Abstract
Eukaryotic up-frameshift 1 (UPF1) is a nucleic acid-dependent ATPase and 5′-to-3′ helicase, best characterized for its roles in cytoplasmic RNA quality control. We previously demonstrated that human UPF1 binds to telomeres in vivo and its depletion leads to telomere instability. Here, we show that UPF1 is present at telomeres at least during S and G2/M phases and that UPF1 association with telomeres is stimulated by the phosphoinositide 3-kinase (PI3K)-related protein kinase ataxia telangiectasia mutated and Rad3-related (ATR) and by telomere elongation. UPF1 physically interacts with the telomeric factor TPP1 and with telomerase. Akin to UPF1 binding to telomeres, this latter interaction is mediated by ATR. Moreover, the ATPase activity of UPF1 is required to prevent the telomeric defects observed upon UPF1 depletion, and these defects stem predominantly from inefficient telomere leading-strand replication. Our results portray a scenario where UPF1 orchestrates crucial aspects of telomere biology, including telomere replication and telomere length homeostasis.
Keywords: telomerase, telomeres, TPP1, replication, UPF1
Introduction
Up-frameshift 1 (UPF1), also known as regulator of non-sense transcripts 1 (RENT1) or suppressor with morphogenetic defects in genitalia 2 (SMG2), is a highly conserved eukaryotic phosphoprotein with nucleic acid-dependent ATPase and 5′-to-3′ helicase activities (Page et al, 1999; Bhattacharya et al, 2000). UPF1 has been best characterized for its central roles in the RNA surveillance pathway ‘non-sense-mediated mRNA decay’ (NMD). NMD recognizes and degrades transcripts harbouring premature termination codons (PTCs) and modulates the expression of numerous physiological mRNA and non-coding RNA molecules (Maquat and Gong, 2009; Nicholson et al, 2010). In addition, UPF1 has been involved in NMD-independent RNA metabolism pathways such as Staufen-1-mediated mRNA decay, replication-dependent histone mRNA degradation and stabilization of HIV-1 genomic RNA (Maquat and Gong, 2009; Nicholson et al, 2010).
Apart from UPF1, the human NMD core machinery also comprises the SMG factors UPF2, UPF3 (A and B), SMG1, hEST1A (also known as SMG6), hEST1B (also known as SMG5) and hEST1C (also known as SMG7) (Nicholson et al, 2010). More recently, SMG8 and SMG9 were added to the list of proteins required for NMD in humans (Yamashita et al, 2009). Several enzymatic activities other than those associated with UPF1 operate within the NMD core machinery. Human SMG1, a member of the phosphoinositide 3-kinase (PI3K)-related protein kinase (PIKK) superfamily, directly phosphorylates UPF1 at specific serine residues in its C-terminal S/T-Q-rich region, thereby ‘licensing’ NMD execution. On the contrary, the three hEST1 proteins promote UPF1 dephosphorylation, presumably through activation and/or recruitment of the protein phosphatase PP2A (Yamashita et al, 2001; Chiu et al, 2003; Kashima et al, 2006). Moreover, the PilT N-terminal domain of hEST1A possesses RNA endonuclease activity capable of initiating cleavage of non-sense messengers in proximity of their PTC (Huntzinger et al, 2008; Eberle et al, 2009).
Although NMD is strictly executed in the cytoplasm (Singh et al, 2007), human UPF1, hEST1A and SMG1 are also directly involved in nuclear processes safeguarding the stability of the genome. A fraction of human UPF1 associates with chromatin, preferentially during S phase, suggesting a role for UPF1 in DNA replication (Azzalin and Lingner, 2006). Consistently, short hairpin RNA (shRNA)-mediated knockdown of UPF1 in human cervical carcinoma HeLa cells leads to cell-cycle arrest at the onset of S phase, to chromosome and chromatid breaks and to accumulation of the DNA damage factor histone H2AX phosphorylated at Serine 139 (γ-H2AX) (Azzalin and Lingner, 2006). UPF1 depletion-induced γ-H2AX accumulation is dependent on the PIKK ataxia telangiectasia mutated and Rad3-related (ATR; Azzalin and Lingner, 2006). Importantly, while downregulation of UPF2 impairs NMD to an extent similar to that observed upon UPF1 depletion, it neither perturbs cell-cycle progression, nor does it lead to detectable DNA damage, indicating that UPF1-associated functions in maintaining genome stability are genetically separable from its function in canonical NMD (Azzalin and Lingner, 2006). Further advocating direct roles for UPF1 in DNA metabolism are its physical interactions with the processive lagging-strand DNA polymerase δ and the DNA-dependent RNA polymerase II (RNAPII) machinery (Carastro et al, 2002; Iborra et al, 2004; Azzalin and Lingner, 2006). hEST1A was originally identified as a putative orthologue of the Saccharomyces cerevisiae protein Est1p, a subunit of the telomerase holoenzyme (Lundblad and Szostak, 1989; Reichenbach et al, 2003; Snow et al, 2003). Telomerase is a specialized reverse transcriptase capable of synthesizing telomeric repeats de novo and adding them to the 3′ end of telomeres, the protective extremities of linear eukaryotic chromosomes (Hug and Lingner, 2006). Similarly to its yeast counterpart, hEST1A physically interacts with active telomerase. In addition, overexpression of hEST1A induces telomere deprotection in human fibrosarcoma-derived HT180 cells and telomere shortening in human embryonic kidney (HEK) 293 cells (Reichenbach et al, 2003; Snow et al, 2003). Finally, the protein kinase activity of human SMG1 is stimulated by ionizing radiation (IR) treatments (Brumbaugh et al, 2004). SMG1 phosphorylates both p53 and UPF1 in vitro and is required for their phosphorylation in vivo upon treatment with IR (Brumbaugh et al, 2004). Moreover, human cancer cells depleted for SMG1 accumulate spontaneous DNA damage and display increased radiosensitivity (Brumbaugh et al, 2004).
An additional twist in the biology of human SMG factors occurred when we discovered that they all bind to telomeres in vivo (Azzalin et al, 2007). Moreover, depletion of UPF1, hEST1A or SMG1 induces severe telomeric aberrations, including loss of entire telomeric tracts, and accumulation of telomeric repeat-containing RNA (TERRA) at telomeres (Azzalin et al, 2007). TERRA is a novel nuclear non-coding RNA originating from RNAPII-mediated transcription of telomeric sequences (Azzalin et al, 2007; Azzalin and Lingner, 2008; Chawla and Azzalin, 2008; Luke et al, 2008; Schoeftner and Blasco, 2008). TERRA accumulation at SMG-depleted telomeres occurs without any apparent perturbation of its degradation rates or total cellular levels, leading us to hypothesize that nuclear SMG proteins might actively displace TERRA from telomeres (Azzalin et al, 2007; Chawla and Azzalin, 2008).
