Abstract
PIWI proteins and their bound PIWI-interacting RNAs (piRNAs) form the core of a gonad-specific small RNA silencing pathway that protects the animal genome against the deleterious activity of transposable elements. Recent studies linked the piRNA pathway to TUDOR biology as TUDOR domains of various proteins bind symmetrically methylated Arginine residues in PIWI proteins. We systematically analysed the Drosophila TUDOR protein family and identified four previously not characterized TUDOR domain-containing proteins (CG4771, CG14303, CG11133 and CG31755) as essential piRNA pathway factors. We characterized CG4771 (Vreteno) in detail and demonstrate a critical role for this protein in primary piRNA biogenesis. Vreteno physically and/or genetically interacts with the primary pathway components Piwi, Armitage, Yb and Zucchini. Vreteno also interacts with the Tdrd12 orthologues CG11133 (Brother of Yb) and CG31755 (Sister of Yb), which are essential for the primary piRNA pathway in the germline and probably replace the function of the related but soma-specific factor Yb.
Keywords: Drosophila , piRNAs, Piwi, transposons, Tudor
Introduction
The PIWI-interacting RNA (piRNA) pathway is an animal-specific small RNA pathway that silences selfish genetic elements such as transposons in gonads (Malone and Hannon, 2009; Khurana and Theurkauf, 2010; Senti and Brennecke, 2010). At the core of this pathway act Argonaute proteins from the PIWI clade and their bound small RNAs, generally referred to as piRNAs. Mutations in PIWI proteins or in factors involved in piRNA biogenesis or piRNA-mediated silencing lead to de-silencing of transposons, to widespread DNA damage and ultimately result in sterility.
The analyses of piRNA populations from vertebrates and invertebrates have provided genuine insight into the genomic origin of piRNAs (Aravin et al, 2006, 2007, 2008; Girard et al, 2006; Lau et al, 2006; Vagin et al, 2006; Brennecke et al, 2007; Li et al, 2009; Malone et al, 2009; Robine et al, 2009; Saito et al, 2009). The three major piRNA sources are long RNAs originating from discrete genomic loci typically enriched in transposon sequences (piRNA clusters), transcripts from active transposons and finally mRNAs from numerous endogenous genes.
The genetic and mechanistic principles of piRNA biogenesis are only poorly understood but sequence analyses of piRNA populations indicated that two modes of piRNA biogenesis exist (reviewed in Senti and Brennecke, 2010). On the one hand, during primary piRNA biogenesis presumably single-stranded precursor transcripts are processed in a seemingly random manner into 23–30 nt primary piRNAs (Lau et al, 2009; Li et al, 2009; Malone et al, 2009; Saito et al, 2009). On the other hand, transposon sense transcripts (typically from active elements) and antisense transcripts (typically from piRNA clusters) participate in the process of secondary piRNA biogenesis: Here, piRNA-mediated cleavage of the target transcript triggers the production of a novel piRNA with the reciprocal polarity (Brennecke et al, 2007; Gunawardane et al, 2007). Hallmarks of this so-called ping-pong amplification of piRNAs are conserved from sponges to mammals (Aravin et al, 2007; Grimson et al, 2008).
The existence of two distinct piRNA biogenesis branches is particularly evident in the Drosophila ovary. Within ovarian germ cells, the three PIWI proteins Piwi, Aubergine and Argonaute 3 (Ago3) are co-expressed and piRNAs are generated via the primary and secondary pathways. The two major players of the secondary ping-pong pathway are Aubergine and Ago3 with Aubergine binding primarily cluster derived antisense piRNAs, while Ago3 is primarily complexed with transposon mRNA-derived sense piRNAs (Brennecke et al, 2007; Gunawardane et al, 2007; Li et al, 2009; Malone et al, 2009). In contrast, the surrounding follicle cells (somatic origin) express exclusively Piwi and piRNAs are produced only via the primary pathway (Lau et al, 2009; Li et al, 2009; Malone et al, 2009; Saito et al, 2009).
As all three PIWI proteins are expressed in germline cells, accurate systems must be in place to guarantee controlled piRNA biogenesis and PIWI loading. Several recent studies indicate that modular interactions between PIWI proteins and TUDOR domain-containing proteins are part of this control system (Chen et al, 2009; Kirino et al, 2009, 2010; Nishida et al, 2009; Reuter et al, 2009; Vagin et al, 2009). The TUDOR domain is a member of the TUDOR ‘royal family’, which among others also contains Chromo, plant Agenet, MBT and PWWP domains (Maurer-Stroh et al, 2003). The core TUDOR domain spans ∼60 amino acids and folds into a strongly bent anti-parallel β-sheet with five strands forming a barrel-like fold (Sprangers et al, 2003; Chen et al, 2009; Friberg et al, 2009; Liu et al, 2010a, 2010b). A key function of this domain is to facilitate protein–protein interactions, which often depend on the post-translational methylation of Lysine or Arginine residues in target proteins. Indeed, several methylated Arginine residues have been identified in PIWI-family proteins and at least in some cases specific interactions between PIWI and TUDOR proteins require the symmetric di-methylation of Arginine residues (sDMAs) in PIWI proteins (Kirino et al, 2009, 2010; Nishida et al, 2009; Reuter et al, 2009; Vagin et al, 2009; Huang et al, 2011b). Based on the observed specificity of PIWI–TUDOR interactions, it is possible that an intricate sDMA code allows the controlled recruitment of selected TUDOR domain-containing proteins at specific points of the life cycle of PIWI–piRNA complexes.
In Drosophila, six (Tudor, Spindle-E, Krimper, Tejas, Yb and Papi) out of the roughly 20 proteins implicated in the piRNA pathway contain TUDOR domains (Boswell and Mahowald, 1985; Gillespie and Berg, 1995; Lim and Kai, 2007; Malone et al, 2009; Nishida et al, 2009; Olivieri et al, 2010; Patil and Kai, 2010; Qi et al, 2010; Saito et al, 2010; Liu et al, 2011). We therefore decided to systematically analyse all fly TUDOR domain-containing proteins for an involvement in the piRNA pathway. This led to the identification of four novel TUDOR proteins as essential piRNA pathway factors. We characterized in detail the role of CG4771 (Vreteno), a tandem TUDOR domain-containing protein. Vreteno localizes to Yb bodies in follicle cells and to nuage in germline cells and is required for primary piRNA biogenesis in both cell types. Vreteno interacts with the three fly Tdrd12 proteins Yb, CG11133 (Brother of Yb) and CG31755 (Sister of Yb), which have partially overlapping functions in the somatic and germline piRNA pathways.
