown extracts was observed for other poly(A) signals, including adenoviral signals IVA2 (Figure 5C) and E1B (data not shown). Overall RNA recovery in the assortment of extracts used in these studies varied by <15%, suggesting that NPM1 levels did not dramatically influence the degradation rates of RNAs in these experiments. In addition, variation in overall polyadenylation efficiency between extracts was relatively minor (<14%), and thus was not sufficient to account for the dramatic differences in the size of poly(A) tails that were observed between control and NPM1-depleted extracts.
Figure 5.
Hyperadenylation can be demonstrated in nuclear extract-based polyadenylation assays when NPM1 is depleted. (A) RNAs containing the pre-cleaved SVL polyadenylation signal were incubated for the indicated times in extracts from either untreated HeLa, LKO.1 transfected control HeLa cells or HeLa cells in which NPM1 was knocked down using a specific shRNA. Ext 1 and 2 denote independent extracts made from each of the indicated cell types. Polyadenylated products were analysed on a 5% denaturing acrylamide gel. The positions of the input, normally polyadenylated and hyperadenylated RNAs are indicated on the right. (B) Same as (A) except that the entire SVL polyadenylation signal was used in the RNA substrates so that transcripts were both cleaved and polyadenylated. (C) Same as the previous panels except RNAs containing the IVA2 polyadenylation signal were used. (D) Time course of polyadenylation using nuclear extracts derived from normal or NPM1 knockdown HeLa cells and the SVL RNA substrate. (E) HeLa nuclear extracts were used untreated (no Ab lane), incubated with protein A Sepharose beads with control IgG before use (control lane), or with increasing amounts of NPM1-specific antisera before removal of antigen–antibody complexes with protein A beads (NPM1 Ab lanes). The SVL RNA substrate was incubated in these treated extracts for the time indicated and polyadenylated products were analysed on a 5% acrylamide gel. (F) Increasing amounts of partially purified NPM1 protein were added back to NPM1-depleted extracts and polyadenylation reactions were performed and assayed as described above.
The hyperadenylation observed in extracts made from NPM1-depleted cells was not simply due to differences in the kinetics of poly(A) tail addition. As seen in Figure 5D, once a maximal poly(A) tail length was obtained in extracts from control HeLa cells at around 10 min, no notable additional length increase occurred in the poly(A) tail. In extracts from NPM1-depleted cells, on the other hand, maximal tail length was clearly increased compared with control cells. These data suggest that NPM1 is likely also influencing the activity CPSF-160 and/or PABPN1 that regulates PAP processivity throughout the cycles of poly(A) addition rather than simply influencing the final termination event. Next, immunodepletion studies performed by adding increasing amounts of NPM1-specific antibodies on a fixed amount of Sepharose beads to HeLa nuclear extracts and testing the supernatant also resulted in a dose-dependent increase in poly(A) tail length (Figure 5E). Control Sepharose beads with non-specific IgG had no influence on polyadenylation. Finally, the addition of partially purified NPM1 protein to NPM1-depleted extracts was able to restore poly(A) tail length to the size found in control extracts (Figure 5F). Therefore, we conclude that the mRNA hyperadenylation observed in NPM1 knockdown cells is likely a direct effect of the reduction in NPM1 levels on the efficiency of poly(A) tail termination.
Knockdown of NPM1 in cells results in increased mRNA accumulation in the nucleus
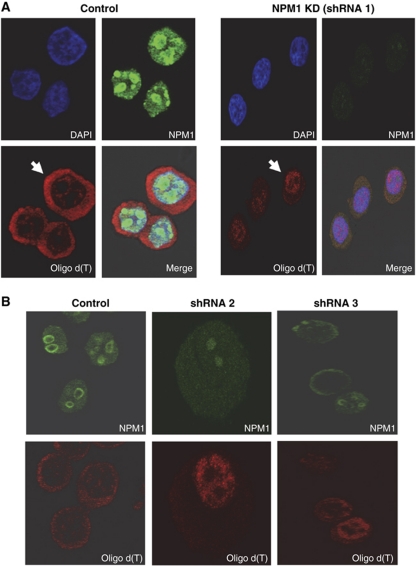
In the yeast Saccharomyces cerevisiae, hyperadenylation is associated with a block in mRNA export from the nucleus (Jensen et al, 2001). In order to assess the overall localization of the poly(A)+ RNA population in NPM1 knockdown HeLa cells, we performed fluorescence in situ hybridization (FISH) analyses using labelled oligo d(T) probes. As seen in Figure 6A, while poly(A)+ mRNA was largely cytoplasmic in control transfected HeLa cells, NPM1 knockdown caused a dramatic accumulation of poly(A)+ RNA in the nucleus. In control cells, 79±3% of the poly(A)+ RNA detected by FISH is cytoplasmic, the remainder being nuclear. In NPM1-depleted cells, 26±4 of the poly(A)+ RNA detected by FISH is cytoplasmic (with 74±4% being nuclear). The phenotype is observed in 85(±9)% of NPM1-depleted cells. Similar data were obtained using two additional shRNAs that targeted independent regions of the NPM1 mRNA (Figure 6B), indicating the effects of NPM1-specific shRNAs were likely direct. The nuclear retention observed in NPM1 knockdown cells was specific for mRNA as neither bulk rRNA (Supplementary Figure S2) nor tRNA (data not shown) showed any change in their relative nuclear/cytoplasmic distribution. Note that in this experiment defects in export of newly synthesized rRNA complexes reported in previous studies (Maggi et al, 2008) would not have been detected since total 25S rRNA was followed as was done for poly(A)+ RNA in Figure 6. We did not detect any dramatic differences on the half-lives of three independent mRNAs tested in NPM1-depleted cells (data not shown), suggesting that NPM1 levels did not have a major impact on mRNA stability. Therefore, we conclude that a reduction in the level of NPM1 in HeLa cells results in a dramatic accumulation of mRNA in the nucleus. This may be due to a block in export as a result of failure to correctly terminate poly(A) synthesis as hyperadenylated mRNAs have been associated with nuclear retention in other organisms (Jensen et al, 2001).