Here, we further characterize the role of human UPF1 in safeguarding telomere integrity. First, we demonstrate that UPF1 is present at telomeres during S and G2/M phases and that it binds to telomeres in a telomere length- and ATR-dependent manner. Second, we demonstrate that UPF1 physically interacts with active telomerase and with TPP1, a component of the mammalian telomere-associated multiprotein complex shelterin (Palm and de Lange, 2008). Third, we reveal that the ATPase activity of UPF1 is required to prevent telomeric instability and that the telomeric defects observed upon UPF1 depletion stem predominantly from incomplete leading-strand telomere replication. Finally, we show that UPF1-depleted cells accumulate DNA damage repair factors at telomeres. Our results shed light on the molecular mechanisms leading to telomere instability in cells depleted for UPF1. We propose that UPF1-associated roles in maintaining the stability of the genome derive, at least in part, from its ability to coordinate semiconservative replication and telomerase-mediated elongation of the telomere.
Results
UPF1 is present at telomeres during S and G2/M phases and it binds to telomeres in a telomere length- and ATR-dependent manner
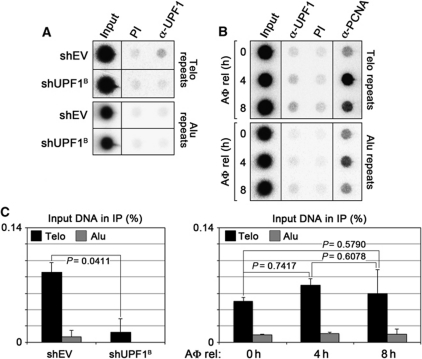
To further confirm the association of UPF1 with telomeric chromatin, we performed chromatin immunoprecipitation (ChIP) experiments using a rabbit polyclonal α-UPF1 antibody and chromatin prepared from HeLa cells transfected with shRNA plasmids directed against UPF1 mRNA (shUPF1B) or with empty shRNA vector controls (shEV). Consistent with our previous results (Azzalin et al, 2007), in control-transfected cells, UPF1 was enriched at telomeres as compared with Alu repeat sequences. The amount of telomeric DNA pulled down with UPF1 antibodies was reproducibly comprised between 0.05 and 0.2% of input telomeric DNA across independent experiments (Figures 1 and 2), suggesting that UPF1 binds to telomeres at low levels or in a transient manner. The amount of telomeric DNA pulled down in UPF1-depleted cells was significantly reduced to background levels (Figure 1A and C; Supplementary Figure S1A), confirming that the telomeric DNA detected in α-UPF1 IP samples was immunoprecipitated through antibody binding to endogenous UPF1 molecules. We next performed ChIP assays using chromatin from HeLa cells synchronized at the G1/S boundary using a single aphidicolin block and released from the block for 4 and 8 h. At these time points, cells accumulated in S and G2/M phases, respectively, as shown by fluorescence-activated cell sorting (FACS) analysis (Supplementary Figure S1F). As expected, the binding of proliferating cell nuclear antigen (PCNA) to both telomeric and Alu repeats peaked in the 4-h sample (Figure 1B). On the other hand, the amount of telomeric DNA immunoprecipitated by UPF1 antibodies did not significantly vary among the different samples (Figure 1B and C). The total cellular levels of both PCNA and UPF1 remained constant among the different samples as well (Supplementary Figure S1B). These results indicate that UPF1 binds to telomeres at least during S and G2/M phases. Whether UPF1 persists at telomeres also in G1 remains to be tested.
Figure 1.
UPF1 is present at telomeres during S and G2/M phases. (A) Chromatin immunoprecipitation (ChIP) analysis of UPF1 binding to telomeres in HeLa cells transfected with the indicated shRNA plasmids or with empty vector control plasmids (shEV). Four days after transfection, cells were harvested and ChIPs were performed with α-UPF1 rabbit polyclonal antibodies or with pre-immune (PI) serum. DNA was dot blotted and hybridized sequentially using probes detecting telomeric (Telo) or Alu repeats. (B) HeLa cells were blocked at the G1/S transition using a single aphidicolin (AΦ) block. Chromatin was prepared 0, 4 and 8 h after release (rel) from the block and ChIPs were performed with α-UPF1 or α-PCNA antibodies or with PI serum. DNA was hybridized as in (A). (C) ChIP quantifications. The amount of immunoprecipitated telomeric and Alu repeats was expressed as fractions of input DNA, after subtraction of the signal obtained for PI samples. Bars and error bars correspond to averages and s.d. from three independent experiments. P-values (two-tailed Student's t-test) are indicated for the relevant samples.
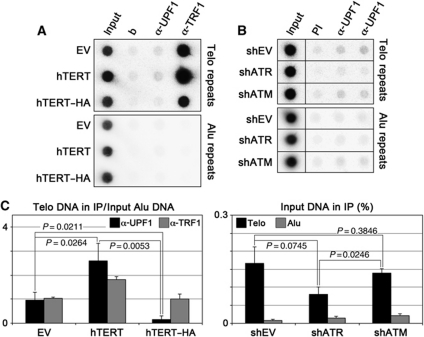
Figure 2.
UPF1 binding to telomeres depends on telomere length and ATR. (A) ChIP analysis of UPF1 binding to telomeres in HeLa cells stably infected with retroviruses expressing untagged hTERT or hTERT C-terminally tagged with HA (hTERT–HA), or with empty vector (EV) retroviruses. ChIPs were performed with rabbit polyclonal antibodies raised against UPF1 and TRF1 or only with beads (b). DNA was dot blotted and hybridized sequentially using probes detecting telomeric (Telo) or Alu repeats. (B) HeLa cells were transfected with the indicated shRNA plasmids and 4 days after transfection ChIPs were performed with α-UPF1 rabbit polyclonal antibodies or with pre-immune (PI) serum. DNA was dot blotted and hybridized sequentially using probes detecting telomeric (Telo) or Alu repeats. (C) ChIP quantifications. For hTERT experiments, the amount of immunoprecipitated telomeric repeats was expressed as a fraction of input Alu DNA, after subtraction of the signal obtained for ChiPs performed only with beads. For shRNA experiments, the amounts of telomeric and Alu repeats were expressed as fractions of input DNA, after subtraction of the signal obtained for PI samples. Bars and error bars correspond to averages and s.d. from at least two independent experiments. P-values (two-tailed Student's t-test) are indicated for the relevant samples.
To assess whether telomere length influences UPF1 binding to telomeres, we performed ChIP assays with chromatin from HeLa cells stably infected with retroviruses expressing either untagged human telomerase reverse transcriptase (hTERT) or an hTERT derivative, C-terminally fused to an influenza haemagglutinin epitope tag (hTERT–HA). hTERT–HA possesses normal catalytic activity in vitro but is unable to elongate telomeres in vivo, likely due to impaired recruitment to telomeres (Counter et al, 1998). As expected, bulk telomere elongation was observed in hTERT- but not in hTERT–HA-infected cells, although the two proteins were expressed at similar levels (Supplementary Figure S1C and E). Expression of hTERT or hTERT–HA did not alter cell-cycle progression or UPF1 total cellular levels (Supplementary Figure S1C and G). The amount of telomeric DNA pulled down by UPF1, when expressed as a fraction of the corresponding input Alu DNA, was significantly increased in the hTERT-expressing cells as compared with EV-infected cells, by a similar factor as for the shelterin component TRF1 (Figure 2A and C). On the contrary, when expressed as a fraction of input telomeric DNA, the amount of telomeric DNA pulled down with UPF1 antibodies did not differ significantly between the two samples (EV: 0.14±0.05%; hTERT: 0.24±0.10%, P=0.1901). These findings indicate that the amount, but not the density of telomere-bound UPF1 is increased upon telomere elongation, presumably due to higher amounts of telomeric substrate available for binding of UPF1. In addition, the telomeric DNA pulled down by α-UPF1 antibodies mostly derives from chromosome ends rather than from intrachromosomal telomeric repeats present in the human genome (Azzalin et al, 2001). Intriguingly, the amount of telomeric DNA pulled down by α-UPF1, but not by α-TRF1 antibodies was significantly decreased in hTERT–HA cells (Figure 2A and C). Because UPF1 physically interacts with telomerase (see below), we speculate that functional hTERT might mediate the interaction of UPF1 with telomeres while mislocalized hTERT–HA might titrate UPF1 away from telomeres.