Results
Identification and classification of TUDOR domain-containing proteins in Drosophila
We mined the Drosophila melanogaster proteome for TUDOR-clan domains (Pfam CL0049) using sensitive sequence–profile (HMMer) and profile–profile comparison methods (Soding et al, 2005). Supplementary Table SI lists all identified proteins and specifies the individual subclasses (see also Figure 1A). For further analysis we focused on the TUDOR-clan domains TUDOR and SMN, which both have been reported to bind sDMA residues (Selenko et al, 2001; Sprangers et al, 2003; Cote and Richard, 2005; Liu et al, 2010a, 2010b). This resulted in 22 proteins containing at least one TUDOR/SMN domain.
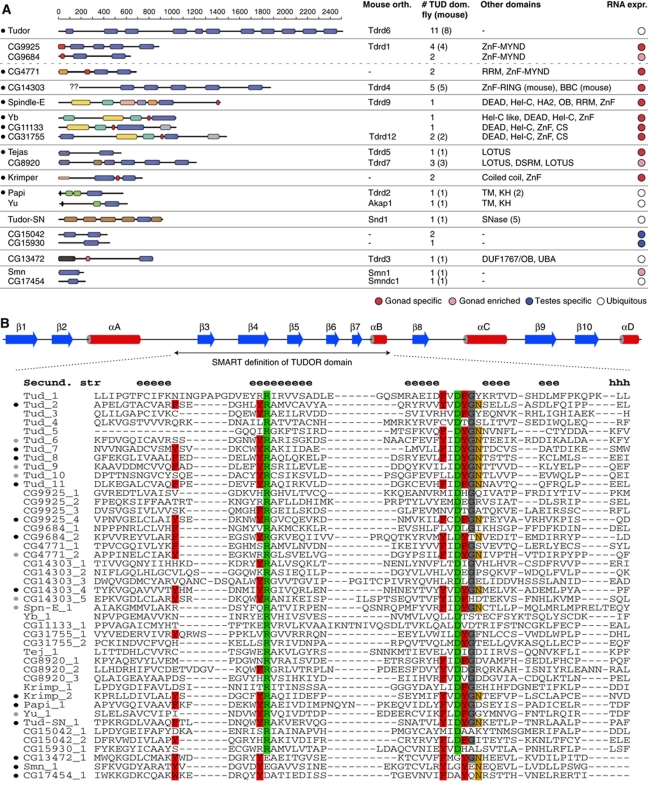
Figure 1.
Characterization of the Drosophila TUDOR proteins. (A) Cartoon showing all Drosophila melanogaster proteins containing TUDOR/SMN domains (blue boxes). All other significant protein domains identified via HHpred searches are indicated with coloured boxes and their identity is given to the right from N to C (ZnF: zinc finger; RRM: RNA recognition motif; BBC: B-Box C-terminal domain; DEAD: DEAD-Box RNA Helicase; Hel-C: Helicase C-terminal; HA2: Helicase associated domain; OB: oligo-nucleotide binding; CS: HSP20-like domain; DSRM: double-stranded RNA binding; TM: trans-membrane domain; KH: K homology; SNase: Staphylococcus nuclease; DUF: domain of unknown function; UBA: ubiquitin-associated domain). TUDOR proteins implicated in the piRNA pathway (including the ones from this study) marked with a black dot (left). The scale indicates amino-acid positions. The identified mouse orthologues (see Supplementary Figure S1), the number of identified TUDOR domains in fly (mouse) and the expression bias towards gonads in adult flies are shown to the right. Proteins with similar domain composition are grouped together. For CG14303, the ‘??’ indicate the non-annotated N-terminus. (B) The secondary structure cartoon (blue indicates β-strands, red α-helices) denotes the extended TUDOR domain and is based on Liu et al (2010a) (see also Supplementary Figure S1). The core TUDOR domain (SMART definition) is shown as an alignment for all identified TUDOR domains (‘e’ and ‘h’ above the alignment indicate β-strands and α-helices, respectively). The conserved Arginine and Aspartate residues present in all extended TUDOR domains are highlighted in green, aromatic cage residues in red, the Asparagine involved in sDMA binding in orange and a strongly conserved glycine in grey. To the left, the predicted likelihood of a domain to bind sDMA residues (based on the aromatic cage residues) is indicated with black (likely binder) and grey (potential binder) circles.
An alignment of all TUDOR/SMN domains contained in this set indicates three subgroups (Supplementary Figure S1). Groups A (Smn, CG13472 and CG17454) and B (Otu, CG3251) show similarity only to the ∼60 amino-acid TUDOR core. All other sequences cluster together in group C and share significant similarity also N- and C-terminal to the TUDOR core. Characteristic for group C are two 100% conserved amino acids, an Arginine in β4 and an Aspartate in the loop linking β5 and β6 of the extended TUDOR structure (Supplementary Figure S1; marked in green in Figure 1B; Liu et al, 2010a). Based on structural studies, group C sequences represent extended TUDOR domains, which are characterized by a core TUDOR domain tightly interacting with an OB-fold that consists of the N-terminal and C-terminal extensions (Liu et al, 2010a, 2010b). So far, every TUDOR domain-containing protein that has been linked to the piRNA pathway belongs to the extended TUDOR group.
To further characterize the set of proteins harbouring extended TUDOR domains, we annotated all additionally contained protein domains and searched for the corresponding mouse orthologues (Figure 1A; Supplementary Figure S2). Most of the fly proteins exhibit strong similarity to their mouse counterparts and the listed pairs in Supplementary Figure S2 are supported by multiple independent orthology assignment methods. CG14303 was linked to Rnf17 based on automated orthology identification (Inparanoid, Compara, OMA) and similarities in the TUDOR domains and an N-terminal B-Box C-terminal domain. We note that the N-terminus of CG14303 is not annotated in FlyBase (lack of EST data), indicating that the similarities between CG14303 and Rnf17 might also include the RING-type zinc finger found in Rnf17. Notably, an N-terminal RING finger could be identified in the Apis and Bombyx CG14303/Rnf17 orthologues. Murine Tdrd12 was assigned to CG11133 and CG31755 based on OrthoMCL (v2:OG2_82474 and v4:OG4_21213). Since both of these proteins share a similar domain composition with the piRNA pathway protein Yb, it appears that Tdrd12 has radiated in Drosophila into three proteins. Indeed, all three fly proteins are more related to each other than to the single mouse or human Tdrd12 proteins. Less obvious was the assignment of Tdrd1, a 4 × TUDOR domain protein with an N-terminal MYND-type zinc finger. Based on domain composition, Tdrd1 might be the single mammalian counterpart of fly CG9925, CG9864 and CG4771, all of which encode besides multiple TUDOR domains also a MYND zinc finger (CG4771 contains in addition an RRM domain). Fly proteins with no assignable mouse counterparts are Krimper as well as the two testes specific proteins CG15042 and CG15930 (the two TUDOR domains of Krimper and CG15042 are highly similar, potentially suggesting a common ancestor). Finally, mouse Tdrd8 seems to lack a detectable fly orthologue.