Figure 6.
Reduction of NPM1 levels results in the accumulation of poly(A)+ RNAs in the nucleus. (A) Control HeLa cells (transfected with an empty pLKO vector) or HeLa cells treated with an NPM1-specific shRNA for 24 h were analysed by immunofluorescence using DAPI to mark nuclei, oligo d(T) to identify poly(A)+ RNA or NPM1 antibodies to visualize nucleophosmin. The arrow highlights a representative cell in which poly(A)+ RNA is largely cytoplasmic (control cells) or is retained in the nucleus (NPM1 KD cells). (B) Same as (A) but cells were treated with two independent shRNAs targeting NPM1 before analysis and the DAPI stained panel to mark nuclei was omitted.
Discussion
The process of mRNA 3′ end formation/polyadenylation is highly regulated and networked with other processes involved in nuclear mRNA biogenesis. This study identifies the oncoprotein NPM1 as a factor that influences the length of the poly(A) tail as well as mRNA export from the nucleus to the cytoplasm. These observations attribute important new functions to this abundant nuclear/nucleolar chaperone which significantly expands our insight into the overall impact of NPM1 on cellular metabolism.
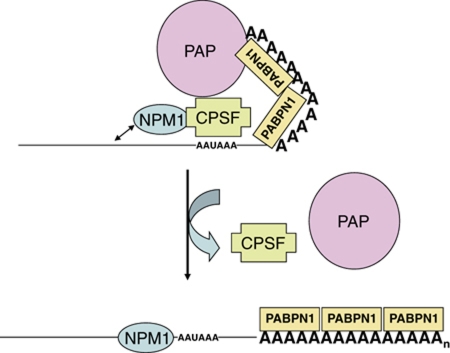
There are several possible models for how NPM1 influences poly(A) tail length. First, by binding CPSF, NPM1 may influence CPSF interactions with PABPN1 and PAP (Figure 7). NPM1 may in some manner anchor CPSF at the AAUAAA element via its ability to interact with nearby RNA sequences. As the poly(A) tail gets longer and interactions between CPSF and the other factors become more difficult to maintain because of distance (Kühn et al, 2009), NPM1 may provide added strain to these interactions and favour dissociation. When CPSF can no longer effectively interact with PAP/PABPN1, poly(A) tail synthesis terminates and NPM1 may simultaneously be released to bind to the 3′ UTR of the mRNA. Thus, binding of NPM1 to the 3′ UTR just upstream of the AAUAAA element (Palaniswamy et al, 2006) may also assist in the release of the core poly(A) factors from the nascent mRNA. Our data indicating a direct interaction between NPM1 and CPSF (Figure 3) as well as a requirement for natural termination of poly(A) tail synthesis for NPM1 deposition (Figure 2) are consistent with this model. Alternative models exist which we consider less likely but cannot formally rule out. NPM1, for example, may influence poly(A) tail size by assisting in the recruitment of a deadenylase to remodel the poly(A) tail. We have not, however, identified any direct interactions between NPM1 and the nuclear localized PARN deadenylase. In addition, to date we have not obtained any clear evidence for a role for deadenylase activity influencing poly(A) tail length in our nuclear extract-based polyadenylation assays where NPM1 regulation of poly(A) tail length could be recapitulated (Figure 5). In another alternative model, NPM1 via its chaperone activity may influence the association of other proteins in the large complex of ∼85 factors that may be involved in polyadenylation (Shi et al, 2009), and thus influence poly(A) tail length indirectly. Given the lack of evidence to date for other factors besides CPSF, PAP and PABPN1 in poly(A) length regulation (Kühn et al, 2009), assessment of this alternative model awaits additional experimental insight into the functions of these other putative poly(A) factors.