Bearing in mind the ATR dependency of UFP1 binding to chromatin at large (Azzalin and Lingner, 2006), we set out to test whether ATR recruits UPF1 to telomeres. We performed ChIP assays in HeLa cells depleted for ataxia telangiectasia mutated (ATM) or ATR proteins, using previously validated shRNA plasmids (Azzalin and Lingner, 2006; Supplementary Figure S1D). At the chosen experimental time point, no obvious alterations of UPF1 total cellular levels or of cell-cycle progression were observed in cells depleted for ATM or ATR (Supplementary Figure S1D and G). Whereas UPF1 binding to telomeres was unaffected in ATM-depleted samples, ATR depletion caused a significant reduction in the amount of telomeric DNA pulled down by α-UPF1 antibodies (Figure 2B and C). We conclude that the diminished telomere binding of UPF1 in ATR-depleted cells reflects, at least in part, a direct role for ATR in recruiting UPF1 to telomeres.
UPF1 physically interacts with active human telomerase in an ATR-dependent manner
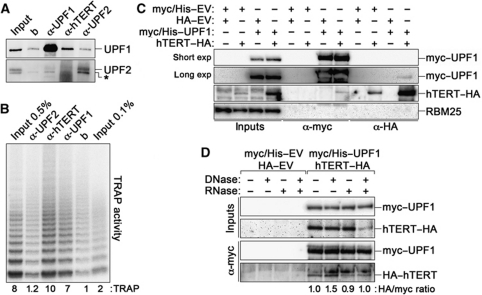
The physical binding of UPF1 to telomeres suggests that UPF1 is part of the telomeric proteome. Given that hEST1A physically interacts with both UPF1 and active telomerase, we set out to test whether UPF1 also interacts with telomerase. We first performed co-immunoprecipitation (CoIP) experiments with HeLa nuclear extracts using antibodies raised against endogenous hTERT, UPF1 and UPF2. α-UPF1 and α-hTERT immunoprecipitates were enriched for UPF1, but not for UPF2 proteins, as compared with control IPs performed using only beads (Figure 3A). Since we were unable to detect endogenous hTERT by western blot, we employed the telomere repeat amplification protocol (TRAP) assay to measure the telomerase activity associated with the immunoprecipitated material. Both α-hTERT and α-UPF1, but not α-UPF2 immunoprecipitates were enriched for telomerase activity as compared with control immunoprecipitates obtained using only beads (Figure 3B). Considering the average UPF1 IP efficiency achieved in our experiments (∼60%), we estimate that ∼16% of nuclear UPF1 is associated with active telomerase. We then ectopically expressed UPF1 variants C-terminally tagged with Myc and Histidine tags (myc/His–UPF1) and hTERT–HA in HEK 293T cells and performed independent CoIPs using antibodies recognizing either tag. Consistent with the results obtained for endogenous proteins, myc/His–UPF1 and hTERT–HA reciprocally co-immunoprecipitated (Figure 3C). The interaction was not affected when we treated extracts with DNase I, RNase A or a combination of both before performing the IPs, although the treatments were effective (Figure 3D; Supplementary Figure S2). Altogether, these results suggest that nuclear UPF1 physically interacts with hTERT in a nucleic acid-independent manner. In addition, UPF1 and its cytoplasmic interacting partner UPF2 seem not to interact within the nucleus.
Figure 3.
Nuclear UPF1 physically interacts with telomerase. (A) Nuclear extracts prepared from HeLa cells were immunoprecipitated using antibodies directed against the indicated proteins or only with beads (b). In all, 2% of input and 33% of immunoprecipitated material were analysed by western blot using antibodies raised against UPF1 and UPF2. The asterisk indicates a crossreacting band revealed by the α-UPF2 antibody. (B) Immunoprecipitations were performed as in (A), and 0.5% or 0.1% of input and 3.5% of immunoprecipitated material were used in TRAP assays. Numbers express TRAP activity in the different samples as fold increase over beads. (C) Myc/His-tagged UPF1 and HA-tagged hTERT plasmids were transfected in HEK 293T cells in different combinations with the corresponding empty vector (EV) plasmids. Immunoprecipitations and western blots were performed with α-myc and α-HA antibodies. In all, 3% of input and 40% of immunoprecipitated material were used for western blots. The unrelated RBM25 protein was not detected in the immunoprecipitates. A long and a short exposure (exp) are shown for α-myc western blots. (D) HEK 293T cells were transfected with the indicated plasmids and extracts were left untreated or treated with DNase I, RNase A, or both enzymes simultaneously before α-myc immunoprecipitations. Ectopically expressed UPF1 and hTERT were detected as in (C). Numbers indicate the ratio between hTERT–HA and myc/His–UPF1 signals in each IP, after normalization through the ratio measured for the untreated sample. Experiments were repeated 2–4 times and only one representative experiment is shown.
Several PIKKs, including ATM, ATR and SMG1, phosphorylate UPF1 at S/T-Q residues (Yamashita et al, 2001; Brumbaugh et al, 2004; Azzalin and Lingner, 2006; Kashima et al, 2006). To test whether the phosphorylation state of UPF1 regulates its interaction with active telomerase, we performed α-UPF1 IPs using nuclear extracts from HEK 293T cells treated with the PIKK inhibitor caffeine. While caffeine treatment did not affect total telomerase activity measured in input samples nor the amount of immunoprecipitated UPF1, it reduced the levels of telomerase activity present in UPF1 immunoprecipitates by ∼10-fold (Figure 4A, C and D). Thus, one or more PIKKs could regulate the interaction between UPF1 and telomerase. We then performed α-UPF1 IPs using nuclear extracts from HEK 293T cells depleted for human ATR or SMG1 using previously validated shRNA plasmids (Azzalin and Lingner, 2006; Figure 4C). While SMG1 depletion did not affect significantly the levels of telomerase activity detected in UPF1 immunoprecipitates, ATR depletion reduced it by ∼10-fold (Figure 4B and D). Again, the levels of immunoprecipitated UPF1 and of input telomerase activity were not affected by ATR or SMG1 depletion (Figure 4B and C). Based on these results, we propose that ATR promotes the interaction of nuclear UPF1 with active telomerase. Nevertheless, we cannot exclude that chemical or shRNA-mediated inhibition of ATR could lead to accumulation of telomerase or TRAP inhibitors, specifically in the UPF1 immunoprecipitates.
Figure 4.