TUDOR/SMN domains often bind peptides with sDMA residues in target proteins. The sDMA-binding pocket resides within the TUDOR core. It consists of four aromatic residues (Figure 1B, marked in red), whose aromatic rings form a cuboid cage and complex the di-methylated guanidine group (Selenko et al, 2001; Sprangers et al, 2003; Cote and Richard, 2005; Liu et al, 2010a, 2010b). An additional conserved Asparagine (Figure 1B, marked in orange) interacts with the sDMA residue via a hydrogen bond (Liu et al, 2010a, 2010b). In sDMA-binding TUDOR domains, the aromatic cage residues are highly conserved and are critical for sDMA binding. We inspected the Drosophila extended TUDOR domains for aromatic cage residues. The alignment in Figure 1B indicates that while a set of TUDOR domains harbours all of these important residues at the exact same position, numerous TUDOR domains seemingly lost the ability to bind sDMA residues due to multiple amino-acid exchanges at critical positions. Nevertheless, many of the identified proteins contain at least one TUDOR domain with an intact aromatic cage and therefore likely interact with sDMA residues.
We finally analysed the RNA expression pattern of all TUDOR/SMN genes via the adult Drosophila Fly Atlas (Chintapalli et al, 2007). This showed a strong bias for genes with extended TUDOR domains to be expressed in ovaries and/or testes, further suggesting a link to piRNA biology (Figure 1A; Supplementary Figure S3).
Defining the set of TUDOR proteins with critical roles in the ovarian piRNA pathway
The implication of several TUDOR proteins in piRNA biology and their often gonad-specific expression prompted us to genetically test all proteins with extended TUDOR domains for their involvement in the piRNA pathway. Defects in the piRNA pathway lead to sterility and to a substantial accumulation of transposon transcripts in ovaries. We therefore assayed these phenotypes in females where individual TUDOR domain-containing proteins were knocked down via RNAi specifically in the ovarian soma (marked in green in Figure 2A and B) or in the germline (marked in beige in Figure 2A and B).
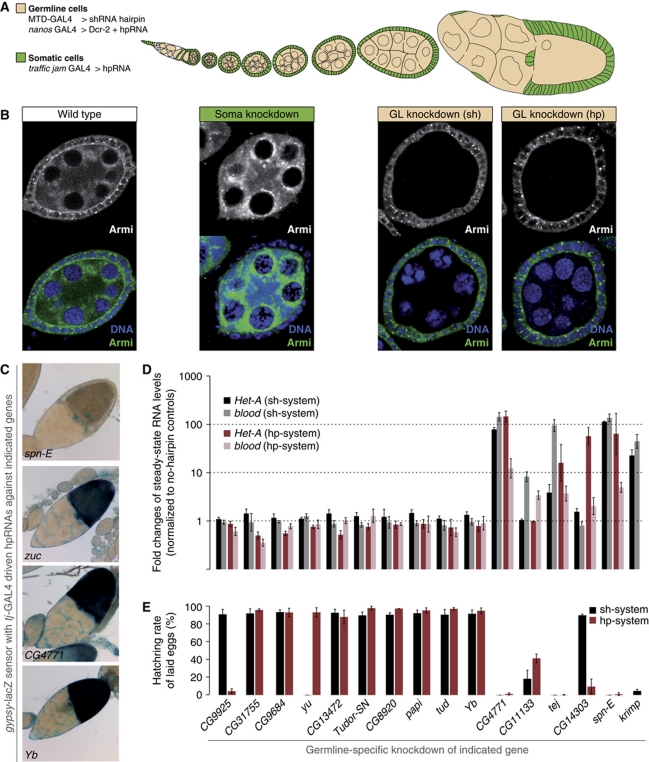
Figure 2.
The set of TUDOR proteins involved in the Drosophila piRNA pathway. (A) Cartoon of a Drosophila ovariole (somatic cells are in green, germline cells are in beige). The RNAi systems used for the two cell types are listed. (B) Immunostaining of Armitage (green) and DNA (blue) in egg chambers expressing RNAi constructs in a tissue-specific manner (left: wild type; middle: soma knockdown via tj-GAL4>hpRNA; right: germline knockdown via MTD-GAL4>shRNA or NGT-GAL4>Dcr-2+hpRNA). Monochrome panels show only the anti-Armitage channel. (C) Bright field images of ovarioles stained for β-GAL activity. The individual genotypes represent soma-specific knockdowns of the indicated genes in the background of the gypsy-lacZ sensor described in Sarot et al (2004). zucchini knockdown serves as a positive control and spindle-E as negative control. Of all TUDOR knockdowns, only those against CG4771 or Yb resulted in sensor de-repression. (D) Changes in steady-state levels of HeT-A and blood transposon transcripts upon knockdown of individual TUDOR proteins in the germline with the shRNA (black/gray) or the hpRNA (red/rose) knockdown systems (normalized to no-hairpin controls via rp49; log scale; n=3; error bars indicate s.d.). Identity of knocked down genes identical to the legend in (E). (E) Fertility rates of females with germline-specific knockdown of indicated TUDOR proteins using the shRNA (black) and the hpRNA (red) systems (∼200 eggs per experiment; n=3; error bars indicate s.d.).
RNAi in the follicular epithelium (soma), where only the primary piRNA pathway is active, was based on tj-GAL4 driven dsRNA-hairpin constructs (hp-lines) from the VDRC (Vienna Drosophila RNAi Centre) library (Dietzl et al, 2007; Olivieri et al, 2010). For the germline we expressed short hairpin constructs (sh-lines) with the germline-specific MTD-GAL4 driver, which allows robust knockdowns (Haley et al, 2008; Ni et al, 2011). In addition, we took advantage of the observation that VDRC hp-lines induce potent RNAi in the germline if expressed in conjunction with Dicer-2 (Sidney Wang and Sarah Elgin, personal communication). Figure 2B illustrates specificity and efficacy of the soma and germline-specific knockdowns using the piRNA biogenesis factor Armitage as an example.
Integrity of the somatic piRNA pathway was monitored via a gypsy-lacZ construct that accurately reports piRNA-mediated silencing in follicle cells (Figure 2C; Sarot et al, 2004; Olivieri et al, 2010). Integrity of the germline piRNA pathway was monitored via the steady-state RNA levels of the two transposons HeT-A and blood (Figure 2D). In addition, we determined female fertility rates (percentage of hatched eggs) for all knockdowns (Figure 2E). The two germline knockdown approaches yielded in nearly all cases identical results. We attribute the three exceptions (CG9925-hp, yu-sh, CG14303-sh; Figure 2D and E) to off-target effects or non-functional RNAi lines.
