Abstract
At the acidic pH of the trans-Golgi and Weibel–Palade bodies (WPBs), but not at the alkaline pH of secretion, the C-terminal ∼1350 residues of von Willebrand factor (VWF) zip up into an elongated, dimeric bouquet. Six small domains visualized here for the first time between the D4 and cystine-knot domains form a stem. The A2, A3, and D4 domains form a raceme with three pairs of opposed, large, flower-like domains. N-terminal VWF domains mediate helical tubule formation in WPBs and template N-terminal disulphide linkage between VWF dimers, to form ultralong VWF concatamers. The dimensions we measure in VWF at pH 6.2 and 7.4, and the distance between tubules in nascent WPB, suggest that dimeric bouquets are essential for correct VWF dimer incorporation into growing tubules and to prevent crosslinking between neighbouring tubules. Further insights into the structure of the domains and flexible segments in VWF provide an overall view of VWF structure important for understanding both the biogenesis of ultralong concatamers at acidic pH and flow-regulated changes in concatamer conformation in plasma at alkaline pH that trigger hemostasis.
Keywords: biosynthesis, endothelium, platelets, shear flow, Weibel–Palade bodies
Introduction
Von Willebrand factor (VWF) has a central role in hemostasis and thrombosis, particularly in the rapid flow of the arteriolar circulation (Wagner, 1990; Sadler, 1998, 2005; Ruggeri and Mendolicchio, 2007). Different domains within VWF have binding sites for clotting factor VIII, collagen, platelet glycoprotein Ib (GPIb), and platelet integrin αIIbβ3 (Figure 1A). VWF multimers are organized as long linear concatamers, in which each monomer disulphide bonds head-to-head at its N-terminus, and tail-to-tail at its C-terminus, with adjacent monomers. A cleavage site for ADAMTS13 (a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13) enables trimming of VWF multimers after secretion. Longer VWF multimers have far greater hemostatic potency than shorter multimers. These differences relate to both the multivalent nature of VWF binding to platelets and vessel walls, and activation of adhesiveness by hydrodynamic force, which scales with the square of the number of VWF monomers (Zhang et al, 2009b). Mutations in VWF in von Willebrand disease (VWD) are the most common cause of heritable bleeding disorders, and often result in a change in the size distribution of circulating multimers.
Figure 1.
VWF schematics and fragments. (A) Sequence homology regions in VWF (Sadler et al, 1985). Cysteines are shown as vertical lines and are connected for chemically determined disulphide bonds (Marti et al, 1987; Katsumi et al, 2000). Longer lines show cysteines in intermonomer disulphides. N- and O-linked glycans are shown as closed and open lollipops, respectively (Titani et al, 1986). (B) Schematic of proVWF dimer at neutral pH. The length of the mature VWF dimer of 120 nm is from Fowler et al (1985). (C, D) Schematics of VWF assembly based on previous work (Huang et al, 2008). (D) Prior to the discovery of dimeric bouquets at trans-Golgi pH, it was unclear how physiologic crosslinking (panel 1) occurred in the absence of intradimer crosslinks (panel 3) or crosslinking between neighbouring VWF tubules that are sufficiently close for a dimer to span between them (panel 2). (E) VWF constructs (left) and their products (right). V8–CK is obtained from V8 protease digestion of A3–CK or D4–CK. (F) Reduced and non-reduced SDS 4–15% gradient PAGE of purified fragments, with Coomassie blue staining.
Early metal-shadowing electron microscopy (EM) experiments visualized the head-to-head and tail-to-tail arrangement of VWF monomers within concatamers (Fowler et al, 1985; Slayter et al, 1985). Monomers, ∼60 nm long, contain two large and one small globule in the N-terminal 22 nm portion and a rod and small final globule in the C-terminal 38 nm portion (Fowler et al, 1985). Both segments are highly flexible (Fowler et al, 1985; Slayter et al, 1985; Siedlecki et al, 1996). Subsequent higher resolution negative stain EM showed that the N-terminal D1, D2, and D′D3 domains are globular when assembled in helices that resemble VWF stored in Weibel–Palade bodies (WPBs) (Huang et al, 2008). Crystal structures of individual A1, A2, and A3 domains show each is globular (Bienkowska et al, 1997; Huizinga et al, 1997; Celikel et al, 1998; Emsley et al, 1998; Zhang et al, 2009a). However, the flexibility and distance between these domains in the N-terminal portion of VWF outside of helical assemblies remain to be defined. More N-terminal globular domains (five) are predicted than the two seen in early EM work (Fowler et al, 1985; Slayter et al, 1985). Moreover, the overall shape and flexibility of the domains in the C-terminal two-thirds of mature VWF remain to be defined by modern negative stain EM particle alignment and averaging methods. VWF transitions from a paracrystalline, highly ordered configuration when stored in WPB (Valentijn et al, 2008; Berriman et al, 2009) to an unordered configuration in the circulation. Within the vasculature, VWF transitions from a compact fuzz ball to an elongated, more active state in increased shear flow and in elongational flow (Slayter et al, 1985; Siedlecki et al, 1996; Schneider et al, 2007; Zhang et al, 2009b; Sing and Alexander-Katz, 2010). Therefore, characterization of the overall shape and flexibility of VWF is important to understand its function.
VWF is the largest known soluble vertebrate protein, ranging in size up to ∼50 × 106 Mr. This unusual size poses difficulties in biosynthesis, storage, and secretion, which are overcome by biological specializations (Wagner, 1990; Zenner et al, 2007; Valentijn et al, 2008; Berriman et al, 2009). VWF glycoprotein is biosynthesized as a preproprotein; the signal sequence is removed during translocation into the endoplasmic reticulum (ER), while cleavage by furin, to generate the propiece and mature fragments (Figure 1A), occurs much later. In the ER, most disulphide bonds are formed, and proVWF is dimerized through formation of intersubunit disulphide bonds in the C-terminal cystine-knot (CK) domain (Figure 1A). ProVWF dimers are then transported to the lower pH environments of the trans-Golgi network (TGN) and nascent WPBs, where VWF is assembled into a helical structure in the tubules characteristic of WPBs (Zenner et al, 2007). Multiple tubules aligned in parallel in WPBs give these organelles their unique cigar-like shape. Individual tubules often extend the entire length of the WPB (Berriman et al, 2009).
The last steps in VWF biosynthesis, interdimer N-terminal disulphide bond formation and cleavage by furin, appear to occur during or after assembly of individual VWF dimers onto the growing ends of tubules in nascent WPBs (Voorberg et al, 1990; Wagner, 1990; Vischer and Wagner, 1994; Haberichter et al, 2000; Rosenberg et al, 2002). Assembly into tubules is required for N-terminal disulphide bond formation, but disulphide bond formation is not required for tubule assembly. The propiece is required for helical assembly and remains associated until secretion; furin cleavage occurs late in biosynthesis, and is not required for assembly.
A series of elegant mutational, cell biological, and EM studies have shown that the propiece and N-terminal D′D3 domains in mature VWF (Figure 1A) are sufficient for assembly into helical tubules that recapitulate the helical structure of the tubules in intact WPBs (Voorberg et al, 1990; Wagner et al, 1991; Huang et al, 2008). Assembly requires Ca2+ and a low pH environment as found in the trans-Golgi and WPBs, and does not occur at neutral pH as found in plasma. From the ability of the D1–D2 propiece but not D′D3–A1 monomeric fragments to dimerize in Ca2+ and low pH, and from the arrangement of globular domains in EM 3D reconstructions of helical tubules, the organization of VWF dimers in WPB tubules has been deduced (Huang et al, 2008) (Figure 1D, panel 1). Based on the studies with purified D1–D2 fragments, homotypic interactions between the D1 and D2 domains are thought to mediate non-covalent dimerization of the N-terminal ends of proVWF dimers (Figure 1C). Assembly of these dimers onto the end of a growing helical tubule places the D′D3 domains within a dimer 13 nm apart from one another, and places one D′D3 domain adjacent to one D′D3 domain from a previously assembled dimer (Figure 1D, panel 1). Helical assembly thus provides a template for disulphide bond formation during N-terminal concatamerization, prevents N-terminal disulphide linkages between monomers within the same dimer (Figure 1D, panel 3), and helps to ensure that covalent linkage between VWF monomers is co-linear with assembly into helical tubules. This mechanism enables a 50-fold compaction in length of tubules compared with extended, secreted VWF (Huang et al, 2008). Orderly coiling of VWF in tubules in WPBs is important for rapid uncoiling during secretion, and prevention of tangling during release of long VWF filaments from endothelial cells that catch platelets at sites of hemostasis (Michaux et al, 2006).