ATR promotes the interaction of nuclear UPF1 with active telomerase. (A, B) Nuclear extracts from HEK 293T cells left untreated or treated with caffeine were immunoprecipitated using antibodies against UPF1 or only with beads (b). TRAP experiments were performed using serial dilutions of inputs and immunoprecipitated material. (B) As in (A), except that extracts were prepared from HEK 293T cells transfected with shRNA plasmids directed against SMG1 or ATR or with empty vector plasmids (shEV). (C) Western blot analysis of UPF1, SMG1 and ATR in the indicated input or immunoprecipitated fractions. CENPA was used as a loading control. (D) Quantification of TRAP activity in the different samples after subtraction of the TRAP signal associated with the beads. Values are expressed as fold increase over untreated or shEV-transfected cells. Bars and error bars correspond to averages and s.d. from at least three independent experiments. P-values (two-tailed Student's t-test) are indicated for the relevant samples.
UPF1 physically interacts with the shelterin factor TPP1
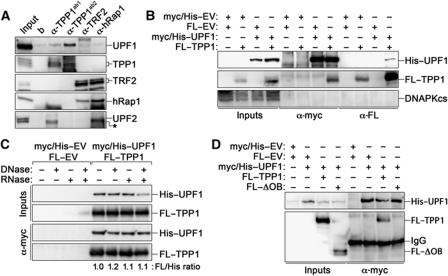
To test whether UPF1 interacts with known telomeric factors other than telomerase, we performed CoIP assays in HeLa nuclear extracts using antibodies against the endogenous shelterin factors TPP1, TRF2 and hRap1. Western blot analysis did not reveal any detectable UPF1 in immunoprecipitates obtained using antibodies against TRF2 or hRap1, although, as expected, the two proteins reciprocally co-immunoprecipitated (Figure 5A). On the contrary, immunoprecipitates obtained using two independent antibodies raised against TPP1 were enriched for endogenous UPF1 protein, as compared with α-TRF2 or α-hRap1 immunoprecipitates or with control immunoprecipitations performed using only beads. UPF2 was not detected in any of the α-TPP1 immunoprecipitates (Figure 5A). To further corroborate the physical interaction between UPF1 and TPP1, we transiently expressed myc/His–UPF1 and Flag-tagged TPP1 (FL–TPP1) alleles in HEK 293T cells and performed CoIP experiments with antibodies against either tag. Myc/His–UPF1 and FL–TPP1 reciprocally co-immunoprecipitated (Figure 5B). Treatment of nuclear extracts with DNase I, RNase A or a combination of both did not affect the ability of UPF1 to co-immunoprecipitate TPP1 (Figure 5C; Supplementary Figure S2), suggesting that the interaction is independent of nucleic acids. Next, we performed CoIP experiments using nuclear extracts from HEK 293T cells overexpressing myc/His–UPF1 in combination with a Flag-tagged version of TPP1 lacking its N-terminal oligonucleotide/oligosaccharide binding (OB)-fold domain (FL–ΔOB). Myc/His–UPF1 failed to co-immunoprecipitate FL–ΔOB (Figure 5D), indicating that the OB-fold of TPP1 mediates the physical interaction between UPF1 and TPP1.
Figure 5.
Nuclear UPF1 physically interacts with TPP1. (A) Nuclear extracts from HeLa cells were immunoprecipitated using antibodies against the indicated proteins or only with beads (b). Two independent α-TPP1 antibodies were used. In all, 3% of input and 40% of immunoprecipitated material were analysed by western blot. The asterisk indicates a crossreacting band revealed by the α-UPF2 antibody. (B) Myc/His-tagged UPF1 and Flag (FL)-tagged TPP1 plasmids were transiently transfected in HEK 293T cells in different combinations with empty vector (EV) plasmids. Immunoprecipitations and western blots were performed with α-myc and α-Flag antibodies. In all, 3% of input and 40% of immunoprecipitated material were used for western blots. The unrelated DNAPKcs protein was not detected in the immunoprecipitates. (C) HEK 293T cells were transfected with the indicated plasmids and extracts were left untreated or treated with DNase I, RNase A or both enzymes simultaneously before α-myc immunoprecipitations. Ectopically expressed UPF1 and TPP1 polypeptides were detected as in (B). Numbers indicate the ratio between FL–TPP1 and myc/His–UPF1 signals in each IP, after normalization through the ratio measured for the untreated sample. (D) Myc/His-tagged UPF1 was co-expressed in HEK 293T cells together with full-length FL–TPP1 or with a FL–TPP1 mutant lacking the N-terminal OB-fold (FL–ΔOB). Immunoprecipitations were performed with α-myc antibodies and western blot analysis was performed with α-myc and α-Flag antibodies. Experiments were repeated 2–4 times and only one representative experiment is shown.
UPF1 ATPase activity sustains leading-strand replication of telomeres
ShRNA-mediated depletion of UPF1 leads to telomeric dysfunctions including chromosomes with telomere-free ends (TFEs; Azzalin et al, 2007). To determine whether UPF1 enzymatic activities are essential to prevent telomere dysfunctions, we concomitantly transfected HeLa cells with shRNA vectors targeting the 3′ UTR of UPF1 (shUPF1C) and vectors carrying 3′ UTR-less cDNAs coding for N-terminally HA-tagged UPF1 proteins, either wild type (wt) or ATPase deficient (ATPD). Confirming its lack of enzymatic activity, HA–UPF1ATPD was unable to restore NMD of the PTC-containing reporter gene β-globinNS39 in cells depleted for endogenous UPF1 (Supplementary Figure S3). On the contrary, HA–UPF1wt fully complemented the NMD defect in the same setting (Supplementary Figure S3). Metaphase spreads were prepared 4 days after transfection, hybridized in situ using telomeric PNA probes and scored for the appearance of TFEs. The same spreads were also scored for the presence of so-called fragile telomeres (FTs), visualized as multiple or shredded telomeric signals thought to originate from defective semiconservative replication of telomeric DNA (Sfeir et al, 2009). Overexpression of HA–UPF1wt or ATPD in cells co-transfected with shEV plasmids did not alter TFE or FT frequencies as compared with control cells. On the contrary, UPF1 depletion led to accumulation of both TFEs and FTs over shEV-transfected controls (Figure 6A and B). Strikingly, ectopically expressed HA–UPF1wt, but not HA–UPF1ATPD, restored TFE and FT frequencies to normal levels in cells depleted for endogenous UPF1 (Figure 6A and B). HA–UPF1wt and HA–UPF1ATPD protein levels were comparable to those of endogenous UPF1 in shEV-transfected cells (Figure 6C). These results indicate that both FTs and TFEs are genuine outcomes of UPF1 depletion and that the ATPase activity of UPF1 is essential to prevent their emergence.
Figure 6.