Seven TUDOR proteins scored as putative piRNA pathway components (CG4771, CG11133, Tejas, CG14303, Spindle-E, Krimper, Yb). All four factors that had previously been shown to be essential pathway members (Spindle-E, Krimper, Tejas, Yb) were identified. In agreement with the literature, Spindle-E, Krimper and Tejas scored only in the germline knockdowns while Yb scored only in the soma assay (Lim and Kai, 2007; Malone et al, 2009; Szakmary et al, 2009; Olivieri et al, 2010; Patil and Kai, 2010). Papi and Tudor—though previously implicated in the pathway—did not result in transposon de-silencing or sterility. This is in agreement with the literature as both proteins are dispensable for fertility and corresponding mutant ovaries contain no or only slightly elevated transposon RNA levels (Nishida et al, 2009; Liu et al, 2011). We note that the grandchild-less phenotype for Tudor (Boswell and Mahowald, 1985) is recapitulated in the Tudor germline knockdowns.
In addition to the known factors, germline knockdowns of three uncharacterized proteins (CG14303, CG4771, CG11133; Figure 2D and E) resulted in sterility and transposon silencing defects. Out of these, CG4771 was also identified as an essential component for the somatic piRNA pathway (Figure 2C) and we therefore decided to characterize this factor in more detail.
Vreteno (CG4771) is an essential piRNA pathway factor
CG4771 is localized on the third chromosome (Figure 3A) and encodes a protein with two extended TUDOR domains (Figure 3B). The C-terminal TUDOR domain might possess sDMA-binding activity (Figure 1B) and the relevant aromatic cage residues are conserved in distantly related Drosophila species (Figure 3B). In addition, CG4771 harbours an N-terminal RRM domain and a highly conserved zinc finger belonging to the MYND family (C2C4HC).
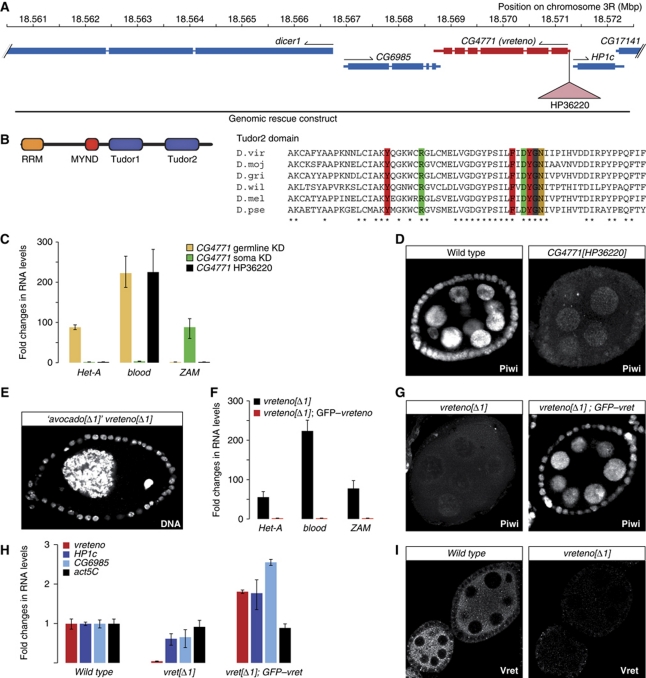
Figure 3.
Vreteno is a novel piRNA pathway member. (A) Overview of the CG4771 (vreteno) genomic locus indicating flanking genes (blue), the HP36220-insertion site (pink triangle) and the extent of the genomic rescue construct. (B) Cartoon of the CG4771 protein domain structure and sequence alignment of the C-terminal TUDOR domain in distantly related Drosophilids (virilis, mojavensis, grimshawi, willistoni, melanogaster, pseudoobscura). Aromatic cage residues and the conserved Arg/Asp residues colour coded as in Figure 1B. (C) Changes in steady-state transposon levels (n=3; s.d.) upon CG4771 knockdown (normalized to no-hairpin controls) in soma (green) or germline (beige) in comparison to those in CG4771[HP36220] mutants (black; normalized to heterozygotes). (D) Immunostaining of Piwi in wild-type and CG4771[HP36220] mutant egg chambers. (E) The occasionally observed egg chamber morphology of CG4771[Δ1] (vreteno) mutants, which originally led us to name the gene ‘avocado’ (DNA stained with DAPI). (F) RNA levels of CG4771, of the flanking genes HP1c and CG6985 and of actin-5C in vreteno[Δ1] mutant ovaries compared with vreteno[Δ1]; GFP–vreteno rescued ovaries (values normalized to w[1118] controls). (G) Immunostaining of Piwi in vreteno[Δ1] mutant egg chambers and in vreteno[Δ1] mutant egg chambers expressing a GFP–vreteno rescue construct. (H) Steady-state RNA levels of the HeT-A, blood and ZAM transposons in vreteno[Δ1] mutant ovaries compared with vreteno[Δ1]; EGFP–vreteno rescued ovaries (values normalized to heterozygous siblings; n=3; error bars indicate s.d.). (I) Immunostaining of Vreteno in wild-type and vreteno[Δ1] mutant egg chambers at identical microscope settings.
To verify that CG4771 is a piRNA pathway factor, we obtained genetic alleles of this gene. Females homozygous for the P-insertion HP36220 (Bloomington), which is inserted into the 5′UTR of CG4771 (Figure 3A) were sterile and laid eggs that exhibited defects in dorso-ventral patterning as evidenced by a high percentage of fused dorsal appendages. This is a common phenotype of piRNA pathway mutants and stems from the activation of the Chk2 DNA damage pathway, presumably caused by widespread DNA damage originating from uncontrolled transposon activity (Chen et al, 2007; Klattenhoff et al, 2007). However, in HP36220 mutants only the blood element was de-repressed, although germline-specific and soma-specific knockdowns of CG4771 clearly de-repressed also HeT-A and ZAM, respectively (Figure 3C). This suggested that HP36220 is a hypomorphic allele. Indeed, nuclear Piwi localization in the mutant was impaired, yet to a lesser degree than in armitage or Yb mutants (Figure 3D; Olivieri et al, 2010).
We therefore generated an additional allele by mobilizing the HP36220 element. Out of 280 analysed excision events, one line (CG4771[Δ1]) exhibited a more pronounced phenotype as homozygous females failed to lay eggs. The ovarian morphology of CG4771[Δ1] mutants strongly resembled those of armitage or zucchini null ovaries (Pane et al, 2007; Olivieri et al, 2010). In some egg chambers, we observed besides the oocyte nucleus a single giant nurse cell nucleus, indicating severe defects in cytokinesis. Based on the morphology of these egg chambers (Figure 3E), we initially named CG4771 ‘avocado’. While this work was under review, CG4771 was named ‘vreteno’ in FlyBase by the Lehmann group (‘vreteno’ means ‘spindle’ in Bulgarian, referring to the spindle class phenotype of eggs laid by CG4771 mutants) and we therefore adopted this name for consistency reasons.