Despite these major advances, important questions remain concerning VWF storage and secretion. What happens to the large C-terminal portion of VWF dimers during helical tubule assembly and secretion? EM results suggest that multiple tubules are assembled simultaneously in one WPB, with their growing ends close to one another at the end of the nascent WPB adjacent to the TGN (Zenner et al, 2007). In crosssections of nascent WPB, the average centre-to-centre spacing of tubules is ∼80 nm (Zenner et al, 2007), but is 28.4±3 nm in mature WPBs (Berriman et al, 2009). These spacings are smaller than that between the D′D3 domains in a mature VWF dimer of ∼120 nm (Fowler et al, 1985). Thus, a VWF dimer could easily span between two nascent tubules and crosslink them (Figure 1D, panel 2). The mechanisms that prevent tangling of VWF within a growing tubule and crosslinking of neighbouring tubules during VWF biosynthesis are incompletely understood.
The secretion process is also incompletely understood. The first step is formation of a narrow constriction between the plasma membrane and the WPB, which then experiences a rapid rise in pH, and a great expansion of width and increase in average spacing between the tubules, which nonetheless retain almost identical helical packing (Berriman et al, 2009). Because VWF is by far the predominant protein constituent of WPBs (Ewenstein et al, 1987), a property of VWF protein itself might provide an explanation for this pH-associated expansion.
Here, we use negative stain EM to study the size, spacing, and arrangement of domains in VWF. We have discovered a pH-regulated organization into a dimeric bouquet of the domains in the C-terminal two-thirds of mature VWF dimer that has important implications for VWF tubule biogenesis and secretion.
Results
Recombinant VWF fragments
N-terminal disulphide linkages of VWF in the D′D3 domain and C-terminal dimerization in the CK domain (Figure 1A) are independent (Sadler, 1998). Therefore, we made a series of overlapping N- and C-terminal fragments (Figure 1E) that could be structurally characterized and then conceptually overlapped to build up a complete picture of the organization of multimeric VWF. All fragments were expressed in HEK293S Gnt1− cells, purified using C-terminal (His)6 tags, and subjected to reduced and non-reduced SDS 4–15% gradient PAGE (Figure 1F). Fragments are named after their N- and C-terminal domains. Assembly of VWF in helices and dimerization in the D′D3 domain requires D1–D2, that is, the prodomain. Therefore, the D1–A1 construct is dimerized in the D′D3 moiety owing to the presence of the D1 and D2 domains (Figure 1E). Furin cleavage during biosynthesis generates His-tagged D′D3–A1 dimer and untagged D1–D2. In contrast, the D′D3–A2 construct generates D′D3–A2 monomer owing to the absence of D1–D2 (Figure 1E and F). The C-terminal fragments A1–CK, A3–CK, and D4–CK are all produced as dimers through disulphide linkage in their CK domains (Figure 1E and F). Each purified fragment is a single peak in gel-filtration. The D4–CK fragment was further cleaved by V8 protease into V8–CK and D4–V8 fragments (Figure 1E).
The C-terminal portion of VWF is flexible and extended at neutral pH
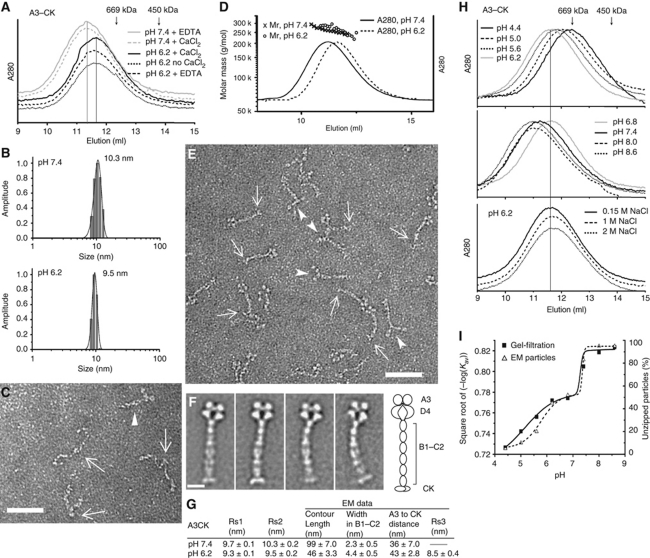
The A3–CK dimer has a theoretical polypeptide chain mass of 248 370 Da. With 14 N-linked and 2 O-linked glycans per dimer (Titani et al, 1986) and assuming complete occupancy of these sites and two GlcNAc and seven mannose per high mannose N-linked site, and four hexoses per O-linked site, the estimated glycoprotein mass is 273 600 Da. However, Superose 6 gel-filtration showed elution prior to the position of thyroglobulin (669 000 Da) (Figure 2A) and a Stokes radius of 9.7 nm at pH 7.4. Furthermore, dynamic light scattering (DLS) showed a Stokes radius of 10.3 nm (Figure 2B). These results suggested an asymmetric, extended conformation. Peak fractions from gel-filtration at pH 7.4 were subjected to negative stain EM (Figure 2C). Particles showed a dimeric organization, with arms of equal length joined at a central globule (arrows, Figure 2C). The arms extending from the central globule were flexible thin rods and ended in thicker, globular regions. These results were in excellent agreement with previous EM studies on the repeating C-terminal dimeric unit within intact VWF at neutral pH (Fowler et al, 1985; Slayter et al, 1985).
Figure 2.
pH-dependent conformational change of the A3–CK VWF fragment. (A) Superose 6 gel-filtration of A3–CK in the indicated conditions. (B) DLS profiles of A3–CK in 20 mM HEPES pH 7.4, 150 mM NaCl, 5 mM EDTA, or 20 mM BisTris pH 6.2, 150 mM NaCl, 10 mM CaCl2. (C) Raw image of negatively stained A3–CK in 20 mM HEPES pH 7.4, 150 mM NaCl. Arrows point to the central globule (CK domain) connecting the dimeric arms. Arrowhead shows a loosely associated stem-like structure. (D) Absolute mass of A3–CK measured by MALS of material eluting from Superose 6 gel-filtration in the indicated buffers. (E) Raw EM image of A3–CK in 20 mM BisTris pH 6.2, 150 mM NaCl, 10 mM CaCl2. Arrows mark the CK domain of typical particles with dimeric bouquets containing a head with four compact globular domains. Arrowheads mark the heads of occasional dimeric bouquets in which the globular head domains are separated. Scale bar is 50 nm. (F) Representative class averages of A3–CK at pH 6.2. Scale bar is 10 nm. Schematic is to right. (G) A3–CK size estimates from different methods. Rs1 and Rs2 are Stokes radii from gel-filtration and DLS, respectively. Rs3 is the Stokes radius estimated from EM measurements as described in Materials and methods. Centre-to-centre distances (x¯ and s.d.) measured by EM between A3 and CK domains at different pH are also shown (15–20 particles for each condition). (H) Superose 6 gel-filtration of A3–CK in 0.15 M NaCl and 20 mM buffers from pH 4.4 to pH 8.6 or at pH 6.2 at different NaCl concentrations. (I) pH titration curves of A3–CK. Partition coefficient in Superose 6 gel-filtration or percentage of unzipped particles at different pH. Smooth lines are drawn through the points.
The C-terminal portion of VWF is a dimeric bouquet at pH 6.2
Whereas in plasma the pH is 7.4, in the trans-Golgi the pH is 6.2 (Seksek et al, 1995), and in mature WPBs the pH decreases further to 5.45 with an s.d. of 0.26 (Erent et al, 2007). At pH 6.2, the A3–CK dimer was more compact than at pH 7.4, as shown by later elution in gel-filtration (Figure 2A) and a decrease in Stokes radius in DLS (Figure 2B). The pH-dependent change in elution position in gel-filtration was not accompanied by any change in mass, as the A3–CK dimer peaks in gel-filtration at pH 6.2 and 7.4 had identical masses of 260 000 Da measured from multiangle light scattering (MALS) (Figure 2D).