The ATPase activity of UPF1 is required to sustain telomere replication. (A) Examples of telomeric DNA-FISH performed on chromosomes from HeLa cells co-transfected with shEV or shRNA plasmids against the 3′-UTR of UPF1 (shUPF1C) and with HA-tag empty vectors (HA–EV). Telomeric sequences are in green and DAPI-stained chromosomes in red. White and black arrowheads point to telomere-free ends (TFEs) and to fragile telomeres (FTs), respectively. Enlarged chromosomes at the bottom are from shUPF1C/HA–EV-transfected cells. (B) Quantifications of TFEs and FTs in HeLa cells transfected with the indicated plasmids. ATPD: ATPase deficient. Frequencies are expressed as numbers of TFE and FT per chromosome. Bars and error bars are averages and s.d. from two independent experiments and n indicates the cumulative number of scored chromosomes. P-values (two-tailed Student's t-test) are indicated for the relevant samples. (C) Western blot analysis of cells used for (B). α-UPF1 (to detect endogenous and ectopically expressed UPF1), α-HA (to detect ectopically expressed UPF1 only) and α-tubulin (loading control) antibodies were used. The two lower bands detected by the α-HA antibody in UPF1wt- and UPF1ATPD-transfected cells correspond to C-terminally truncated UPF1 proteins not detected by α-UPF1 antibody. (D) Examples of leading- and lagging-strand telomeres detected by CO-FISH analysis. Chromosomes are from HeLa cells transfected with shUPF1C plasmids. Leading-strand telomeres are in red; lagging-strand telomeres are in green; DAPI-stained chromosomes are in blue. White and black arrowheads point to a TFE and a FT, respectively. (E) Quantifications of leading- and lagging-strand TFEs and FTs per chromosome. Bars and error bars are averages and s.d. from three independent experiments and n indicates the cumulative number of scored chromosomes. P-values (two-tailed Student's t-test) are indicated for the relevant samples.
Given the presence of UPF1 at telomeres during S phase (Figure 1B), its ATR-dependent binding to telomeres (Figure 2B), its interaction with polymerase δ (Carastro et al, 2002; Azzalin and Lingner, 2006), as well as the early S-phase arrest (Azzalin and Lingner, 2006) and emergence of FTs and TFEs (Figure 6A) observed upon UPF1 depletion, we reasoned that UPF1 could sustain semiconservative telomere replication. We, therefore, performed chromosome-orientation fluorescence in situ hybridization (CO-FISH) (Bailey et al, 2001; Dimitrova and de Lange, 2009) on metaphases prepared from HeLa cells depleted for UPF1 using the same shUPF1C vector as in the above-described complementation experiment. CO-FISH analysis revealed that, in UPF1-depleted cells, both TFEs and FTs preferentially occurred at telomeres replicated by leading-strand DNA synthesis, although the lagging-strand telomeres were also slightly affected (Figure 6D and E). Because UPF1ATPD is completely unable to prevent the accumulation of TFEs and FTs in cells depleted for endogenous UPF1 (Figure 6A and B), we conclude that catalytically active UPF1 sustains telomere stability primarily by assuring completion of leading-strand telomere replication.
γ-H2AX and RPA32 accumulate at UPF1-depleted telomeres
The telomeric dysfunctions induced by UPF1 depletion imply activation of a DNA damage response at telomeres. Preliminary indirect immunofluorescence (IF) experiments performed on HeLa cells depleted for UPF1 did not consistently detect accumulation of γ-H2AX at telomeres (unpublished observations), possibly due to the limited length of telomeres in these cells and/or to the low number of telomeres becoming simultaneously dysfunctional within the same cell. This second possibility is supported by the relatively low frequencies of TFEs and FTs recorded in DNA-FISH experiments (see Figure 6). We, therefore, exploited human HeLa-derived cell lines (HeLa-E1) bearing very long (20–50 kb) telomeres owing to stable overexpression of both hTERT and the telomerase RNA moiety hTR (Cristofari and Lingner, 2006). We depleted these cells for UPF1 with two independent shRNA vectors (shUPF1A/B) and performed indirect IF using antibodies against γ-H2AX and the single-stranded DNA-binding protein subunit RPA32. Consistent with previously published data (Azzalin and Lingner, 2006), UPF1 depletion-induced accumulation of cells containing discrete nuclear γ-H2AX foci (Supplementary Figure S4). Interestingly, UPF1-depleted cells also accumulated discrete nuclear RPA32 foci (Supplementary Figure S4). Double staining with antibodies directed against γ-H2AX or RPA32 and the shelterin component hRap1 revealed that ∼33–35% of hRap1 foci co-localized with γ-H2AX foci in cells depleted for UPF1, as opposed to ∼12% in shEV-transfected cells (shEV: 12.5±6.1%; shUPF1A: 35.0±1.2%, P=0.0014; shUPF1B: 33.1±6.9%, P=0.0039; Figure 7A and B). Similarly, ∼27–37% of hRap1 foci co-localized with RPA32 foci in UPF1-depleted cells, as opposed to ∼8% in control cells (shEV: 8.3±4.1%; shUPF1A: 27.6±5.1%, P=0.0025; shUPF1B: 36.9±6.9%; P=0.0012; Figure 7A and B). These observations disclose that at least a subset of telomeres are recognized as sites of DNA damage when UPF1 is depleted and suggest accumulation of single-stranded telomeric DNA, possibly at the same telomeres carrying γ-H2AX. Notably, only a fraction of the γ-H2AX and RPA32 foci detected in UPF1-depleted cells co-localized with hRap1, indicating that telomeres are not the sole genomic regions experiencing DNA damage in these cells.
Figure 7.
Dysfunctional telomeres in cells depleted for UPF1. (A) HeLa-E1 cells transfected with the indicated shRNA plasmids were subjected to indirect immunofluorescence experiments 4 days after transfection. HRap1 is in green, γ-H2AX and RPA32 are in red and DAPI-stained DNA is in grey. Colocalization of green and red signals generates yellow signals in the merged panels (examples are indicated by the yellow arrows). The bottom panels show enlarged examples of signal colocalization from cells transfected with shUPF1B. (B) Quantifications of hRap1 foci colocalizing with γ-H2AX or with RPA32 foci in cells transfected with the indicated shRNA plasmids. Bars and error bars represent averages and s.d. from three independent experiments where a cumulative number of 300–650 hRap1 foci were scored. P-values (two-tailed Student's t-test) are indicated for the relevant samples.
UPF1 depletion affects neither shelterin composition nor the heterochromatic state of telomeres
Incomplete telomere replication occurs in mouse cells deficient for the shelterin component TRF1 (Sfeir et al, 2009). This raises the possibility that the telomere replication defects observed upon UPF1 depletion might derive from altered shelterin composition at telomeres. We depleted HeLa cells for UPF1 or UFP2 and analysed shelterin density at telomeres by ChIP using antibodies directed against each of the six shelterin factors TRF1, TRF2, TPP1, hRap1, POT1 and TIN2 (Palm and de Lange, 2008). UPF1 depletion did not alter total protein levels of any of the shelterin components, nor did it affect shelterin density at telomeres (Supplementary Figure S5). Similarly, depletion of UPF2 had no effect on shelterin abundance and binding to telomeres (Supplementary Figure S5). We also tested whether UPF1 depletion induces defective heterochromatin establishment at telomeres, which, in turn, could cause the observed telomeric defects. ChIP experiments performed with antibodies against different histone modifications (acetylated histone H3K9, trimethylated histone H3K9 and trimethylated histone H4K20) and against heterochromatin protein 1 (HP1) γ did not reveal any substantial alteration in their density at telomeres or at Alu repeats upon UPF1 depletion (Supplementary Figure S6A and B). Similarly, no substantial alteration of the total levels of these marks was detected in western blot experiments (Supplementary Figure S6C). Finally, UPF1 depletion did not compromise the methylation state of subtelomeric CpG islands associated with TERRA promoters (Nergadze et al, 2009; Supplementary Figure S6D). We conclude that, at least in our experimental conditions, UPF1 depletion impacts neither on shelterin composition nor on the heterochromatic state of telomeres. In addition, canonical NMD appears not to regulate the cellular abundance of shelterin components and of the tested heterochromatic marks.