In vreteno[Δ1] homozygous ovaries, germline- and soma-specific transposons were severely de-repressed, indicating the stronger nature of this allele (Figure 3F). This was paralleled by a more pronounced defect in nuclear Piwi accumulation (compare Figure 3G and D). To verify that the vreteno[Δ1] phenotype is due to defects in the CG4771 locus, we restored fertility (not shown), transposon silencing (Figure 3F) and nuclear Piwi localization (Figure 3G) to wild-type levels by introducing a genomic rescue construct that expresses GFP-tagged Vreteno under its endogenous regulatory regions (Figure 3A). As this rescue construct also contained the complete loci for HP1c and CG6985, we measured steady-state RNA levels of all three genes in ovaries of vreteno[Δ1] mutants and of the GFP–vreteno rescued animals (Figure 3H). This confirmed the specificity of the Δ1 allele for the vreteno locus. Immunofluorescence analysis with an antibody recognizing the Vreteno N-terminus further indicated that also the protein is essentially not detectable in ovaries from vreteno[Δ1] mutants (Figure 3I).
We also analysed the requirement of vreteno for the piRNA pathway in males. Towards this end, we measured steady-state levels of the transposons mdg1 and copia as well as of the repetitive Stellate locus that is under control of the piRNA pathway in testes. This indicated a requirement of vreteno for copia and Stellate silencing, supported by the observation that silencing was fully restored in males expressing a GFP–vreteno rescue construct (Supplementary Figure S4). Taken together, vreteno encodes a novel piRNA pathway factor that is essential for the ovarian and testes piRNA pathways.
Vreteno is required for primary piRNA biogenesis in soma and germline
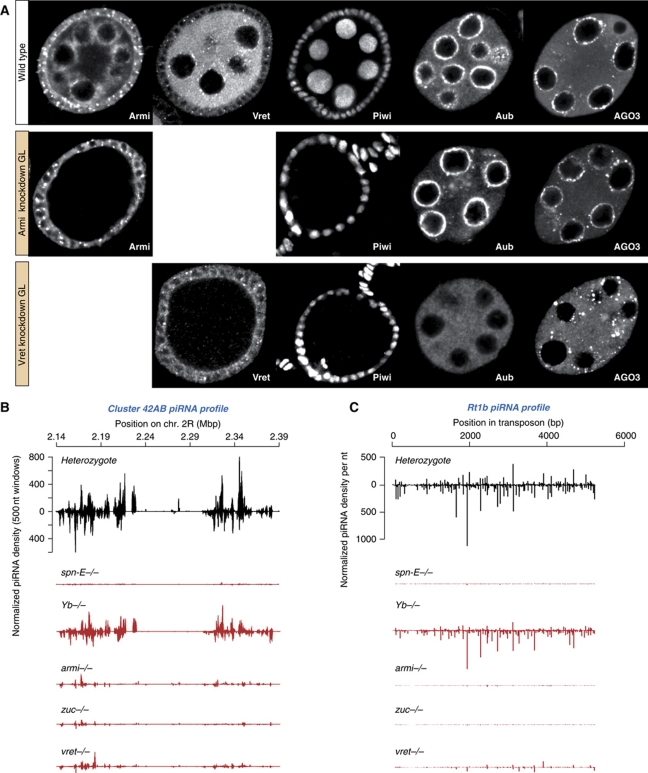
Defects in primary piRNA biogenesis (e.g. in armitage or zucchini mutants) result in a collapse of piRNA populations in follicle cells, in a severe reduction of most germline piRNA species and in defects in Piwi's nuclear accumulation accompanied by a significant loss of Piwi protein (Pane et al, 2007; Malone et al, 2009; Haase et al, 2010; Olivieri et al, 2010; Saito et al, 2010). Delocalization and decreased levels of Piwi were also observed in the vreteno[Δ1] mutant (Figure 3D and G). We therefore analysed the effects of loss of Vreteno on PIWI-family proteins and on piRNA populations in the ovarian soma and germline.
Similar to an RNAi-mediated Armitage knockdown, knockdown of Vreteno in follicle cells led to an almost complete loss of Piwi protein in these cells (Figure 4A), indicating defects in primary piRNA biogenesis. Indeed, analysis of small RNA populations obtained from vreteno[Δ1] mutant ovaries showed that piRNAs originating from the flamenco cluster, which gives rise to primary piRNAs in follicle cells (Lau et al, 2009; Li et al, 2009; Malone et al, 2009), were almost entirely lost in vreteno[Δ1] mutants (Figure 4B). Highly similar profiles were obtained from ovaries mutant for armitage, zucchini or Yb, the only known factors involved in primary piRNA biogenesis. In contrast, piRNA populations from spindle-E mutant ovaries showed no impact, in agreement with spindle-E functioning exclusively in the germline pathway (Figure 4B; Malone et al, 2009). Very similar results were obtained when piRNAs mapping to individual soma-specific transposons such as Tabor or ZAM were analysed (Supplementary Figure S5).
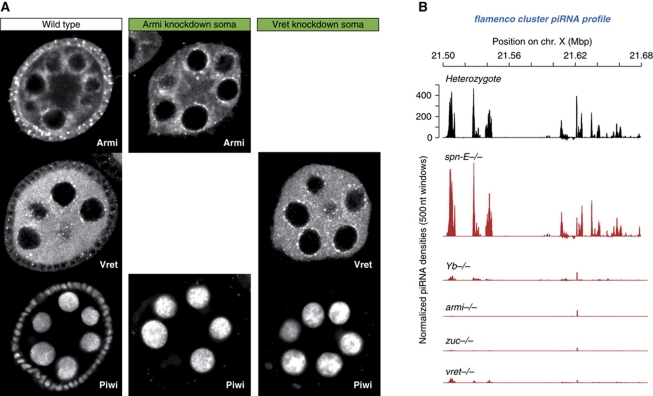
Figure 4.
Vreteno is essential for primary piRNA biogenesis in the soma. (A) Immunostaining of Piwi (lower panels) in wild-type egg chambers (left) in comparison to egg chambers expressing hpRNAs against armitage (centre) or vreteno (right) specifically in somatic cells. Armitage and Vreteno stainings indicate the knockdown efficiency. (B) Normalized piRNA profiles (23–30 nt small RNAs) obtained from control ovaries (vret heterozygote; black) in comparison to profiles obtained from indicated mutant ovaries (red) mapping uniquely to the soma-specific piRNA cluster flamenco. The y axis for the heterozygote plot is representative for all plots.