Formation of WPB-like helical tubules by the N-terminal domains of VWF requires both Ca2+ and pH 6.2, as does formation of D1–D2 propeptide homodimers (Huang et al, 2008). By contrast, we found no effect of Ca2+ on zipping up of A3–CK dimers as shown by Stokes radius. Elution position at pH 6.2 was unaffected by the presence or absence of 10 mM Ca2+ or 5 mM EDTA (Figure 2A). The elution position at pH 7.4 was also identical in the presence of 5 mM EDTA or 10 mM Ca2+ (Figure 2A).
Negative stain EM revealed that at pH 6.2, the A3–CK dimer zipped up into a dimeric bouquet (Figure 2E). Representative class averages are shown in Figure 2F; the complete sets of all class averages in this manuscript are summarized in Supplementary Table SI and shown in Supplementary Figure S1. Most particles showed a long stem with four globular domains in a flower-like arrangement at one end and the CK domains extending like a base for the stem at the other (Figure 2E and F). Occasionally, the four globules were less closely associated while the stem remained dimeric (arrowheads, Figure 2E). Infrequently, a less closely associated stem-like structure was also visible at pH 7.4 (arrowhead, Figure 2C). The gel-filtration, DLS, and EM results summarized in Figure 2G showed that the A3–CK dimers undergo pH-regulated conformational changes.
The pH dependence of dimeric bouquet formation was examined by gel-filtration at different pH (Figure 2H) and by EM (Figure 2I). The titration curves plotting elution position (partition coefficient) and percentage unzipped particles showed very similar pH dependence (Figure 2I). Shifts in elution position and zipping were each half-maximal near pH 6.5. However, each measure showed two transitions: one around pH 5.5 and the other near pH 7.5. pH-dependent dimeric bouquet formation was not abolished by high salt (Figure 2H).
The D4–CK fragment (Figure 1E) also formed a pH-dependent dimeric bouquet (Figure 3). Raising the pH from 6.2 to 7.4 increased Stokes radius, and there was little effect of raising the salt concentration at pH 6.2 (Figure 3A), as seen with A3–CK. The presence of four flowers in A3–CK dimeric bouquets, and only two in D4–CK dimeric bouquets (Figure 3B and C) showed that the stem-distal and stem-proximal globules correspond to the A3 and D4 domains, respectively.
Figure 3.
Conformational changes of D4–CK and V8–CK fragments. (A) DLS of D4–CK in 20 mM BisTris pH 6.2, 150 mM NaCl, 10 mM CaCl2 (upper panel), with addition of 1 M NaCl to the sample (middle panel), or with addition of 0.1 M HEPES pH 7.4 to the sample (lower panel). (B) Raw EM image of D4–CK in 20 mM BisTris pH 6.2, 150 mM NaCl, 10 mM CaCl2. (C) Representative class averages of D4–CK at pH 6.2 (left) and schematic (right). (D) Superose 6 gel-filtration of V8 protease-digested A3–CK in 20 mM buffers from pH 5.0 to pH 8.6 and 150 mM NaCl, 10 mM CaCl2 (solid lines). Purified V8–CK was also rechromatographed in the same buffers with 1 M NaCl (dashed curves). (E) Raw EM image of V8–CK in 20 mM BisTris pH 6.2, 150 mM NaCl, 10 mM CaCl2. Arrowheads mark occasional dimeric bouquets. Most particles are unzipped. Scale bars are 50 nm in (B, E), and 10 nm in (C).
The A3–CK and D4–CK dimeric bouquets revealed remarkable fine structure. In A3–CK (Figure 2F), the D4 domains were larger than the A3 domains, consistent with content of 380 and 190 amino-acid residues, respectively. The two D4 domains were further apart from one another than the two A3 domains, and appeared to be separated by solvent from one another as shown by a stain-rich region in between (Figure 2F). Remarkably, the same separation between the two D4 domains was seen in the D4–CK (Figure 3C) and A3–CK (Figure 2F) constructs. Each D4 domain had the shape of a bulbous crescent. The regular spacing between the two D4 crescents appears to be dictated by pairwise association at each tip of the crescents. A weak density bridging the A3-proximal tip of each D4 crescent was visible in most D4–CK class averages (Figure 3C). This density was stronger in A3–CK class averages, in which a bridge between the two monomers was clearly evident between the positions of the A3 and D4 domains (Figure 2F).
Six to eight, smaller, dyad-associated domains were resolved throughout the length of the stem region (Figures 2F and 3C). The stem region corresponds to the region containing the VWF B and C repeats (Figure 1A). The two stem domains closest to D4 were particularly well resolved, and each of the two domains appeared as a single globule. The domains distal to the D4 domain often appeared paired side-by-side, but in some class averages appeared as single globules. It should be kept in mind that whether each pair of stem domains appeared as a single globule or separate domains paired side-by-side reflects how they lie on the grid and are seen in projection. A preferred orientation on the grid dictated by the A3 and D4 domains may result in the most nearby stem domains lying on top of one another, so they are seen as a single globule in projection. Some variation in whether more C-terminal pairs of domains appear side-by-side or on top of one another may result from twisting in the axis of the stem. The overall conclusion is that the VWF stem consists of as many as eight discrete domains that are closely paired at pH 6.2, and that in each pair the two domains are either non-covalently associated or brought into very close association by nearby pairs of domains. Although the stem was overall straight, some bending of the stem was apparent between different particles and class averages. The density at the end of the stem projected laterally like an upside–down tee or base (Figure 3C). This density clearly corresponded to the CK domain dimer, where VWF monomers are disulphide linked, as shown by the similar density present in unzipped dimers (arrows, Figure 2C).
V8 protease cleaves within the D4 domain, after residue 2128, and generates N- and C-terminal fragments that are not disulphide linked to one another (Girma et al, 1986), as confirmed by non-reducing SDS–PAGE (Figure 1E) and separation of the two fragments by gel-filtration (Figure 3D, solid curves). The C-terminal V8–CK fragment showed a lower proportion (∼20%) of zipped up dimers at pH 6.2 than D4–CK (arrowheads, Figure 3E). However, zipping was still apparent as shown by later elution in gel-filtration at lower pH (Figure 3D). Similarly to A3–CK and D4–CK, the pH-dependent change in V8–CK elution position was not affected by high salt (Figure 3D).
The N-terminal extent of the dimeric bouquet
EM studies at pH 6.2 of the A1–CK fragment revealed three pairs of flower-like globules corresponding to the A2, A3, and D4 domains (Figure 4A–D). This type of elongated arrangement of flowers along a central axis is known as a raceme. Because of some flexibility at both ends of the construct, density for the A2 domain was weaker than for the A3 domain, and the lower half of the stem tended to be averaged out of A1–CK class averages (Figure 4B). To overcome the effect of flexibility of the stem, particles were selected using smaller boxes that centred on the flower domains and omitted the distal stem. Class averages of this smaller portion of particles showed strong density for the A2 domains (Figure 4C). The A2 domains were located as close to or even closer to the dyad axis than the A3 domains, and were somewhat less ordered than the A3 domains. Density for the A1 domain, sometimes only in one monomer, and varying from strong to weak, was present in some class averages (arrowheads, Figure 4C). This A1 domain density was more stem-distal than the A2 domains, and this location supported the identity of the A2 domains as the third pair of flowers from the stem (Figure 4D).
Figure 4.
Conformation of the A1–CK fragment and native VWF multimers. (A) Raw EM image of A1–CK fragments in 20 mM BisTris pH 6.2, 150 mM NaCl, 10 mM CaCl2. (B) Representative class averages of A1–CK at pH 6.2, with boxes centring on the middle of the whole molecule, and a radius for alignment and averaging of 37 nm. (C) Representative class averages of A1–CK, with boxes centring on the A1–D4 domains, and a radius for alignment and averaging of 17 nm. Arrowheads mark weak densities for A1. (D) Representative diagram of A1–CK dimeric bouquet. Variability in A1 domain position is symbolized by use of dashed circles. (E) Raw EM image of VWF multimers in 20 mM BisTris pH 6.2, 150 mM NaCl, 10 mM CaCl2. (F) Raw EM image of VWF multimers in 20 mM HEPES pH 7.4, 150 mM NaCl. (G, H) Same as in (E, F), with added D1–D2 prodomain. Scale bars are 50 nm in (A, E–H), and 10 nm in (B, C). Arrows point to CK dimers and arrowheads point to N-terminal globules.