Discussion
Reinforcing our previously published data on the in vivo binding of UPF1 to telomeres (Azzalin et al, 2007), we show that UPF1 is present at telomeres at least in S and G2/M and that UPF1 binding to telomeres depends on telomere length and ATR. Yet, UPF1 co-immunoprecipitates only a minor fraction of telomeric DNA, similarly to several non-shelterin proteins involved in DNA replication and repair at telomeres (Verdun et al, 2005; Verdun and Karlseder, 2006). The ATR dependency of UPF1 binding to telomeres as well as the presence of UPF1 at telomeres during S phase suggests a function for UPF1 in telomere replication. After all, ATR is essential for genome maintenance during S phase, likely by stabilizing and restarting replication forks stalled at genomic regions difficult to replicate, including telomeres (Brown and Baltimore, 2003; Cimprich and Cortez, 2008; Martínez et al, 2009; McNees et al, 2010). In addition, ATR is found at telomeric chromatin during S phase (Verdun and Karlseder, 2006). The molecular mechanism used by ATR to promote UPF1 binding to telomeres remains to be clarified. Nevertheless, ATR directly phosphorylates UPF1 in vitro and sustains UPF1 phosphorylation in vivo (Azzalin and Lingner, 2006). Moreover, chromatin-bound UPF1 is hyperphosphorylated as compared with its soluble counterpart (Azzalin and Lingner, 2006). It is, therefore, likely that ATR-mediated phosphorylation of UPF1 is necessary for UPF1 loading onto or stabilization at telomeres.
We have also revealed an intriguing connection between UPF1 and two major proteins involved in telomere stability processes, TPP1 and telomerase. As is the case for UPF1, TPP1 itself has been linked to both ATR and telomerase functions at telomeres. Functional inactivation of mammalian TPP1 leads to an ATR-dependent telomeric DNA damage response, due to impaired POT1 recruitment to telomeres, and to rare telomeric aberrations (Liu et al, 2004; Ye et al, 2004; Kibe et al, 2010; Tejera et al, 2010). Based on these observations, it would be tempting to speculate that TPP1 recruits UPF1 to telomeres and that the ATR-dependent DNA damage and the telomeric instability observed in TPP1-deficient cells could stem in part from defective association of UPF1 with telomeres. However, ChIP experiments performed in cells depleted for TPP1 failed to reveal any consistent alteration in the amounts of UPF1 bound to telomeric chromatin, although a telomeric DNA damage response was activated in the same cells (unpublished observations). Therefore, more sophisticated hypotheses need to be conceived. TPP1 directly interacts with and recruits telomerase to telomeres (Wang et al, 2007; Abreu et al, 2010; Tejera et al, 2010). Interestingly, TPP1 interacts with UPF1 and telomerase through the same OB-fold domain (Abreu et al, 2010), suggesting a competitive interaction mode. The recruitment of UPF1 to telomeres could coincide with its association with TPP1 and consequent release of TPP1-bound telomerase, thus raising the possibility that UPF1 might negatively regulate telomerase function at telomeres. On the other hand, the ATR-regulated interaction of UPF1 itself with telomerase might reflect a coordination of the telomeric functions exerted by the two enzymes during S phase (see model in Figure 8). Further adding to the complexity of this model, results published by independent laboratories indicate that hEST1A directly interacts with both UPF1 and telomerase, likely through independent domains on hEST1A (Fukuhara et al, 2005; Redon et al, 2007). It is, therefore, possible that hEST1A physically mediates the interaction of UPF1 and telomerase. Alternatively, hEST1A, as well as ATR, could influence this interaction by regulating the phosphorylation status of UPF1 (see model in Figure 8).
Figure 8.
Speculative model on UPF1 association and function at telomeres. The white arrows indicate the direction of replication fork progression. The black arrow indicates the ATR-mediated regulation of UPF1. The black inhibitory symbol indicates the UPF1-mediated displacement of TERRA from telomeres. The dark region on TPP1 indicates the OB-fold domain that mediates the interaction with UPF1 and with telomerase. The question mark indicates one or more so far unknown factors possibly mediating the interaction between UPF1 and TPP1. See text for details.
While a participation of UPF1 in telomere replication was previously speculated upon (Chawla and Azzalin, 2008), we now validate this hypothesis for the first time and offer a mechanistic explanation for the telomere instability observed upon functional inactivation of UPF1. Our CO-FISH experiments strongly suggest that leading-strand replication is severely impaired in the absence of UPF1. Because an interaction between UPF1 and the lagging-strand polymerase δ has previously been documented (Carastro et al, 2002; Azzalin and Lingner, 2006), we were surprised that lagging-strand telomere synthesis was only marginally affected in UPF1-depleted cells. Nevertheless, a putative interaction of UPF1 with the leading-strand replication machinery has so far not been directly tested and cannot be ruled out. In addition, the milder defect in lagging-strand replication observed upon UPF1 depletion could also reflect more efficient repair mechanisms or the existence of redundant helicase activities, including that of Werner syndrome protein (Crabbe et al, 2004). In any case, in UPF1-depleted cells, stalling of the replication fork on leading-strand telomeres seems to be the most significant defect leading to the observed FTs, which are expected to contain γ-H2AX and RPA32 (Ward and Chen, 2001; Zou and Elledge, 2003; Sfeir et al, 2009). FTs could then degenerate into TFEs upon replication fork collapse and DNA double-strand breakage.
The replication defects induced by UPF1 depletion are not secondary to altered shelterin composition or heterochromatinization of telomeres. Hence, how does UPF1 activity sustain replication of the telomere? Since the ATPase activity of UPF1 is required for its 5′-to-3′ helicase activity (Bhattacharya et al, 2000), UPF1 could work as a canonical replicative helicase in front of the replication fork by unwinding the double-stranded telomeric DNA or by stripping telomeric proteins off the DNA. Nevertheless, the dramatic accumulation of telomere-bound TERRA molecules induced by UPF1 depletion (Azzalin et al, 2007) rather suggests that improper displacement of TERRA from a replicating telomere might induce the observed telomere instability. This notion is further substantiated by recent observations from independent laboratories, indicating that TERRA levels are progressively downregulated throughout S and G2/M phases (Porro et al, 2010; Flynn et al, 2011). A recent report from the Lykke-Andersen laboratory elegantly demonstrated that UPF1 ATPase activity is essential for NMD completion by allowing disassembly of cytoplasmic ribonucleoprotein complexes containing non-sense mRNAs (Franks et al, 2010). The ATPase (and perhaps helicase) activity of telomere-associated UPF1 could perform similarly by freeing TERRA from telomeric proteins such as TRF1 and TRF2, which have been shown to interact with TERRA molecules in human cultured cells and in vitro (Deng et al, 2009), thus enabling TERRA displacement and perhaps degradation during S phase (see model in Figure 8). In UPF1-depleted cells, the lack of a timely removal of TERRA molecules from a replicating telomere would allow TERRA G-rich telomeric repeats to hybridize with the single-stranded C-rich telomeric DNA, thereby specifically concealing the template strand for leading-strand telomere synthesis. Alternatively, the aberrant accumulation of TERRA molecules at replicating telomeres may restrain heterogeneous nuclear ribonucleoprotein A1 from regulating the RPA-to-POT1 switch on single-stranded DNA at the end of S phase (Flynn et al, 2011).