Knockdown of Vreteno in the germline resulted in a near complete loss of Piwi protein, again pheno-copying the Armitage germline knockdown (Figure 5A). In addition, piRNA populations mapping to the germline-specific piRNA cluster 42AB or to the transposon Rt1b, whose piRNA populations depend on primary piRNA biogenesis (Malone et al, 2009), were severely reduced in vreteno mutant ovaries as well as in spindle-E, armitage or zucchini mutant ovaries (Figure 5B and C). No significant changes were observed in Yb mutant ovaries consistent with this gene acting in somatic follicle cells only. We also observed a severe disruption of the nuage localization of Aubergine and Ago3 in germline cells depleted of Vreteno (Figure 5A). This is significant, as loss of the primary biogenesis factor Armitage does not impact the subcellular localization of Aubergine and Ago3 (Figure 5A). Also in armitage null mutants, Aubergine and AGO3 localization to nuage was not significantly perturbed (not shown), indicating that the observed differences cannot be attributed to insufficient knockdowns.
Figure 5.
Vreteno is essential for piRNA biogenesis in the germline. (A) Immunostaining of Piwi, Aubergine and Ago3 in wild-type egg chambers (top row) in comparison to egg chambers expressing shRNAs against armitage (centre row) or vreteno (lower row) specifically in germline cells. Armitage and Vreteno stainings indicate the knockdown efficiency. (B) Normalized piRNA profiles (23–30 nt small RNAs) obtained from control ovaries (vret heterozygote; black) in comparison to profiles obtained from indicated mutant ovaries (red) mapping uniquely to the germline-specific piRNA cluster 42AB (sense and antisense piRNAs are indicated with peaks pointing up- and downwards). (C) Normalized piRNA profiles obtained from control ovaries (black) in comparison to profiles obtained from indicated mutant ovaries (red) mapping to the germline-dominant Rt1b transposon.
To characterize piRNA populations from vreteno mutants in more detail, we mapped small RNAs isolated from ovaries mutant for vreteno, armitage, zucchini, Yb or spindle-E as well as small RNAs isolated from their respective heterozygote controls to annotated transposon families (Jurka et al, 2005; Supplementary Table S2). While the spindle-E libraries were taken from the literature (Malone et al, 2009), armitage, zucchini and Yb libraries were prepared for this study (previous libraries were based on unfavourable genetic alleles in the case of armitage, or suffered from inaccurate genotyping in the case of zucchini; Malone et al, 2009). All heterozygote libraries were scaled to 1 million repeat-derived small RNAs (23–30 nt in length). Each mutant library was then normalized to its respective heterozygote library using non-transposon-derived endo-siRNA populations.
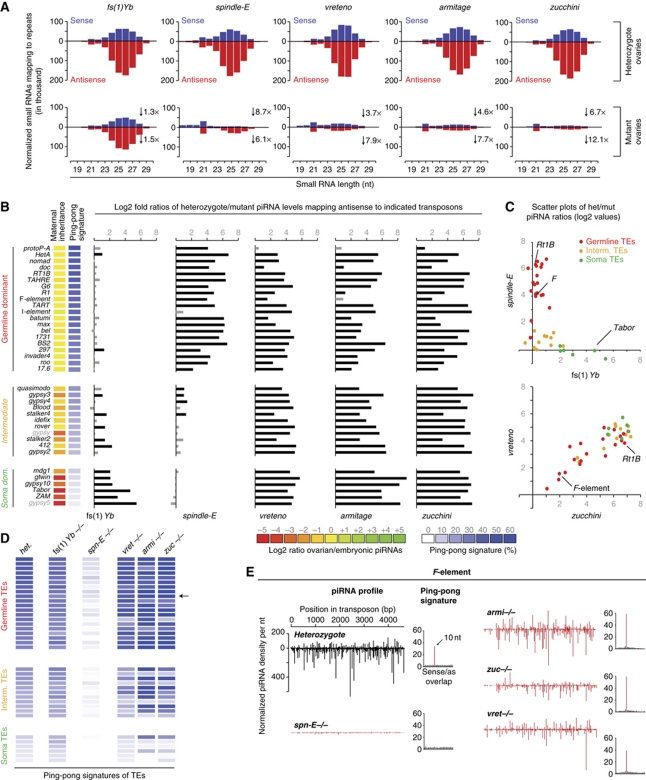
Figure 6A displays the levels of small RNAs that mapped to annotated transposons in sense or antisense orientation as a function of their length. The mutant small RNA profiles could be grouped into three classes: (1) Yb is a soma-specific factor essential for primary piRNA biogenesis (Olivieri et al, 2010; Qi et al, 2010; Saito et al, 2010). Accordingly, the majority of ovarian piRNAs were unchanged in this mutant. Only a slight decrease (preferentially in antisense piRNAs) was seen, in agreement with soma piRNAs exhibiting an extreme antisense bias (Malone et al, 2009). (2) Spindle-E is required for the germline-specific ping-pong cycle but is dispensable for primary piRNA biogenesis (Malone et al, 2009). Accordingly, spindle-E mutant piRNA profiles showed a severe collapse in piRNA populations, with a subpopulation of piRNAs (nearly exclusively antisense) remaining. These showed a pronounced bias towards a 5′ terminal Uridine, lacked the characteristic ping-pong signal (see below) and therefore represent primary piRNAs (not shown). (3) The vreteno, armitage and zucchini mutant libraries strongly resembled each other. They exhibited a severe depletion of piRNAs, with more pronounced losses in antisense populations, indicative of these factors being required for primary piRNA biogenesis.
Figure 6.
Vreteno, Zucchini and Armitage are essential primary piRNA biogenesis factors but are dispensable for the ping-pong cycle. (A) Length profiles of all repeat-derived (transposon and satellite repeats) small RNAs (18–30 nt) isolated from ovaries of the indicated mutants and their respective heterozygous controls (all heterozygote libraries normalized to 1 million repeat-derived 23–30 nt RNAs). Sense populations are in blue and antisense populations are in red. The fold decrease in the respective populations (23–30 nt only) is indicated. (B) Bar diagram indicating the changes of normalized piRNAs mapping antisense to the indicated transposons (left) in the indicated mutant ovaries compared with the respective heterozygous control ovaries (het/mut ratios are given as log2 values). Grey bars indicate values below 1 (less than two-fold changes). Identity of the analysed mutants is given at the bottom. Transposons are grouped into germline-dominant (red), intermediate (yellow) and soma-dominant (green) based on Malone et al (2009). The heatmaps indicate degree of maternal piRNA inheritance (yellow: strong; red: weak) and ping-pong signature (blue: strong; white: absent) of each individual element. (C) Scatter plots of the log2 values plotted in (B), where individual transposons are colour coded as in (B). (D) Ping-pong signatures of the individual transposons (classification and order as in (B) in the average heterozygote (het.) and the indicated mutants as a heatmap ranging from strong signals (dark blue) to no signal (white). The F-element is indicated with an arrow. (E) Normalized piRNA profiles (sense and antisense) mapping to the F-element. Compared are populations from heterozygotes (black) to the indicated mutants (red). Ping-pong signatures (basis for the heatmap in (D) are shown to the right of each plot.