Native VWF multimers
For comparison, we examined size-fractionated native VWF concatamers. For clarity, a VWF fraction containing a small number of monomers/concatamer was chosen for EM. Concatamers at pH 6.2 showed dimeric zipped up stem-like structures emanating from a VWF necklace (arrows, Figure 4E). Concatamers at pH 7.4 formed more extended necklaces, with few or no stem-like structures (Figure 4F). The previously reported head-to-head and tail-to-tail arrangement of monomers in concatamers was found with globular domains in the heads (arrowheads, Figure 4F) and little extra density at C-terminal linkages (arrows, Figure 4F).
When D1–D2 prodomain was added to native multimers, necklaces at pH 6.2 were more compact, and zipped up C-terminal stem-like structures were found extending radially away from the centre of multimers (Figure 4G). The N-terminal domains were located centrally in ring-like structures with diameters of 12–20 nm. Such structures did not form at pH 7.4 (Figure 4H) and monomers were seen linked tail-to-tail (arrow, Figure 4H) and head-to-head (arrowhead, Figure 4H), as in the absence of prodomain.
Arrangement of the N-terminal domains of VWF
In all, 90% of documented O-glycosylation sites in VWF (Titani et al, 1986) are located adjacent to the A1 and A2 domains (Figures 1A and 5A). O-glycosylation is added in the Golgi, after protein folding in the ER, and is associated with flexible regions rich in Ser, Thr, and Pro (Van den Steen et al, 1998). Two ∼30 amino-acid O-glycosylated linkers flank the A1 domain (Figure 5B). Raw images of A1–A3 at pH 7.4 (Figure 5C) revealed that the three A domains adopted random orientations, with variable distances in between. Relative to the central of the three globules (A2), one outer globule was generally markedly closer than the other, and for the purpose of calculating distances (Figure 6A), we have assumed that the A1 domain is further away than the A3 domain, based on the greater length and O-glycosylation of the A1–A2 than A2–A3 linker (Figure 5A). This assumption is supported by the identical lengths of the A1–A2 linker measured in D′D3–A2 monomers and A1–A3 (Figure 6A).
Figure 5.
The VWF N-terminal globular domains. (A) Diagram of triplet VWA domains. N- and O-linked glycans are shown as closed and open lollipops, respectively. (B) Loops flanking the VWA domains. O-glycosylation sites are labelled with open lollipops. (C) Raw EM image of A1–A3 in 20 mM HEPES pH 7.4, 150 mM NaCl. (D) Raw EM image of D′D3–A2 monomer in 20 mM HEPES pH 7.4, 150 mM NaCl. (E) Representative class averages of linear (top row) and more compact (bottom row) D′D3–A2 monomer at pH 7.4, with a schematic on the right. (F) Raw EM image of D′D3–A1 dimer in 20 mM BisTris pH 6.2, 150 mM NaCl, 10 mM CaCl2. Arrowheads mark a thin density seen in some particles. (G) Representative class averages of linear (top row) and more compact (bottom row) D′D3–A1 dimers, with schematics on the right. Arrowheads mark thin densities present in some monomers in some class averages. (H) MALS profiles of D′D3–A1 dimer eluting from Superose 6 gel-filtration in 20 mM HEPES pH 7.4, 5 mM EDTA, or 20 mM BisTris pH 6.2, 10 mM CaCl2, and 0.15 M NaCl. Scale bars are 50 nm in (C, D, F) and 10 nm in (E, G).
Figure 6.
Dimensions of VWF and schematics of putative intermediates in biosynthesis and secretion. (A) Dimensions of fragments. Measurements are shown to the right of each fragment, and are between centres of domains (indicated with points in the schematic). The only exception is A3–CK, in which contour length was measured by tracing the connections between A3 and CK (large points) through intervening domains (small points). (B) ProVWF dimer in ER. (C) ProVWF dimer in the trans Golgi. All dimensions are averages from class averages. D1–D2 is dashed because it was not studied here, and the arrangement of the D1D2D′D3 dimer is based on Huang et al (2008). (D) Assembly of proVWF dimers in helical tubules, using data from both this manuscript and Huang et al (2008). Helical assembly templates disulphide bond formation between D′D3 domains of adjacent proVWF dimers. (E) Model of VWF secretion. The dimensions shown for an intact D′D3–CK mature monomer are from adding maximal and average dimensions of fragments at pH 7.4.
Studies on fragments containing the D4 domain described above showed that the D4 domain is larger (6.1±0.4 nm), and more asymmetric with a bulbous crescent shape, than the globular A2 and A3 domains (4.5±0.3 nm). Similarly, in most class averages of D′D3–A2 monomers, the D′D3 domain could be tentatively assigned based on its larger and more asymmetric shape compared with the A1 and A2 domains (Figure 5D and E). The VWD and VWA domains were 6.7±0.5 and 4.6±0.3 nm in diameter, respectively. These domain assignments allowed the distances between each pair of domains to be measured (Figure 6A).
D′D3–A1 dimer class averages clearly revealed a dimeric structure (Figure 5F and G). Most class averages revealed a dyad axis and a significant interface between the two D′D3 monomers (Figure 5G). The A1 monomers were distal from one another in D′D3–A1 dimers, showing an anti-parallel arrangement of the D′D3–A1 tandems in each dimer. The arrangement of the two A1 and the D′D3 dimer could be extended and quasi-linear (Figure 5G, upper class averages) or more compact (Figure 5G, lower class averages). Distances between the D′D3 and A1 domains were similar in D′D3–A1 dimers and D′D3–A2 monomers (Figure 6A). A thin density that extended from D′D3 that was clearly distinct from A1 was often seen in particles (Figure 5F, arrowheads), and sometimes was seen on one of the D′D3 monomers in class averages (Figure 5G, arrowheads). Its disappearance (averaging out) in most class averages is ascribable to dominance in class averaging by the variably positioned and larger A1 density and flexibility of the thin density. D3 must be at the dimer interface because it contains the interdimer disulphide bonds (Figure 1A); therefore, the thin density may correspond to the D` domain. No change in Stokes radius for the D′D3–A1 dimer was evident between pH 6.2 and 7.4 (Figure 5H).
Discussion
Our results are important not only for understanding the behaviour of VWF in shear and elongational flow, but also its biosynthesis, assembly into long concatamers in WPBs, and secretion into the vasculature. Although fragments in this study were expressed with high mannose-type N-linked carbohydrates to reduce carbohydrate heterogeneity, there is no reason to expect an effect on domain organization, and similarly zipped dimeric bouquets were seen with intact VWF isolated from plasma. The structures of overlapping N- and C-terminal VWF fragments at neutral pH can be conceptually overlapped to understand the structure of VWF multimers in the circulation. Previous EM work had shown globules in the N-terminal end and a smaller globule in the C-terminal end of VWF; however, the number, spacing between and fine structure of the globules was not defined. Our work suggests that the D′D3 and D4 domains are not only larger (∼6.5 nm), but also more irregular or granular in shape than the A1, A2, and A3 domains (4.5 nm in diameter). The D4 domain has a bulbous crescent shape, and a similar bulbous crescent shape for the D′D3 domain was visible in some D′D3–A1 dimer class averages. The approximate shape and size of domains and the minimum, average, maximum, and s.d. measurements of distances between domains are summarized in Figure 6A.
The reconstructed overall structure of mature VWF in the vasculature may be discussed from N- to C-terminus. When disulphide linked in dimers, the D′D3 domain monomers are closely associated with one another across a significant interface that often appears dyad-like, and must bear the disulphide bonds responsible for N-terminal covalent linkages of VWF. The linker to the A1 domain is flexible, and appears to connect to D′D3 distal from the multimerization site. A thin density extending from D3 may correspond to D′.