In conclusion, we have uncovered so far unappreciated molecular mechanisms underlying the protective role of UPF1 in maintaining the stability of human telomeres. By doing so, we have added a new member to the team of players essential to guarantee faithful replication of the telomere. We have also shown that ATR has a crucial role in regulating UPF1-associated functions at telomeres and that TPP1 and telomerase enter the game as well, although through avenues that still need to be fully illuminated. It will be fascinating to thoroughly dissect the common pathways followed by these different factors to protect the integrity of one of the most essential and delicate structures of our chromosomes, the telomere.
Materials and methods
Cell lines, cell culture procedures and cell-cycle analysis
HeLa, HeLa-E1 (Cristofari and Lingner, 2006), HeLa β-globinNS39 (a kind gift from Oliver Mühlemann) and HEK 293T cells were cultured in high-glucose D-MEM supplemented with 10% v/v bovine calf serum and non-essential amino acids (Invitrogen). Transfections were performed using Lipofectamine 2000 reagent (Invitrogen). For CoIP experiments, plasmids were transfected in a ratio of 1:1. For rescue experiments, shRNA plasmids and UPF1 expression plasmids were transfected in a ratio of 1:4. Retroviruses were produced in HEK 293T cells according to the standard procedures. Twenty-four hours after transfection or infection, cells were trypsinized and passed into medium containing 1.5 μg/ml puromycin, 10 μg/ml blasticidin and/or 200 μg/ml hygromycin B and cultured for at least three additional days. Selective medium was replaced with normal medium 24 h before performing the experiments. Where indicated, 10 mM caffeine (Sigma-Aldrich) was added to the cells for 4 h before harvesting. For cell-cycle synchronization, cells were incubated in medium containing 1 μg/ml aphidicolin for 24 h and released in normal medium after two washes with warm 1 × PBS. FACS analysis was performed as previously described (Azzalin and Lingner, 2006).
Protein and ChIPs
Cells were harvested and incubated in cytoplasm lysis buffer (50 mM Tris–HCl (pH 8.0), 140 mM NaCl, 1.5 mM MgCl2, 0.5% v/v Nonidet P-40) for 5 min on ice. Nuclei were centrifuged at 2000 r.p.m. for 3 min and resuspended in nuclei lysis buffer (50 mM Tris–HCl (pH 7.4), 1 mM EDTA, 400 mM NaCl, 1% v/v Triton X-100, 0.1–0.3% w/v SDS) supplemented with 1 mM DTT, protease inhibitors and phosphatase inhibitors (Sigma-Aldrich). After 30 min on ice, lysates were mixed 1:1 with cold water and centrifuged at 14 000 r.p.m. for 10 min. Where indicated, extracts were incubated with 83 U/ml DNase I (New England Biolabs) and/or 0.1 mg/ml RNase A (Roche) at room temperature (RT) for 30 min. For immunoprecipitations, extracts were incubated with 2–6 μg of antibody at 4°C for 5 h. In all, 25 μl of protein A/G beads was added and samples were left overnight at 4°C with constant rotation. Beads were washed four times with 1:1 diluted nuclei lysis buffer. For western blot analysis, samples were boiled in 2 × Laemmli buffer at 99°C for 10–20 min and analysed by SDS–PAGE immunoblotting. For DNase I and RNase A treatment controls, DNA and RNA were extracted from input fractions and subjected to PCR and RT–PCR experiments. For DNase controls, we amplified DNA fragments corresponding to sequences from human XpYp (oligonucleotides: 5′-TTGTCTCAGGGTCCTAGTG-3′ and 5′-TCTGAAAGTGGACCTATCAG-3′) and 17p (oligonucleotides: 5′-GAATCCACGGATTGCTTTGTGTACTT-3′ and 5′-CCTCAGCCTCTCAACCTGCTTGG-3′) subtelomeres. For RNase controls, we reverse transcribed RNA with random hexamer oligonucleotides and amplified DNA fragments corresponding to sequences from human ARF1 (oligonucleotides: 5′-GGAAACCGTGGAGTACAAGAAC-3′ and 5′-GGAGTACCAGAAACTGCCTCAT-3′) and CHK1 (oligonucleotides: 5′-GATTCTGCTCCTCTAGCTCTGC-3′ and 5′-CAATCCATCACCCTTAGAAAGC-3′) mRNAs. TRAP and ChIP assays were performed as described previously (Reichenbach et al, 2003; Azzalin et al, 2007). Radioactive signals were detected using a Storm PhosphoImager (GE Healthcare) and quantified with Quantity One software (Bio-Rad). For ChIP analysis, the signal associated with negative control samples (only beads or pre-immune serum) was subtracted from the one obtained for each experimental sample. Immunoprecipitated DNA in each sample was expressed as fraction of the corresponding input DNA. For statistical analysis of TRAP and ChIP experiments, averages, standard deviations and P-values (two-tailed Student's t-test) were calculated using the Microsoft Excel software.
Indirect IF, telomeric DNA-FISH and CO-FISH
For indirect IF, cells grown on glass coverslips were fixed in methanol for 15 min at −20°C. Cells were incubated in blocking solution (5% w/v BSA, 0.05% v/v Tween-20, 1 × PBS) for 45 min at RT and successively with primary antibodies diluted in blocking solution for 50 min at RT. After extensive washes, cells were incubated with secondary antibodies conjugated to Alexa Fluor 488 or Alexa Fluor 568 (Invitrogen) for 45 min at RT. Nuclei were stained with 100 ng/ml DAPI (Sigma-Aldrich) in 1 × PBS and images were acquired and analysed using the LMS510 confocal microscope and software (Zeiss) and Adobe Photoshop. DNA-FISH was performed as previously described (Azzalin et al, 2007) using Cy3-OO-(CCCTAA)5 PNA probes (Panagene). Images were acquired using the Axiovert 200M epifluorescence microscope (Zeiss). Strand-Specific CO-FISH was performed as previously described (Dimitrova and de Lange, 2009) using the following PNA probes: Fluorescein-OO-(CCCTAA)3 and TAMRA (6-carboxytetramethylrhodamine)-OO-(TTAGGG)3 (Bio-Synthesis Inc) or Cy3-OO-(CCCTAA)5, and Alexa488-OO-(TTAGGG)5 (Panagene). After hybridization and washes, chromosomes were stained with DAPI and imaged using a Zeiss Axio-Imager Z1 microscope and the Metasystems Isis Software (Zeiss). For statistical analysis of IF and FISH results, averages, standard deviations and P-values (two-tailed Student's t-test) were calculated using the Microsoft Excel software.