Figure 7.
Model for the regulation of poly(A) tail length by NPM1. NPM1 is directly associated with CPSF-160 which influences poly(A) polymerase activity on the growing tail in conjunction with the poly(A) binding processivity factor PABPN1. The propensity for NPM1 to bind nearby nucleic acids possibly places an additional strain on the CPSF-160 interactions with PAP and PABPN1, perhaps helping the complex dissociate when the poly(A) tail reaches a specific size. In the absence of the constraints imposed by NPM1 interaction, the CPSF−PAP−PABPN1 complex allows for additional rounds of poly(A) synthesis on the growing tail.
Since NPM1 could directly associate with the AAUAAA-binding factor CPSF, we investigated whether NPM1 influences alternative polyadenylation. Given the overexpression of NPM1 in cancer cells and the altered use of upstream poly(A) signals on many genes (Sandberg et al, 2008; Ji et al, 2009; Mayr and Bartel, 2009), we considered this an attractive possibility. However, to date we have not obtained evidence for a role for NPM1 in alternative polyadenylation as poly(A) site choice was not altered in NPM1 knockdown cell lines for a variety of genes tested (data not shown). This observation would be consistent with our mechanistic data that indicates NPM1 is not deposited during the mRNA cleavage event (Palaniswamy et al, 2006), but rather at later times in the process of 3′ end formation, long after the choice of polyadenylation signals on the pre-mRNA has been made.
We can envision several possible explanations as to why NPM1 was not detected previously as a factor involved in the termination/sizing of poly(A) tails. First, previous studies that identified CPSF, PAP and PABPN1 as factors involved in poly(A) tail sizing were done via a biochemical reconstitution approach (Bienroth et al, 1993; Kühn et al, 2009). While these three factors are undoubtedly required for proper sizing of the poly(A) tail, they may not be sufficient for poly(A) tail sizing in biological systems. Second, NPM1 is extremely abundant in mammalian tissue culture cells and could theoretically have been overlooked in some reconstitution experiments as a presumably minor and unimportant contaminant. Finally, the size of the poly(A) tail on individual mRNAs has not been very well characterized to date. While the field often uses a value of 150–200 bases for the length of a poly(A) tail in mammalian systems, this represents more of a generalization than an accurate measurement. Reconstitution assays using CPSF, PAP and PABPN1 often give longer tails (∼250 bases) rather than the generalized value of 150–200 bases for poly(A) tails in biological systems (Kühn et al, 2009). This length increase in reconstituted assays could, we hypothesize, be due to the limiting amount/lack of NPM1. Unfortunately, we have not been able to obtain purified biologically active NPM1 from recombinant sources to date that can be deposited onto mRNA in reconstituted polyadenylation reactions. This is perhaps due to a need for specific post-translational modification (Haindl et al, 2008) or additional unidentified factors involved in the deposition of NPM1 onto mRNAs. Thus, reconstitution studies to test this hypothesis remain for future experimentation.
The apparent block in mRNA export observed in NPM1-depleted cells demonstrates the biological relevance of NPM1 to mRNA biology and could be related to the role of NPM1 in mRNA polyadenylation. In addition to the possible association between hyperadenylation and a block in mRNA export (Jensen et al, 2001), there are several hypotheses for how NPM1 might directly influence mRNA movement from the nucleus to the cytoplasm. First, NPM1 is a known nucleo-cytoplasmic shuttling protein (Maggi et al, 2008) and could have a direct role in chaperoning the movement of mature mRNAs from the nucleus to the cytoplasm when it is deposited on transcripts that have been properly polyadenylated. Along these lines, it is important to note that while NPM1 is readily found associated with nuclear mRNAs, the interaction appears to be largely lost on mRNAs isolated from the cytoplasm (Palaniswamy et al, 2006). NPM1 is known to have a key role in ribosome export from the nucleus (Maggi et al, 2008) and has recently been implicated in miRNA export from cells (Wang et al, 2010b); thus, it may be generally involved in RNP movement in the cell. Second, NPM1 is known to serve as a shuttle for HIV Rev protein accumulation in the nucleus (Fankhauser et al, 1991). Thus, NPM1 could be involved in the nuclear import of other shuttling proteins and its effect on mRNA export could formally be independent of its deposition onto mRNAs as a polyadenylation mark. Third, NPM1 has been shown to associate with the nuclear pore, specifically through interactions with Nup98 (Crockett et al, 2004). NPM1 could, therefore, directly assist in the loading of mRNA export complexes at the pore. Fourth, there have been a variety of connections between polyadenylation and mRNA export (Rougemaille et al, 2008; Johnson et al, 2009; Qu et al, 2009; Ruepp et al, 2009), and exploring the connections between NPM1 and these export factors could provide significant insight. Finally, NPM1-influenced mRNA export could be a regulated process in the cell. The tumour suppressor ARF blocks NPM1 shuttling, affects ribosome biogenesis and reduces polysome formation (Rizos et al, 2006). It will be informative to assess whether activation of ARF also affects mRNA export from the nucleus, which could contribute to the translational effects that have been observed.