Eight of the 10 documented O-glycosylation sites in VWF occur adjacent to the A1 domain, four in the D′D3 linker and four in the A2 linker. Independent measurements on D′D3–A2 monomers, D′D3–A1 dimers, and A1–A3 monomers showed an average and maximal distance, respectively, in projections of 7.2–7.4 and 10–11 nm between the D′D3 and A1 domains, and 7.9–8.0 and 11 nm between the A1 and A2 linker domains (Figure 6A). In contrast, the average and maximal distance between the A2 and A3 domains was 5.0 to 6.4 nm. If we use an estimate of 4.5 nm for A domain diameter, and 6.5 nm for D′D3 diameter, the maximal lengths of the D′D3–A1, A1–A2, and A2–A3 linkers in projections would be 5.6, 6.5, and 1.9 nm, respectively. The mucin-like PSGL-1 molecule extends 0.2 nm/residue in EM (Li et al, 1996). This metric predicts lengths for the D′D3 and A1–A2 linkers of 6.4 and 6.2 nm, respectively, and is in good agreement with the maximal linker length of the mucin-like segments on either side of the A1 domain (the A2–A3 linker is less mucin-like with only one O-linked site).
At the neutral pH characteristic of plasma, we also found flexibility between the A3 and D4 domains, and throughout the length of the connection through smaller domains to the CK domain (Figure 6). By tracing the connections through these domains in individual A3–CK and D4–CK dimers, we measure the average contour length as 50±3.5 nm between A3 and CK.
These measurements yield a maximal extension of 82 nm per monomer and an average extension of 70 nm per monomer within VWF multimers. This estimate is higher than the previous estimate of 60 nm per monomer from rotary shadowed VWF multimers. A higher value is expected from the higher granularity of the current measurements, in which more domains are visualized, and hence more domain–domain measurements are included in the contour length estimate. Accurate estimation of the extended length of VWF is important, because hydrodynamic force on VWF in flow regulates its function, and force scales with the square of length (Zhang et al, 2009b).
pH-regulated ordering of VWF monomers in dimeric bouquets substantially advances our knowledge of VWF ultrastructure. The A2, A3, and D4 domains form a raceme-like flower head, whereas the C-terminal domains are stem-like. The C-terminal segment of dimeric VWF, previously described as rod-like and flexible when monomeric, is here revealed to contain six to eight small globules between the D4 and CK domains. These globules correspond to the B and C sequence repeat region in VWF; however, it is not yet possible to associate each globule with a specific sequence. Although these smaller domains are only well resolved in dimeric bouquets at acidic pH, they are likely to be present at neutral pH as well. We speculate that six to eight small domains, with flexible hinges in between, are present in each monomer, and that pairwise association between them at acidic pH favours an overall straighter, extended conformation, greater order, and greater contrast in negative stain for the paired than single domains, enabling their visualization in EM for the first time.
pH-dependent dimerization involves specific pairing between multiple independent modules between the A1 and CK domains. Treatment of the D4–CK fragment with V8 protease showed that after cleavage of D4, the V8–CK fragment could completely zip up throughout its length at pH 6.2, although fewer dimers were zipped. Studies with D4–CK, A3–CK, and A1–CK fragments showed that zipping could extend N-terminally all the way through the D4, A3, or A2 domains, without requirement for more N-terminal domains. pH titration between 5 and 8 implicates histidine sidechains and the requirement of histidine protonation for dimeric bouquet formation. Insensitivity to salt suggests that electrostatic interactions are not sufficient to mediate dimerization, and that more specific non-covalent bonds, such as hydrogen bonds involving histidine sidechains, are required.
The long-range ordering of VWF in a dimeric bouquet at acidic pH has important implications for biosynthesis, storage, and secretion of VWF. pH-dependent interactions are frequently employed to regulate association–dissociation reactions between receptors and ligands in the alkaline extracellular and acidic endocytic compartments, for example, in the low-density lipoprotein receptor (Rudenko et al, 2002). However, to our knowledge, the pH-dependent ordering of such a large protein segment, encompassing >1200 residues in each monomer, is unprecedented.
Elegant studies with the VWF prodomain (D1–D2) and N-terminal D′D3 or D′D3–A1 disulphide-linked dimer fragments have shown that these spontaneously form helical assemblies in Ca2+ at pH 6.2 with the same dimensions and 120 Å helical pitch as the tubules in WPBs in vivo (Huang et al, 2008; Berriman et al, 2009). These in vitro assembly studies omit the C-terminal 2/3 of mature VWF, and use predimerized D′D3 fragments. In vitro assembly studies therefore do not address the dilemma of how disulphide crosslinks between neighbouring tubules are avoided, and how crosslinking in the D′D3 domain occurs exclusively between monomers that are not crosslinked in their CK domains. Tubulation is highly processive, because single VWF tubules can be traced along the entire length of WPBs (Berriman et al, 2009).
The formation of a dimeric bouquet in the C-terminal portion of VWF at the same trans-Golgi pH that enables assembly of the N-terminal portion into helical tubules solves many of the dilemmas in VWF concatamer biogenesis (Figure 6C and D). One issue is ensuring that homodimerization between D1–D2 monomers occurs intramolecularly within a proVWF dimer (Figures 1D, panel 1, 6C and D). At neutral pH, the distance between D1–D2 monomers in a proVWF dimer would be comparable to that between D3 domains, or on average within 140 nm of one another. By contrast, at acidic pH, the dimeric bouquet is zipped up to the A2 domains, which ensures the D3 domains in a dimer are no more than 30 nm away from one another (Figure 6C). Dimerization has second-order dependence on monomer concentration, and this increase in proximity of D1–D2 monomers within a proVWF dimer will thus significantly contribute to D1–D2 association within a dimer (Figure 6C and D) rather than between different dimers (Figure 1D, panels 2 and 3). Within the dimeric bouquet, the centre-to-centre distance between A2 domains is 6 nm (Figure 6C), whereas that between the two D3 domains in the same dimer should be 13 nm in the assembling helical VWF tubule (Figure 6D). The transition between the 13-nm D′D3 spacing in the helical VWF and the 6-nm spacing between the A2 domains in the dimeric bouquet falls to the two A1 domains and their linkers (Figure 6C and D).
Dimeric bouquets help ensure that covalent crosslinking occurs between D′D3 domains of proVWF dimers in neighbouring positions in the helical tubule (Figure 1D, panel 1), rather than within proVWF dimers (Figure 1D, panel 3), between proVWF monomers on different tubules (Figure 1D, panel 2) or in non-neighbouring positions in the same helical tubule (not shown). All of these non-native assemblies would be possible in the absence of dimeric bouquets, given the 140-nm separation between the N-terminal ends of VWF at pH 7.4, the 80-nm centre-to-centre distance between adjacent growing ends of tubules in nascent WPBs, the 25-nm diameter of tubules, and the 12-nm rise per helical turn (Zenner et al, 2007; Berriman et al, 2009). Dimeric bouquets help ensure that disulphide linkage between VWF monomers is co-linear with assembly in tubules. The lack of crosslinking between tubules promotes orderly unfurling of VWF from the ends of helices similarly to orderly uncoiling of rope (Michaux et al, 2006).
Dimeric bouquets are no doubt also important in compaction of WPBs during maturation and their expansion during secretion. The dimeric bouquets visualized here are constrained to lie flat on an EM grid, and may be less elongated in 3D. However, if we use their length in 2D as an approximation, dimeric bouquets, 51 nm in length from the A2 to CK domains, could not extend radially from the sides of 25 nm diameter helices spaced 80 nm on centre without almost complete interdigitation of bouquets extending from neighbouring nascent tubules (Zenner et al, 2007). It is more likely that linkers on either side of the A1 domain act as hinges (Figure 6C and D) and enable the dimeric bouquet region to pack against the cylindrical side of helical tubules.
We propose that the A1 domain and its neighbouring linkers, which are located between the N-terminal VWF helical region and the C-terminal bouquet region, are important for breaking symmetry between these regions. In contrast to DNA, with two molecules per helix, the VWF helices are one-start; that is, there is one VWF concatamer per helix (Huang et al, 2008). However, like DNA there is two-fold symmetry, so that the helical structure remains identical when flipped end-for-end. In VWF, this is a consequence of the two-fold dyad axis about the D1, D2, D′D3 dimer in the helix (Huang et al, 2008). We propose that the A1 domain and its mucin-like N and C linkers act as hinges between the N-terminal helical assembly and the C-terminal dimeric bouquet. The A1 domain is not ordered in dimeric bouquets, and does not contribute strong density to in vitro-assembled tubules (Huang et al, 2008). Thus, the ∼51 nm long dimeric bouquets need not obey helical two-fold symmetry and extend radially from the 25-nm diameter tubules, but could instead extend more axially and avoid interdigitation during biogenesis when the centre-to-centre tubule spacing is 80 nm (Zenner et al, 2007).