The described classification of the analysed pathway mutants was supported by the changes in transposon-specific piRNA populations (Figure 6B and C). We focused on a set of 37 transposon families that has been well characterized previously (Malone et al, 2009). These transposons can be grouped into three cohorts (Figure 6B) depending on whether piRNAs targeting them are predominantly found in germline cells (germline dominant), in somatic cells (soma dominant) or in both cell types (intermediate). Transposons shown in Figure 6B were ranked according to their ping-pong signature in the average heterozygote library (blue heatmap). This correlated strongly with the previously reported extent of maternal piRNA inheritance (yellow/red heatmap) as maternal deposition is only possible for germline piRNAs, which typically participate in the ping-pong cycle (Malone et al, 2009).
We mapped piRNAs to the selected transposon families and calculated the log2 fold ratios of the respective heterozygote/mutant pairs. In agreement with the genetic data, piRNA populations from Yb and spindle-E mutants were essentially anti-correlated (Figure 6B). While Yb mutations affected soma-dominant elements, spindle-E mutations affected germline-dominant elements. A scatter plot of the log2 fold het/mut ratios underlines this further (Figure 6C; r=−0.52; Pearson correlation). In contrast, libraries obtained from vreteno, armitage or zucchini mutants exhibited nearly identical losses of piRNA populations mapping to soma, intermediate or germline-dominant elements (Figure 6B; Pearson correlations: r(armi/zuc)=0.80; r(armi/avo)=0.76; r(zuc/avo)=0.87). The soma-dominant elements were the most consistently affected group, in agreement with all of them depending on primary piRNA biogenesis. Within the germline-dominant group, some transposons showed only very mild losses of piRNAs (e.g. protoP-A, Doc, F-element). This pattern was observed in all three libraries (see e.g. the bottom scatter plot in Figure 6C), further underlining the hypothesis that all three factors participate in a common step of piRNA biogenesis.
We finally analysed the impact of the five mutants on the ping-pong cycle (Figure 6D). While ping-pong collapsed in spindle-E mutants (Malone et al, 2009), it was unaffected in Yb mutants, consistent with this factor being soma specific. Interestingly, vreteno, armitage or zucchini mutants showed no defects in ping-pong signatures. On the contrary, for many elements, the ping-pong signal was elevated in comparison to the wild-type situation. This trend was particularly evident for the intermediate group of transposons, especially in armitage and zucchini mutants. Of note, ping-pong was still efficient in vreteno mutants despite the fact that the key players Aubergine and Ago3 were delocalized from nuage (Figure 5A). This indicates that nuage localization per se is not required for ping-pong.
Figure 6E shows sense and antisense piRNA populations mapping to the F-element, which is a particularly interesting case. piRNA populations for this element were highly sensitive to perturbations in the ping-pong cycle (spindle-E mutant), but not to defects in primary piRNA biogenesis (vreteno, armitage, zucchini mutants). The increased ping-pong signal in primary pathway mutants suggests that in these mutants, the pool of primary piRNAs (lacking ping-pong signatures) is eliminated, thereby leaving only ping-pong pairs. For the Rt1B element (also a germline-dominant element), this is different as piRNA populations in this case evidently depend on ping-pong amplification as well as on primary piRNA biogenesis (Figure 5C). Nevertheless, the small pool of remaining Rt1B piRNAs in primary pathway mutants still displays strong ping-pong signatures. We speculate that these element-specific differences are related to the maternally transmitted piRNA pool (Brennecke et al, 2008). For unknown reasons, the F-element ping-pong might be maintained by maternally transmitted piRNAs, whereas this is not the case for elements such as Rt1B.
In summary, our analysis shows that Vreteno is essential for primary piRNA biogenesis and that the ping-pong cycle can operate independently of primary piRNA biogenesis in Drosophila as suggested previously (Malone et al, 2009).
Vreteno accumulates in Yb bodies and physically interacts with Piwi, Armitage and Yb
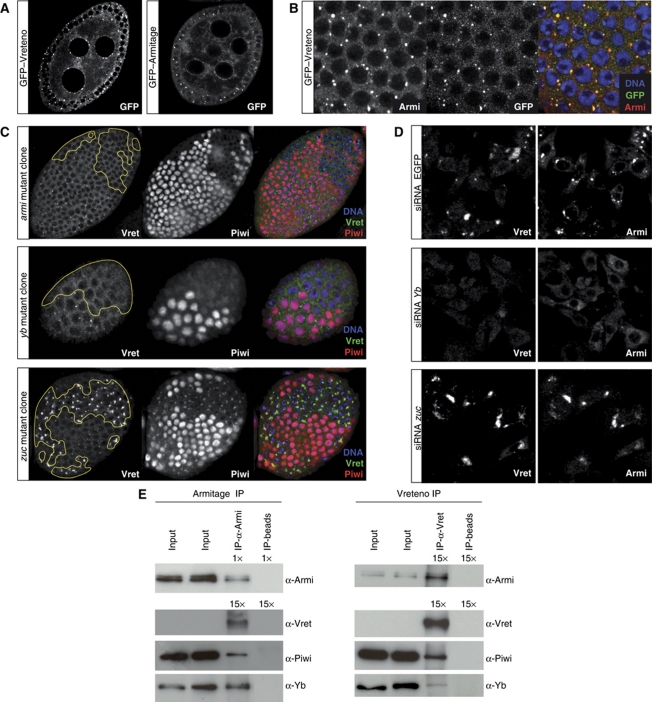
In follicle cells and in cultured somatic stem cells (OSCs; Niki et al, 2006; Saito et al, 2009), Armitage and Yb are enriched in discrete foci termed Yb bodies, which presumably are sites of primary piRNA biogenesis as Piwi transits through these bodies (Olivieri et al, 2010; Qi et al, 2010; Saito et al, 2010). To better understand at which step Vreteno acts during primary piRNA biogenesis, we performed genetic epistasis experiments in ovarian follicle cells as well as in OSCs. A GFP–Vreteno fusion protein expressed under the endogenous control regions localized to the cytoplasm of somatic and germline cells in the ovary (Figure 7A and B). Intriguingly, Vreteno was enriched in discrete foci in follicle cells and in nuage in germline cells, highly reminiscent of an Armitage–GFP fusion protein. A more detailed analysis of the subcellular localization in follicle cells revealed that the majority of Vreteno and Armitage foci precisely overlapped (Figure 7B). Also in OSCs, Vreteno localized to the cytoplasm and accumulated in distinct foci, which typically were Armitage positive (Supplementary Figure S6). In some cases, however, Vreteno and Armitage foci seemed to directly flank each other and in several cells we observed accumulation of Vreteno in large cytoplasmic domains that showed no Armitage accumulation (Supplementary Figure S6). Typically, these cells lacked discernable Armitage foci, potentially suggesting that Yb bodies undergo remodelling at specific stages.