Could NPM1 be involved in the networking of polyadenylation to other aspects of nuclear mRNA biogenesis? Tarapore et al (2006) have demonstrated that NPM1 phosphorylated on Thr 199 can localize to nuclear speckles and repress mRNA splicing. It will be interesting to investigate the role of NPM1 deposition by polyadenylation in the connections between splicing and 3′ end formation.
There are numerous interesting potential implications in the observations made in this study to cancer-related cellular processes. First, DNA damage results in increased NPM1 synthesis as well as accumulation of NPM1 in the nucleoplasm (Wu et al, 2002). Given the changes in polyadenylation that have been noted in response to DNA damage (Cevher et al, 2010), it will be worthwhile to determine whether NPM1 contributes to any alterations in 3′ end processing seen under these conditions. Second, NPM1 undergoes significant alterations in cancer cells and has been implicated both as an oncogene and as a tumour suppressor (Grisendi et al, 2006). Many if not all cancer cells show a dramatic increase in NPM1 expression (Pianta et al, 2010). As the experiments we have performed to date are in tissue culture cells (HeLa and 293T cells) that express high levels of NPM1, it may be informative to compare NPM1 deposition, poly(A) tail length regulation and mRNA export in normal human cells. These studies could provide important insight into novel roles for NPM1 in the regulation and quality control of mRNA biogenesis and the promotion of tumourogenic phenotypes. Third, the N-terminal half of NPM1 is a common target for chromosomal translocations that generate oncogenic fusion proteins (Grisendi et al, 2006). Whether and how these NPM1 fusion proteins function as polyadenylation marks will be interesting fodder for future studies. Finally, about a third of adult acute myeloid leukaemias (AML) possess a frameshift in the C-terminal portion of NPM1 that results in an aberrant cytoplasmic accumulation of the protein (Grisendi et al, 2006; Lindström, 2011). How this altered NPM1 subcellular localization influences mRNA poly(A) tails and mRNA export regulation in AML cells will also be an interesting question for future studies.
Materials and methods
Cell culture and transfection
HeLa cell lines were grown at 37°C in 5% CO2 in Dulbecco's modification of Eagle's medium supplemented with 10% fetal bovine serum, L-glutamine and penicillin/streptomycin. HeLa suspension cells were grown in Eagle's minimum essential medium supplemented with 10% horse serum. All plasmid DNA was treated with the MiraCLEAN endotoxin removal kit (Mirus Bio) before being used for transfection with Lipofectamine 2000 (Invitrogen). For establishment of the LKO.1 and NPM1 KD HeLa cell lines, either empty LKO.1 plasmid or LKO.1 plasmid containing an shRNA against the coding sequence of human NPM1 (shRNA 1: Sigma MISSION clone ID NM_002520.4-169s1c1) was transfected into cells. Two days post-transfection, cells were switched to media containing puromycin (5 μg/ml). Single colonies were selected, and expression of NPM1 was assessed by qRT–PCR using primers 5′-GGTCTGAAAATGGAGGT-3′ and 5′-GGCGCTTTTTCTTCAGCTT-3′ and corroborated by western blot. For growth analysis, cells were trypsinized and counted using a hemocytometer. For transient shRNA-mediated knockdown of NPM1 expression, either empty LKO.1 plasmid or NPM1 shRNA LKO.1 plasmids (shRNA 2: NM_002520.4-664s1c1; shRNA 3: NM_002520.4-819s1c1) were transfected into HeLa cells. Twenty-four hours post-transfection, cells were processed for FISH assays.