These ideas are supported by 3D reconstruction of VWF tubules in vivo from cryoelectron tomograms of paracrystalline regions of WPBs that reveal a structure very similar to in vitro-assembled helices containing only the D1, D2 and D′D3 domains (Berriman et al, 2009). The lack of contribution to helical density of dimeric bouquets in vivo demonstrates that they do not have a well-defined packing geometry with respect to the N-terminal helical segment. Compaction of the dimeric bouquet region of VWF must become extreme in mature WPBs, in which the 25-nm tubules pack with a centre-to-centre spacings of 28.4±3 nm (Berriman et al, 2009). No doubt this is assisted by the reduction in pH in mature WPBs to 5.45±0.26, which approaches the isoelectric point of VWF protein (Erent et al, 2007).
Orderly coiling of VWF in tubules in WPBs is important for rapid unfurling of VWF during secretion. Experimental elevation of pH within WPBs, which results in their rounding, inhibits subsequent release of VWF fibrils. Thus, tangling is thought to inhibit VWF secretion (Michaux et al, 2006). The first step in secretion is formation of a narrow constriction between the plasma membrane and the WPB, which then experiences a rapid rise in pH. Alkalinization rapidly propagates from one end of a WPB to the other (Erent et al, 2007). Secretion occurs from one end of a WPB, as shown by EM (Berriman et al, 2009).
Early after fusion with the plasma membrane, there is a great expansion in width of WPBs and increase in average spacing between tubules (Berriman et al, 2009). VWF is by far the predominant protein constituent of WPBs (Ewenstein et al, 1987). Therefore, we propose that the alkalinization of WPBs causes dissociation of the dimeric bouquet, and together with charge repulsion between VWF domains above their pI's, causes the observed increase in spacing between WPB tubules (Berriman et al, 2009). Maintenance of the helical patterns in Fourier transforms of cryoelectron tomograms after expansion (and presumed pH increase to ∼7.4) demonstrates that the interdimer disulphide crosslinks stabilize VWF helical structure, and suggests that the D1–D2 propiece remains associated (Berriman et al, 2009).
These findings further suggest that secretion begins by unfurling of VWF from the ends of tubules, which are adjacent to the secretion pore (Berriman et al, 2009), and where packing interactions are the weakest. Thus, both dissociation of the D1–D2 propiece, and unfurling of VWF concatamers are likely to proceed in parallel from the ends of the VWF tubules closest to the secretion pore (Figure 6E).
In conclusion, studies on dimeric bouquets and the N-terminal region of VWF not only provide novel insights into the ultrastructure of domains in VWF, but also into pH-regulated changes in the organization of these domains during biogenesis, storage, and secretion. VWF belongs to a larger family of proteins that include the mucins MUC5AC and MUC5B (Perez-Vilar, 2007); otogelin, a component of the acellular membranes of the inner ear required for hearing (Cohen-Salmon et al, 1997); and hemolectin, an insect hemolymph protein important in coagulation and immunity (Goto et al, 2001). The mucins are stored in granules and expand explosively upon secretion. The above proteins have N-terminal regions homologous to the region forming helical tubules in VWF (Perez-Vilar, 2007). They further contain C-terminal regions corresponding to the dimeric bouquet region of VWF containing a D domain followed by a varying number of VWF B and C repeats and a C-terminal CK domain. In contrast, the domains in the middle corresponding to the A domains in VWF are not shared; mucins, otogelin, and hemolectin each contain distinctive types of middle domains. The N- and C-terminal domains appear to enable construction of long concatamers displaying many copies each of different types of ‘middle’ domains, and concatamer formation has been verified for MUC5AC (Perez-Vilar, 2007). It will be interesting to learn whether the C-terminal regions of these other proteins also form pH-regulated dimeric bouquets.
Most of the length of the mature VWF monomer (55 nm of 70 nm) is derived from the dimeric bouquet A2–CK region. The key to VWF function is its length, which enables increased shear flow and conversion of shear flow to elongational flow at sites of haemorrhage to exert substantial hydrodynamic force on it, convert it from a compact to an extended conformation, and enable multivalent binding of its A1 domains to platelet GPIbα to form a hemostatic plug. Elongational force on VWF scales with the square of its length (Zhang et al, 2009b) and hemostatic potency of VWF is highly length dependent (Sadler, 1998). The pH-regulated dimeric bouquet region provides most of VWF's length at plasma pH and thus is key to the biogenesis of VWF's function in length-dependent regulation of hemostasis.
Materials and methods
Constructs and protein expression
VWF fragments with preproprotein residue numbering shown in Figure 1 were PCR amplified from pSVH-VWF (Matsushita and Sadler, 1995) and subcloned into the in-house ExpressTag-8 (ET8) vector. ET8 is similar to ET1 (Mi et al, 2008), except it utilizes ligation-independent cloning and contains only a (His)6 tag at the C-terminus. Constructs were confirmed by DNA sequencing. HEK293S Gnt1− cells (Reeves et al, 2002) were cultured in suspension in Freestyle 293 SFM medium (Invitrogen), and transiently transfected using a 1:2.5 DNA:polyethylenimine mass ratio (Boussif et al, 1995). Supernatants were harvested 7 days after transfection.
Protein purification
Purification and gel-filtration experiments were at 4°C. Supernatants were filtered and adjusted to pH 8.0 by adding 1 M Tris buffer to 20 mM final concentration. In all, 0.3 M NaCl and 0.5 mM NiCl2 were added to the media before adding Ni-NTA beads (15 ml beads slurry per litre medium), except for D1–A1, no NaCl was added, since untagged D1–D2 fragment bound to Ni-NTA beads in the absence of extra NaCl. After washing with 20 mM imidazole, 20 mM Tris pH 8.0, 0.5 M NaCl, His-tagged fragments and untagged D1–D2 were eluted with 300 mM imidazole, 20 mM Tris pH 8.0, 0.5 M NaCl.
Eluates were buffer exchanged to 20 mM Tris pH 8.0. A1–CK and D′D3–A1 fragments only were bound to a 5-ml Hitrap heparin column, and were eluted with 10 column volumes of a gradient of 0–1 M NaCl in 20 mM Tris pH 8.0. All proteins in 20 mM Tris pH 8.0 were loaded onto a monoQ 5/50 GL column (GE Healthcare), and eluted with a gradient of 20–40 column volumes of 0–0.5 M NaCl in 20 mM Tris pH 8.0. The final step was gel-filtration with a Superose 6 10/300 GL column (GE Healthcare). Purified fragments were identified by reducing and non-reducing SDS–PAGE followed by either Coomassie blue staining or western blotting using polyclonal antibodies. Fractions were also subjected to ELISA with anti-human VWF polyclonal antibodies.
For V8 protease digestion, D4–CK or A3–CK fragment in 0.1 M NH4HCO3 pH 8.0 was cleaved at a V8 protease:VWF fragment mass ratio of 1:20 at room temperature (22°C) for 40 h as previously described (Girma et al, 1986; Titani et al, 1986). Fragments were separated by Superose 6 gel-filtration in 20 mM BisTris pH 6.2, 0.15 M NaCl, 10 mM CaCl2.
Purified human VWF multimer, free of Factor VIII, was a kind gift from Dr Cecile V Denis, INSERM U770, Le Kremlin-Bicetre, France. The preparation was size fractionated on Sepharose CL-2B (GE Healthcare) in 20 mM BisTris pH 6.2, 0.15 M NaCl, 10 mM CaCl2. A later eluting fraction of VWF containing lower molecular weight multimers was used in EM.