Supplementary Material
Acknowledgments
We thank J Lingner, O Mühlemann, T de Lange and H-M Jäck for reagents and the ETHZ Light Microscopy Centre (LMC) for microscope services. We are also grateful to the members of the Azzalin laboratory and to J Lingner for helpful discussions. CMA's laboratory is funded by ETH Zürich (ETH-15 08-1 and ETH-03 08-3), the Swiss National Science Foundation (3100A0-120090 and PP00P3-123356), the European Research Council (BFTERRA) and Fondazione Cariplo (2008-2507). SG's laboratory is supported by the Biomedical Research Foundation of the Academy of Athens (BRFAA) and the General Secretariat of Research and Technology/Greek Ministry of Development (05NON-EU-449). RC received a fellowship from the Swiss National Science Foundation (323500-115044). SR was supported in part by the Swiss National Science Foundation in the laboratory of J Lingner at EPFL.
Author contributions: CMA, RC and SG designed the experiments. RC, CMA, SR, CR and HW performed the experiments. CMA, RC, SR, CR and SG analysed the data. CMA and RC wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP (2010) TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol 30: 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Lingner J (2006) The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol 16: 433–439 [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Lingner J (2008) Telomeres: the silence is broken. Cell Cycle 7: 1161–1165 [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Nergadze SG, Giulotto E (2001) Human intrachromosomal telomeric-like repeats: sequence organization and mechanisms of origin. Chromosoma 110: 75–82 [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801 [DOI] [PubMed] [Google Scholar]
- Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH (2001) Strand-specific postreplicative processing of mammalian telomeres. Science 293: 2462–2465 [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW (2000) Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6: 1226–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D (2003) Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev 17: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbaugh KM, Otterness DM, Geisen C, Oliveira V, Brognard J, Li X, Lejeune F, Tibbetts RS, Maquat LE, Abraham RT (2004) The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell 14: 585–598 [DOI] [PubMed] [Google Scholar]
- Carastro LM, Tan C-K, Selg M, Jack H-M, So AG, Downey KM (2002) Identification of delta helicase as the bovine homolog of HUPF1: demonstration of an interaction with the third subunit of DNA polymerase delta. Nucleic Acids Res 30: 2232–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla R, Azzalin CM (2008) The telomeric transcriptome and SMG proteins at the crossroads. Cytogenet Genome Res 122: 194–201 [DOI] [PubMed] [Google Scholar]
- Chiu S-Y, Serin G, Ohara O, Maquat LE (2003) Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA 9: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter CM, Hahn WC, Wei W, Caddle SD, Beijersbergen RL, Lansdorp PM, Sedivy JM, Weinberg RA (1998) Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc Natl Acad Sci USA 95: 14723–14728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe L, Verdun RE, Haggblom CI, Karlseder J (2004) Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 306: 1951–1953 [DOI] [PubMed] [Google Scholar]
- Cristofari G, Lingner J (2006) Telomere length homeostasis requires that telomerase levels are limiting. EMBO J 25: 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM (2009) TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell 35: 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, de Lange T (2009) Cell cycle-dependent role of MRN at dysfunctional telomeres: ATM signaling-dependent induction of nonhomologous end joining (NHEJ) in G1 and resection-mediated inhibition of NHEJ in G2. Mol Cell Biol 29: 5552–5563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Mühlemann O, Jensen TH (2009) SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55 [DOI] [PubMed] [Google Scholar]
- Flynn RL, Centore RC, O’Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L (2011) TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 471: 532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Singh G, Lykke-Andersen J (2010) Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense- mediated mRNA decay. Cell 143: 938–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E (2005) SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 17: 537–547 [DOI] [PubMed] [Google Scholar]
- Hug N, Lingner J (2006) Telomere length homeostasis. Chromosoma 115: 413–425 [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Saulière J, Izaurralde E (2008) SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14: 2609–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra FJ, Escargueil AE, Kwek KY, Akoulitchev A, Cook PR (2004) Molecular cross-talk between the transcription, translation, and nonsense-mediated decay machineries. J Cell Sci 117: 899–906 [DOI] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S (2006) Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibe T, Osawa GA, Keegan CE, de Lange T (2010) Telomere protection by TPP1 is mediated by POT1a and POT1b. Mol Cell Biol 30: 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Safari A, O’Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z (2004) PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol 6: 673–680 [DOI] [PubMed] [Google Scholar]
- Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J (2008) The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell 32: 465–477 [DOI] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW (1989) A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643 [DOI] [PubMed] [Google Scholar]
- Maquat LE, Gong C (2009) Gene expression networks: competing mRNA decay pathways in mammalian cells. Biochem Soc Trans 37: 1287–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez P, Thanasoula M, Munoz P, Liao C, Tejera A, McNees C, Flores JM, Fernández-Capetillo O, Tarsounas M, Blasco MA (2009) Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev 23: 2060–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNees CJ, Tejera AM, Martínez P, Murga M, Mulero F, Fernandez-Capetillo O, Blasco MA (2010) ATR suppresses telomere fragility and recombination but is dispensable for elongation of short telomeres by telomerase. J Cell Biol 188: 639–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nergadze S, Farnung BO, Wischnewski H, Khoriauli L, Vitelli V, Chawla R, Giulotto E, Azzalin CM (2009) CpG-island promoters drive transcription of human telomeres. RNA 15: 2186–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Mühlemann O (2010) Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci 67: 677–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page MF, Carr B, Anders KR, Grimson A, Anderson P (1999) SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol 19: 5943–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T (2008) How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Porro A, Feuerhahn S, Reichenbach P, Lingner J (2010) Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol Cell Biol 30: 4808–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon S, Reichenbach P, Lingner J (2007) Protein RNA and protein protein interactions mediate association of human EST1A/SMG6 with telomerase. Nucleic Acids Res 35: 7011–7022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach P, Höss M, Azzalin CM, Nabholz M, Bucher P, Lingner J (2003) A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol 13: 568–574 [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA (2008) Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 10: 228–236 [DOI] [PubMed] [Google Scholar]
- Sfeir AJ, Kosiyatrakul ST, Hockemeyer D, Macrae SL, Karlseder J, Schildkraut CL, de Lange T (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Jakob S, Kleedehn MG, Lykke-Andersen J (2007) Communication with the exon-junction complex and activation of nonsense-mediated decay by human Upf proteins occur in the cytoplasm. Mol Cell 27: 780–792 [DOI] [PubMed] [Google Scholar]
- Snow BE, Erdmann N, Cruickshank J, Goldman H, Gill RM, Robinson MO, Harrington L (2003) Functional conservation of the telomerase protein Est1p in humans. Curr Biol 13: 698–704 [DOI] [PubMed] [Google Scholar]
- Tejera AM, Stagno d’Alcontres M, Thanasoula M, Marion RM, Martinez P, Liao C, Flores JM, Tarsounas M, Blasco MA (2010) TPP1 is required for TERT recruitment, telomere elongation during nuclear reprogramming, and normal skin development in mice. Dev Cell 18: 775–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun RE, Crabbe L, Haggblom C, Karlseder J (2005) Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell 20: 551–561 [DOI] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J (2006) The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 127: 709–720 [DOI] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M (2007) The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445: 506–510 [DOI] [PubMed] [Google Scholar]
- Ward IM, Chen J (2001) Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem 276: 47759–47762 [DOI] [PubMed] [Google Scholar]
- Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, Kurata R, Hirano H, Anderson P, Ohno S (2009) SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev 23: 1091–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S (2001) Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev 15: 2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye JZ-S, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T (2004) POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev 18: 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.