Plasmids and RNAs
pSVL contains a 241-base pair (bp) BamHI-BclI fragment containing the SVL polyadenylation signal inserted into the BamHI site of pSP65. RNA substrates containing the full polyadenylation signal were generated by DraI cleavage. Pre-cleaved SVL RNA was generated by SP6 transcription of pSVL that had been linearized with HpaI. pIVA2 contains the 155-bp BamHI-PvuII fragment of adenovirus pE1B cloned into pGem4 at the HincII and BamHI sites. SP6 transcription of BglI-linearized template yielded a 156-base RNA (IVA2). A 148 base fragment of the BGH poly(A) signal-containing RNA was PCR amplified from pcDNA3.1 using the primers 5′-ATTTAGGTGACACTATAGAAAGTCTAGAGGGCCCGTTTAAAC-3′ and 5′-GCAATTTCCTCATTTTATTAGGAAGGACACTGGG-3′ and used directly in transcription reactions to produce a pre-cleaved BGH poly(A) signal. A 154 base fragment of the CFIm 68-kDa subunit was PCR amplified from pCINeo (a gift from Dr G Gilmartin, University of Vermont) using the primers 5′-ATTTAGGTGACACTATAGAACTTGTTAAAAATGTTTATATGAT-3 and 5′-TTGCTGAACACAAGACTTTCCCTGAG-3 and used directly in transcription reactions to produce a pre-cleaved CFIm poly(A) signal. Transcription reactions using SP6 polymerase were performed in the presence of [32P]UTP and m7GpppG as described previously (Wilusz and Shenk, 1988). All RNAs were gel purified before use.
Polyadenylation assays
HeLa nuclear extracts were prepared and cell-free polyadenylation assays were performed as previously described (Wilusz and Shenk, 1988). A typical reaction contained 3% (w/v) polyvinyl alcohol, 1 mM ATP, 20 mM phosphocreatine, 12 mM HEPES (pH 7.9), 12% (v/v) glycerol, 60 mM KCl, 0.12 mM EDTA, 0.3 mM DTT and 60% (v/v) nuclear extract. Cordycepin 5′-triphosphate and AMPP(CH2)P (Sigma-Aldrich) were dissolved in water and added to polyadenylation assays where indicated. Immunodepleted extracts were obtained by incubating nuclear extract with Sepharose-Protein A beads containing either NPM1-specific antibody or control normal rabbit IgG. Partially purified NPM1 protein was prepared by fractionation of HeLa nuclear extracts over Q-Sepharose, S-Sepharose and poly(A)-Sepharose 4B columns using linear salt gradients. Fractions containing NPM1 were identified by western blot, concentrated using Microcon filters and dialysed against buffer D (Wilusz and Shenk, 1988). Reactions were incubated at 30°C for the indicated times and products were analysed on 6% (w/v) acrylamide gels containing 7 M urea.
Protein–protein co-immunoprecipitation analysis
HeLa cells were lysed by douncing, purified nuclei were suspended in NET-2 buffer (150 mM NaCl, 50 mM Tris–Cl pH 7.4, 0.05% NP-40 and 1 mM PMSF) and disrupted by freeze thawing. The lysate was pre-cleared using Pansorbin cells (Calbiochem). In some samples, 10 units of RNase ONE (Promega) was added to the pre-cleared lysate and incubated at 37oC for 15 min. NPM1-associated complexes were purified with anti-NPM1 antibody H-106 (Santa Cruz Biotechnology) and Pansorbin. Following washing with NET-2 buffer, protein samples were separated on a 10% SDS-polyacrylamide gel and blotted onto polyvinylidene difluoride membrane (Millipore). NPM1 was detected using mouse monoclonal antibody FC8791 (Santa Cruz Biotechnology). The loading control GAPDH was detected using mAB374 (Millipore). CPSF-160 was detected using a rabbit polyclonal serum. CPSF-100 (A301-581A) and CPSF-30 (A301-585A) were detected using the indicated antibodies from Bethyl laboratories. CPSF-73 antibodies were the kind gift from Dr David Bentley (University of Colorado, Denver). CstF-64 and -77 were detected using purified rabbit IgG obtained from Dr Christine Milcarek (University of Pittsburgh). PABPN1 was detected using rabbit anti-PABPN1 sera generously provided by Dr David G Bear (University of New Mexico). PAP was detected using antibody ab72492 (Abcam).
UV crosslinking and immunoprecipitations
In a typical reaction, 50–200 fmol of 32P-labelled RNA was incubated in the nuclear extract-based polyadenylation system for the time indicated. Reaction mixtures were irradiated for 6 min on ice using a UV Stratalinker 2400 (Stratagene). RNase A was added to a final concentration of 1 mg/ml and reaction mixtures were incubated at 37°C for 15 min. Crosslinked proteins were separated by SDS–PAGE and visualized by phosphorimaging. In crosslinking and immunoprecipitation experiments, samples were pre-cleared after RNase treatment and incubated with antisera at 4°C. Protein–antibody complexes were purified using Pansorbin cells.