Light scattering experiments and molecule size calculation
MALS was measured with a triple angle detector at room temperature (miniDAWN, Wyatt Technology Corporation, Santa Barbara, CA) connected in line with a Superose 6 10/300 column at 4°C. Absolute mass was calculated using Wyatt's ASTRA software. The same Superose 6 column was also used for determination of Stokes radius (Rs) after calibration with protein standards with known Rs. The partition coefficient was calculated as Kav=(Ve–Vo)/(Vt–Vo), where Ve is the elution volume, Vt is the total volume determined by acetone, and Vo is the void volume determined with Blue dextran 2000. Plot of Rs versus square root of (−log(Kav)) gave correlation coefficient R2>0.99.
Rs was also determined by DLS at 20°C (802 DLS, Viscotek Corporation, Houston, TX). Samples were concentrated to 0.5–1 mg/ml after gel-filtration, and stored in 20 mM BisTris pH 6.2, 0.15 M NaCl, 10 mM CaCl2 or 20 mM HEPES pH 7.4, 0.15 M NaCl, 5 mM EDTA. To raise the salt or pH of samples in 20 mM BisTris pH 6.2, 0.15 M NaCl, 10 mM CaCl2, 4.0 M NaCl or 1.0 M HEPES pH 7.4 was added to give final concentrations of 1.0 M NaCl or 0.1 M HEPES pH 7.4, respectively. Samples were microcentrifuged and 0.2 μm filtered prior to DLS. Stokes radius was calculated with the OmniSIZE software from Viscotek.
The Stokes radius of A3–CK at pH 6.2 was also estimated from its dimensions in EM. The dimeric bouquet was approximated as a long rod. At low concentration, the translational diffusion coefficient D is given by D=kT/f, where k is the Boltzmann constant, T is the absolute temperature, and f is the translational frictional coefficient. For rod-like molecules, f=3πηL/(lnp+γ), where η is the viscosity of the solution, L is the length of the rod, p is the axial ratio, and γ is a correction for end effects (Tirado and Garcia de la Torre, 1979). Substitution of the Stokes–Einstein equation, Rs=kT/6πηD, yields Rs=L/2(lnp+γ). Rs and its error were estimated with EM dimensions of length L=46±3.3 nm and diameter=4.4±0.5 nm. The axial ratio p was approximated to length/diameter, 11±1.2. γ=0.35 was taken from previously published calculations (Tirado and Garcia de la Torre, 1979). The calculated Rs was 8.5±0.4 nm.
EM and image processing
Purified fragments (10–30 μg) were subjected to one additional gel-filtration on Superose 6 10/300 GL in 20 mM BisTris pH 6.2, 0.15 M NaCl, 10 mM CaCl2 or 20 mM HEPES pH 7.4, 0.15 M NaCl. Peak fractions were adsorbed to grids. Size-fractionated VWF multimers (20 μl) were mixed with 2 μl 1 M stock solution of BisTris pH 6.2 or HEPES pH 7.4, with or without 0.1 μl 2 mg/ml D1–D2. Mixtures were gently vortexed for 3 s, and grids were made 30 min later. EM was performed using conventional negative staining with 0.75% (w/v) uranyl formate (Ohi et al, 2004). Image collection on a Tecnai T12 electron microscope (FEI, Hillsboro, OR, USA) and alignment of 3551–5604 particles and class averaging were as previously described (Chen et al, 2010). A1–CK particles were picked with a 90 × 90 nm2 box centring on the middle of the whole fragment or a 54 × 54 nm2 box centring on the A1–D4 head region. The radius for alignment and averaging was 17 nm for head centring and 37 nm for the whole fragment. Further details of particle numbers and class averaging are summarized in Supplementary Table SI, and all class averages are shown in Supplementary Figure S1.
Distances in particles and class averages were measured in ImageJ (National Institutes of Health) using segmented lines. N=15–20 particles of A3–CK and A1–A3 at pH 7.4 or all well-resolved class averages of other fragments were included in measurements. Distances measured in class averages were weighted by the numbers of particles per class.
pH titration of A3–CK
Purified A3–CK at 10 mg/ml in 20 mM BisTris pH 6.2, 0.15 M NaCl, 10 mM CaCl2 was diluted 10-fold in 0.15 M NaCl, 10 mM CaCl2, and 20 mM of Na-acetate pH 4.4, 5.0, or 5.6, BisTris pH 6.2 or 6.8, HEPES pH 7.4, or Tris pH 8.0 or 8.6, and subjected to Superose 6 10/300 gel-filtration in the same buffer.
For EM, purified A3–CK at 10 mg/ml in 20 mM BisTris pH 6.2, 0.15 M NaCl, 10 mM CaCl2 was diluted 100-fold in water, and then diluted a further 20-fold in 0.15 M NaCl and 50 mM of Na-acetate pH 4.4, 5.0, or 5.6, BisTris pH 6.2 or pH 6.8, HEPES pH 7.4, or Tris pH 8.0 or 8.6. Dilutions were left overnight at 4°C before making EM grids. Ten random image fields were recorded on a CM10 electron microscope at each pH. All particles (n=40–182, there were fewer particles at pH 8.0 and 8.6 due to aggregation) were counted and assigned as zipped or unzipped, similarly to the particles with arrows or arrowheads in Figure 2C and E.
Different pH conditions were also tested for the V8–CK fragment. V8 protease-digested products from A3–CK were subjected to Superose 6 gel-filtration in buffers containing 0.15 M NaCl, 10 mM CaCl2, and 20 mM of Na-acetate pH 5.0, BisTris pH 6.2 or 6.8, HEPES pH 7.4, or Tris pH 8.0. To study the effect of salt concentration on conformation, 0.15 M NaCl versus 1 M NaCl in gel-filtration buffer was compared at pH 6.2 and 5.0.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant HL-48675. TW is an investigator in the Howard Hughes Medical Institute.
Author contributions: Y-FZ prepared constructs, designed and carried out experiments, and wrote the manuscript. CL prepared constructs, designed and carried out experiments, and discussed the write-up. NN and EE performed the EM experiments. TW discussed and supervised the EM experiments and strategy and wrote the manuscript. TAS designed the overall experimental approach, supervised experiments, and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Berriman JA, Li S, Hewlett LJ, Wasilewski S, Kiskin FN, Carter T, Hannah MJ, Rosenthal PB (2009) Structural organization of Weibel-Palade bodies revealed by cryo-EM of vitrified endothelial cells. Proc Natl Acad Sci USA 106: 17407–17412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienkowska J, Cruz M, Atiemo A, Handin R, Liddington R (1997) The von Willebrand factor A3 domain does not contain a metal ion-dependent adhesion site motif. J Biol Chem 272: 25162–25167 [DOI] [PubMed] [Google Scholar]
- Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr J (1995) A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA 92: 7297–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celikel R, Varughese KI, Madhusudan, Yoshioka A, Ware J, Ruggeri ZM (1998) Crystal structure of the von Willebrand factor A1 domain in complex with the function blocking NMC-4 Fab. Nat Struct Biol 5: 189–194 [DOI] [PubMed] [Google Scholar]
- Chen X, Xie C, Nishida N, Li Z, Walz T, Springer TA (2010) Requirement of open headpiece conformation for activation of leukocyte integrin αXβ2. Proc Natl Acad Sci USA 107: 14727–14732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Salmon M, El-Amraoui A, Leibovici M, Petit C (1997) Otogelin: a glycoprotein specific to the acellular membranes of the inner ear. Proc Natl Acad Sci USA 94: 14450–14455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley J, Cruz M, Handin R, Liddington R (1998) Crystal structure of the von Willebrand factor A1 domain and implications for the binding of platelet glycoprotein Ib. J Biol Chem 273: 10396–10401 [DOI] [PubMed] [Google Scholar]
- Erent M, Meli A, Moisoi N, Babich V, Hannah MJ, Skehel P, Knipe L, Zupancic G, Ogden D, Carter T (2007) Rate, extent and concentration dependence of histamine-evoked Weibel-Palade body exocytosis determined from individual fusion events in human endothelial cells. J Physiol 583 (Part 1): 195–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewenstein BM, Warhol MJ, Handin RI, Pober JS (1987) Composition of the von Willebrand factor storage organelle (Weibel-Palade body) isolated from cultured human umbilical vein endothelial cells. J Cell Biol 104: 1423–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler WE, Fretto LJ, Hamilton KK, Erickson HP, McKee PA (1985) Substructure of human von Willebrand factor. J Clin Invest 76: 1491–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girma JP, Chopek MW, Titani K, Davie EW (1986) Limited proteolysis of human von Willebrand factor by Staphylococcus aureus V-8 protease: isolation and partial characterization of a platelet-binding domain. Biochemistry 25: 3156–3163 [DOI] [PubMed] [Google Scholar]
- Goto A, Kumagai T, Kumagai C, Hirose J, Narita H, Mori H, Kadowaki T, Beck K, Kitagawa Y (2001) A Drosophila haemocyte-specific protein, hemolectin, similar to human von Willebrand factor. Biochem J 359(Part 1): 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberichter SL, Fahs SA, Montgomery RR (2000) von Willebrand factor storage and multimerization: 2 independent intracellular processes. Blood 96: 1808–1815 [PubMed] [Google Scholar]
- Huang RH, Wang Y, Roth R, Yu X, Purvis AR, Heuser JE, Egelman EH, Sadler JE (2008) Assembly of Weibel-Palade body-like tubules from N-terminal domains of von Willebrand factor. Proc Natl Acad Sci USA 105: 482–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizinga EG, Martijn van der Plas R, Kroon J, Sixma JJ (1997) Crystal structure of the A3 domain of human von Willebrand factor: imlications for collagen binding. Structure 5: 1147–1156 [DOI] [PubMed] [Google Scholar]
- Katsumi A, Tuley EA, Bodo I, Sadler JE (2000) Localization of disulfide bonds in the cystine knot domain of human von Willebrand factor. J Biol Chem 275: 25585–25594 [DOI] [PubMed] [Google Scholar]
- Li F, Erickson HP, James JA, Moore KL, Cummings RD, McEver RP (1996) Visualization of P-selectin glycoprotein ligand-1 as a highly extended molecule and mapping of protein epitopes for monoclonal antibodies. J Biol Chem 271: 6342–6348 [DOI] [PubMed] [Google Scholar]
- Marti T, Rosselet SJ, Titani K, Walsh KA (1987) Identification of disulfide-bridged substructures within human von Willebrand factor. Biochemistry 26: 8099–8109 [DOI] [PubMed] [Google Scholar]
- Matsushita T, Sadler JE (1995) Identification of amino acid residues essential for von Willebrand factor binding to platelet glycoprotein Ib. Charged-to-alanine scanning mutagenesis of the A1 domain of human von Willebrand factor. J Biol Chem 270: 13406–13414 [DOI] [PubMed] [Google Scholar]
- Mi LZ, Grey MJ, Nishida N, Walz T, Lu C, Springer TA (2008) Functional and structural stability of the epidermal growth factor receptor in detergent micelles and phospholipid nanodiscs. Biochemistry 47: 10314–10323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaux G, Abbitt KB, Collinson LM, Haberichter SL, Norman KE, Cutler DF (2006) The physiological function of von Willebrand's factor depends on its tubular storage in endothelial Weibel-Palade bodies. Dev Cell 10: 223–232 [DOI] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T (2004) Negative staining and image classification -- powerful tools in modern electron microscopy. Biol Proced Online 6: 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Vilar J (2007) Mucin granule intraluminal organization. Am J Respir Cell Mol Biol 36: 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PJ, Callewaert N, Contreras R, Khorana HG (2002) Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci USA 99: 13419–13424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg JB, Haberichter SL, Jozwiak MA, Vokac EA, Kroner PA, Fahs SA, Kawai Y, Montgomery RR (2002) The role of the D1 domain of the von Willebrand factor propeptide in multimerization of VWF. Blood 100: 1699–1706 [DOI] [PubMed] [Google Scholar]
- Rudenko G, Henry L, Henderson K, Ichtchenko K, Brown MS, Goldstein JL, Deisenhofer J (2002) Structure of the LDL receptor extracellular domain at endosomal pH. Science 298: 2353–2358 [DOI] [PubMed] [Google Scholar]
- Ruggeri ZM, Mendolicchio GL (2007) Adhesion mechanisms in platelet function. Circ Res 100: 1673–1685 [DOI] [PubMed] [Google Scholar]
- Sadler JE (1998) Biochemistry and genetics of von Willebrand factor. Annu Rev Biochem 67: 395–424 [DOI] [PubMed] [Google Scholar]
- Sadler JE (2005) New concepts in von Willebrand disease. Annu Rev Med 56: 173–191 [DOI] [PubMed] [Google Scholar]
- Sadler JE, Shelton-Inloes BB, Sorace JM, Harlan JM, Titani K, Davie EW (1985) Cloning and characterization of two cDNAs coding for human von Willebrand factor. Proc Natl Acad Sci USA 82: 6394–6398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider SW, Nuschele S, Wixforth A, Gorzelanny C, Alexander-Katz A, Netz RR, Schneider MF (2007) Shear-induced unfolding triggers adhesion of von Willebrand factor fibers. Proc Natl Acad Sci USA 104: 7899–7903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seksek O, Biwersi J, Verkman AS (1995) Direct measurement of trans-Golgi pH in living cells and regulation by second messengers. J Biol Chem 270: 4967–4970 [DOI] [PubMed] [Google Scholar]
- Siedlecki CA, Lestini BJ, Kottke-Marchant KK, Eppell SJ, Wilson DL, Marchant RE (1996) Shear-dependent changes in the three-dimensional structure of human von Willebrand factor. Blood 88: 2939–2950 [PubMed] [Google Scholar]
- Sing CE, Alexander-Katz A (2010) Elongational flow induces the unfolding of von Willebrand factor at physiological flow rates. Biophys J 98: L35–L37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slayter H, Loscalzo J, Bockenstedt P, Handin RI (1985) Native conformation of human von Willebrand protein. J Biol Chem 260: 8559–8563 [PubMed] [Google Scholar]
- Tirado M, Garcia de la Torre J (1979) Translational friction coefficients of rigid, symmetric top macromolecules. Application to circular cylinders. J Chem Phys 71: 2581–2588 [Google Scholar]
- Titani K, Kumar S, Takio K, Ericsson LH, Wade RD, Ashida K, Walsh KA, Chopek MW, Sadler JE, Fujikawa K (1986) Amino acid sequence of human von Willebrand factor. Biochemistry 25: 3171–3184 [DOI] [PubMed] [Google Scholar]
- Valentijn KM, Valentijn JA, Jansen KA, Koster AJ (2008) A new look at Weibel-Palade body structure in endothelial cells using electron tomography. J Struct Biol 161: 447–458 [DOI] [PubMed] [Google Scholar]
- Van den Steen P, Rudd PM, Dwek RA, Opdenakker G (1998) Concepts and principles of O-linked glycosylation. Crit Revs Biochem Mol Biol 33: 151–208 [DOI] [PubMed] [Google Scholar]
- Vischer UM, Wagner DD (1994) von Willebrand factor proteolytic processing and multimerization precede the formation of Weibel-Palade bodies. Blood 83: 3536–3544 [PubMed] [Google Scholar]
- Voorberg J, Fontijn R, van Mourik JA, Pannekoek H (1990) Domains involved in multimer assembly of von willebrand factor (vWF): multimerization is independent of dimerization. EMBO J 9: 797–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DD (1990) Cell biology of von Willebrand factor. Annu Rev Cell Biol 6: 217–246 [DOI] [PubMed] [Google Scholar]
- Wagner DD, Saffaripour S, Bonfanti R, Sadler JE, Cramer EM, Chapman B, Mayadas TN (1991) Induction of specific storage organelles by von Willebrand factor propolypeptide. Cell 64: 403–413 [DOI] [PubMed] [Google Scholar]
- Zenner HL, Collinson LM, Michaux G, Cutler DF (2007) High-pressure freezing provides insights into Weibel-Palade body biogenesis. J Cell Sci 120(Part 12): 2117–2125 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zhou Y-F, Zhang C-Z, Springer TA (2009a) Structural specializations of A2, a force-sensing domain in the ultralarge vascular protein von Willebrand factor. Proc Natl Acad Sci USA 106: 9226–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Halvorsen K, Zhang CZ, Wong WP, Springer TA (2009b) Mechanoenzymatic cleavage of the ultralarge vascular protein, von Willebrand factor. Science 324: 1330–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.