LLM-PAT poly(A) tail assays
LLM-PAT assays were performed as described (Garneau et al, 2008). Total RNA was prepared from wild-type, pLKO.1 control and NPM1 KD HeLa cell lines using TRIzol reagent (Invitrogen). The A0 marker was generated by hybridizing total RNA with oligo d(T) (Integrated DNA Technologies), followed by RNase H (Fermentas) digestion. Both total RNA and RNase H/d(T) RNA were ligated to a 5 pppRNA linker (5′-rApppTTTAACCGCGAATTCCAG-3′ddC) at 16°C for 2 h in 50 mM Tris–Cl, pH 7.5, 10 mM MgCl2, 20 mM DTT and 0.1 mg/ml bovine serum albumin. The ligated RNA was then reverse transcribed using a reverse transcription primer specific to the RNA linker (5′-CTGGAATTCGCGGT-3′). The resulting cDNA was then amplified by PCR using the primer (5′-CTGGAATTCGCGGTTAAATTT-3′) and one of the following mRNA-specific primers: β-actin, 5′-GAATGATGAGCCTTCGTGCC-3′; rps5, 5′-CTGAGTGCCTGGCAGATGAC-3′: hnRNP H, 5′-TGAGACGCAATACCAATACT-3′. PCR products were separated on a 5% non-denaturing polyacrylamide gel. Following electrophoresis, the gel was soaked in 1 × Tris-Borate-EDTA buffer with ethidium bromide and visualized using a Typhoon Trio imager.
Immunofluorescence and FISH assays
Cells grown on cover slips were fixed using 1% paraformaldehyde, methanol and then 70% ethanol. Poly(A)+ RNAs and 25S rRNA were visualized by an oligo d(T)-Cy3 probe or an Alexa 647-rRNA probe (5′-ATCAGAGTAGTGGTATTTC-3′; IDT), respectively. Mouse anti-NPM1 antibodies and Cy2-goat anti-mouse antibodies (Jackson ImmunoResearch) were used to stain NPM1. Prolong Gold antifade mounting reagent with DAPI (Invitrogen) was used to stain nuclei. Images were obtained using a Zeiss Laser Scanning Axiovert Confocal microscope and a Nikon inverted epifluorescence microscope.
Analysis of mRNPs in HeLa cells
HeLa cells were washed with PBS, resuspended in a 0.1% formaldehyde solution in PBS and incubated for 15 min at room temperature. The reaction was quenched using 0.25 M glycine. Cell pellets were washed with PBS and resuspended in RIPA buffer (50 mM Tris–HCl (pH 7.5), 1% (v/v) NP-40, 0.5% (w/v) sodium deoxycholate, 0.05% (w/v) SDS, 1 mM EDTA and 150 mM NaCl). Cells were disrupted by sonication on ice, and insoluble materials were removed via centrifugation. Aliquots received anti-NPM1 or control antibodies and were incubated at 4°C for 2 h. Antibody-bound complexes were recovered using Pansorbin cells and washed seven times with RIPA buffer containing 1 M urea. Formaldehyde crosslinks were reversed by heating at 70°C for 45 min. Isolated RNAs were analysed by RT–PCR using random hexamers in the RT step to assess the amount of an individual RNA that was precipitated using the following primer sets: Schnurri-2: 5′-GGGAAAAGGAGATGGAGACC-3′ and 5′-CAGACATCCTCCCACGAGTT-3′; PAPOLG: 5′-TTTCCAGATTGGCACATGAA-3′ and 5′-GCATTTCAAAGGCCTCCAT-3′; H2A 5′-AGCTCAACAAGCTTCTGGGCAA-3′ and 5′-TTGTGGTGGCTCTCGGTCTTCTT-3′; MC4R: 5′-GATTACCTTGACCATCCTGA-3′ and 5′-AGTGAGACATGAAGCACACA-3′; JunB: 5′-TTAACAGGGAGGGGAAGAGG-3′ and 5′-TGCGTGTTTCTTTTCCACAG-3′; hnRNP H: 5′-GGGAAAAATTTGAGACGCAAT-3′ and 5′-GACAAGTTTCACTTAGCGCAAT-3′; CFIm (CPSF6): 5′-TGAACCTGTAAGGATTCATGG-3′ and 5′-TGCACATCATAATGGCCAAA-3′.
Supplementary Material
Acknowledgments
We wish to thank Drs David Bear, David Bentley, Chris Milcarek and Greg Gilmartin for providing reagents and members of the Wilusz Laboratories for critical comments. We acknowledge funding from a Core Infrastructure Grant for Microscope Imaging from Colorado State University. This work was supported by grant GM072481 to JW.
Author contributions: FS performed experiments for Figures 1–6 and Supplementary Figures S1 and S2. HI performed experiments for Figures 4 and 5; Supplementary Figure S1. ALM performed experiments for Figure 1. CJW participated in the conceptual design of the study and data interpretation. JW participated in conceptual design of the study, data interpretation and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abad X, Vera M, Jung SP, Oswald E, Romero I, Amin V, Fortes P, Gunderson SI (2008) Requirements for gene silencing mediated by U1 snRNA binding to a target sequence. Nucleic Acids Res 36: 2338–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienroth S, Keller W, Wahle E (1993) Assembly of a processive messenger RNA polyadenylation complex. EMBO J 12: 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SN, Maggi LB Jr, Winkeler CL, Toso EA, Gwinn AS, Pelletier CL, Weber JD (2009) Nucleophosmin protein expression level, but not threonine 198 phosphorylation, is essential in growth and proliferation. Oncogene 28: 3209–3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CE, Tarun SZ Jr, Boeck R, Sachs AB (1996) PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol Cell Biol 16: 5744–5753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevher MA, Zhang X, Fernandez S, Kim S, Baquero J, Nilsson P, Lee S, Virtanen A, Kleiman FE (2010) Nuclear deadenylation/polyadenylation factors regulate 3′ processing in response to DNA damage. EMBO J 29: 1674–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke C, Alwine JC (1996) The cap and the 3′ splice site similarly affect polyadenylation efficiency. Mol Cell Biol 16: 2579–2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crockett DK, Lin Z, Elenitoba-Johnson KS, Lim MS (2004) Identification of NPM-ALK interacting proteins by tandem mass spectrometry. Oncogene 23: 2617–2629 [DOI] [PubMed] [Google Scholar]
- Falini, B Nicoletti I, Bolli N, Martelli MP, Liso A, Gorello P, Mandelli F, Mecucci C, Martelli MF (2007) Translocations and mutations involving the nucleophosmin (NPM1) gene in lymphomas and leukemias. Haematologica 92: 519–532 [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Izaurralde E, Adachi Y, Wingfield P, Laemmli UK (1991) Specific complex of human immunodeficiency virus type 1 rev and nucleolar B23 proteins: dissociation by the Rev response element. Mol Cell Biol 11: 2567–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty SM, Fortes P, Izaurralde E, Mattaj IW, Gilmartin GM (1997) Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc Natl Acad Sci U S A 94: 11893–11898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Sokoloski KJ, Opyrchal M, Neff CP, Wilusz CJ, Wilusz J (2008) The 3′ untranslated region of sindbis virus represses deadenylation of viral transcripts in mosquito and mammalian cells. J Virol 82: 880–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin EC, Rottman FM (1992) The 3′-flanking sequence of the bovine growth hormone gene contains novel elements required for efficient and accurate polyadenylation. J Biol Chem 267: 16330–16334 [PubMed] [Google Scholar]
- Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, Manova K, Pandolfi PP (2005) Role of nucleophosmin in embryonic development and tumorigenesis. Nature 437: 147–153 [DOI] [PubMed] [Google Scholar]
- Grisendi S, Mecucci C, Falini B, Pandolfi PP (2006) Nucleophosmin and cancer. Nat Rev Cancer 6: 493–505 [DOI] [PubMed] [Google Scholar]
- Gu H, Das Gupta J, Schoenberg DR (1999) The poly(A)-limiting element is a conserved cis-acting sequence that regulates poly(A) tail length on nuclear pre-mRNAs. Proc Natl Acad Sci U S A 96: 8943–8948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Garrett M, Micklem G, Brogna S (2011) PolyA signals located near the 5′ of genes are silenced by a general mechanism that prevents premature 3′ end processing. Mol Cell Biol 31: 639–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haindl M, Harasim T, Eick D, Muller S (2008) The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep 9: 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Pogar T, Liang S, Hague LK, Lutz CS (2007) Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3′-UTR. RNA 13: 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisaoka M, Ueshima S, Murano K, Nagata K, Okuwaki M (2010) Regulation of nucleolar chromatin by B23/nucleophosmin jointly depends upon its RNA binding activity and transcription factor UBF. Mol Cell Biol 30: 4952–4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TH, Patricio K, McCarthy T, Rosbash M (2001) A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol Cell 7: 887–898 [DOI] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B (2009) Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A 106: 7028–7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Cubberley G, Bentley DL (2009) Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol Cell 33: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G (2010) U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn U, Gündel M, Knoth A, Kerwitz Y, Rüdel S, Wahle E (2009) Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J Biol Chem 284: 22803–22814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hann SR (2009) The Myc-nucleophosmin-ARF network: a complex web unveiled. Cell Cycle 8: 2703–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Tan BC, Liu H, Shih CJ, Chien KY, Lin CL, Yung BY (2010) Dephosphorylation of nucleophosmin by PP1â facilitates pRB binding and consequent E2F1-dependent DNA repair. Mol Biol Cell 21: 4409–4417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström MS (2011) NPM1/B23: a multifunctional chaperone in ribosome biogenesis and chromatin remodeling. Biochem Res Int 2011: 195209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Reichow SL, Kim M, Suh H, Leeper TC, Yang F, Mutschler H, Buratowski S, Meinhart A, Varani G (2010) Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nat Struct Mol Biol 17: 1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi LB Jr, Kuchenruether M, Dadey DY, Schwope RM, Grisendi S, Townsend RR, Pandolfi PP, Weber JD (2008) Nucleophosmin serves as a rate-limiting nuclear export chaperone for the mammalian ribosome. Mol Cell Biol 28: 7050–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH (2010) Crosstalk between mRNA 3′ end processing and transcription initiation. Mol Cell 40: 410–422 [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP (2009) Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meani N, Alcalay M (2009) Role of nucleophosmin in acute myeloid leukemia. Expert Rev Anticancer Ther 9: 1283–1294 [DOI] [PubMed] [Google Scholar]
- Millevoi S, Vagner S (2010) Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res 38: 2757–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes NM, Li W, Tian B, Furger A (2010) A functional human Poly(A) site requires only a potent DSE and an A-rich upstream sequence. EMBO J 29: 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuwaki M (2008) The structure and functions of NPM1/Nucleophsmin/B23, a multifunctional nucleolar acidic protein. J Biochem 143: 441–448 [DOI] [PubMed] [Google Scholar]
- Palaniswamy V, Moraes KC, Wilusz CJ, Wilusz J (2006) Nucleophosmin is selectively deposited on mRNA during polyadenylation. Nat Struct Mol Biol 13: 429–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pianta A, Puppin C, Franzoni A, Fabbro D, Di Loreto C, Bulotta S, Deganuto M, Paron I, Tell G, Puxeddu E, Filetti S, Russo D, Damante G (2010) Nucleophosmin is overexpressed in thyroid tumors. Biochem Biophys Res Commun 397: 499–504 [DOI] [PubMed] [Google Scholar]
- Qu X, Lykke-Andersen S, Nasser T, Saguez C, Bertrand E, Jensen TH, Moore C (2009) Assembly of an export-competent mRNP is needed for efficient release of the 3′-end processing complex after polyadenylation. Mol Cell Biol 29: 5327–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizos H, McKenzie HA, Ayub AL, Woodruff S, Becker TM, Scurr LL, Stahl J, Kefford RF (2006) Physical and functional interaction of the p14ARF tumor suppressor with ribosomes. J Biol Chem 281: 38080–38088 [DOI] [PubMed] [Google Scholar]
- Rougemaille M, Dieppois G, Kisseleva-Romanova E, Gudipati RK, Lemoine S, Blugeon C, Boulay J, Jensen TH, Stutz F, Devaux F, Libri D (2008) THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell 135: 308–321 [DOI] [PubMed] [Google Scholar]
- Ruepp MD, Aringhieri C, Vivarelli S, Cardinale S, Paro S, Schümperli D, Barabino SM (2009) Mammalian pre-mRNA 3′ end processing factor CF I m 68 functions in mRNA export. Mol Biol Cell 20: 5211–5223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB (2008) Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, Frank J, Manley JL (2009) Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 33: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarapore P, Shinmura K, Suzuki H, Tokuyama Y, Kim SH, Mayeda A, Fukasawa K (2006) Thr199 phosphorylation targets nucleophosmin to nuclear speckles and represses pre-mRNA processing. FEBS Lett 580: 399–409 [DOI] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS (2005) A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 33: 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman K, Brown KM, Gilmartin GM (2005) Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev 19: 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E (1995) Poly(A) tail length control is caused by termination of processive synthesis. J Biol Chem 270: 2800–2808 [DOI] [PubMed] [Google Scholar]
- Wang K, Zhang S, Weber J, Baxter D, Galas DJ (2010b) Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res 38: 7248–7259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fairley JA, Roberts SG (2010a) Phosphorylation of TFIIB links transcription initiation and termination. Curr Biol 20: 548–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Proudfoot NJ (2009) Transcriptional termination enhances protein expression in human cells. Mol Cell 33: 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J, Shenk T (1988) A 64 kd nuclear protein binds to RNA segments that include the AAUAAA polyadenylation motif. Cell 52: 221–228 [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Beemon KL (2006) The negative regulator of splicing element of Rous sarcoma virus promotes polyadenylation. J Virol 80: 9634–9640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MH, Chang JH, Chou CC, Yung BY (2002) Involvement of nucleophosmin/B23 in the response of HeLa cells to UV irradiation. Int J Cancer 97: 297–305 [DOI] [PubMed] [Google Scholar]
- Yun C, Wang Y, Mukhopadhyay D, Backlund P, Kolli N, Yergey A, Wilkinson KD, Dasso M (2008) Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J Cell Biol 183: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.