Figure 7.
Vreteno is a novel Yb-body component. (A) Subcellular localization of GFP-tagged Vreteno or Armitage (optical section of egg chambers) expressed under the respective endogenous regulatory regions. (B) Confocal section through the follicular epithelium of a GFP–Vreteno (green) expressing egg chamber stained for Armitage (red) and DNA (blue). (Right panel) Merge of all three channels (co-localization of Vreteno and Armitage results in yellow). (C) Immunostaining of Vreteno (green), Piwi (red) and DNA (blue) in egg chambers, where clones of cells mutant for the indicated genes (left) have been induced in the follicular epithelium (clone borders are indicated with a yellow line). (D) Co-immunostaining of Vreteno (left) and Armitage (right) in OSCs transfected with siRNAs against EGFP (top), Yb (middle) or zucchini (bottom). (E) Co-IPs of Armitage and Vreteno from OSC lysate. Western blots against Armitage, Vreteno, Piwi and Yb. For the Armitage western blot (Armi-IP), only 1/15th compared with all other blots was loaded in the IP-lanes. Beads lacking antibody (IP-beads) served as control.
We next analysed the subcellular localization of Vreteno in cells lacking known primary biogenesis factors using mitotic follicle cell clones in flies or RNAi in OSCs. Vreteno localization to Yb bodies was dependent on Armitage and Yb (Figure 7C). Strikingly, loss of Zucchini, a phospho-lipase or putative nuclease localizing to the outer mitochondrial membrane (Pane et al, 2007; Saito et al, 2010; Watanabe et al, 2011; Huang et al, 2011a) resulted in the accumulation of Vreteno in massive Yb bodies, similar to what has been observed previously for Armitage (Figure 7C; Olivieri et al, 2010). In contrast, Vreteno localization was unperturbed in cells lacking Piwi (Supplementary Figure S7). Furthermore, Armitage and Yb still localized to Yb bodies in cells lacking Vreteno (Supplementary Figure S7). Comparable results were obtained in OSCs upon knockdown of individual factors (Figure 7D shows Yb and Zucchini knockdowns; Yb and zuc mRNA levels were reduced to 7.2% and 13.6%, respectively, resulting in 19-fold (Yb) or eight-fold (zuc) elevated mdg1 transposon levels).
We obtained further support for a role of Vreteno in primary piRNA biogenesis by co-immunoprecipitation (co-IP) experiments from OSC lysate. This indicated that Armitage and Vreteno reside in a common complex (supported by reciprocal co-IP experiments). In addition, both proteins were also found to physically interact with Yb and Piwi (Figure 7E).
Taken together, Vreteno is a novel Yb-body component that interacts with Armitage, Yb and Piwi. Vreteno localization depends on Armitage and Yb and in zucchini mutant cells it accumulates together with Armitage and Yb in large cytoplasmic aggregates.
The Tdrd12 proteins CG11133 and CG31755 interact with Vreteno and are essential primary piRNA pathway factors in the germline
Vreteno is besides Piwi, Armitage and Zucchini the fourth known factor required for primary piRNA biogenesis in the ovarian soma and germline. In contrast, the Tdrd12-like protein Yb is an essential piRNA biogenesis factor but acts only in somatic cells (Olivieri et al, 2010; Qi et al, 2010; Saito et al, 2010). The Drosophila genome contains two additional Tdrd12-like proteins (CG11133, CG31755; Figure 8A), both of which are also specifically expressed in gonads (Supplementary Figure S3; FlyAtlas). Since Vreteno physically interacts with Yb, we reasoned that it might also interact with CG11133 and CG31755. We performed IP experiments from ovaries expressing GFP–Vreteno instead of the endogenous protein and from wild-type control ovaries using a monoclonal GFP antibody followed by quantitative mass spectrometry analysis. Strikingly, CG11133 and CG31755 were among the five most enriched proteins in the GFP–Vreteno IP (besides Vreteno, Hsp27 and CG9281; Supplementary Table S3).
Figure 8.
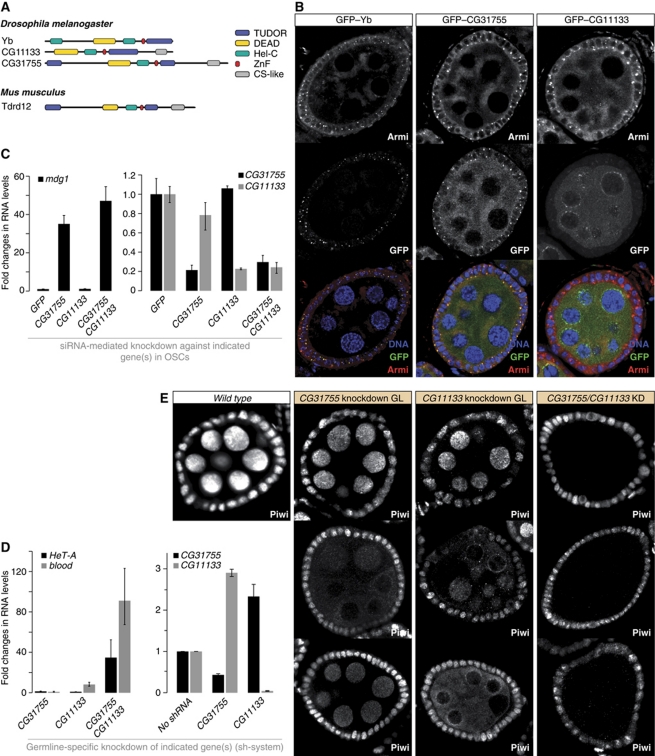
The Tdrd12 orthologues CG11133 and CG31755 are essential primary piRNA pathway factors in the germline. (A) Cartoon overview of the Tdrd12 proteins in fly and mouse (Tudor domains: blue; other domains as indicated). (B) Immunofluorescence staining for Armitage and GFP-tagged Yb, CG31755 or CG11133. (Lower panels) Merge of the three channels (DNA, GFP, Armitage). (C) The left chart shows changes in steady-state levels of the mdg1 transposon upon knockdown of CG11133, CG31755 or both together in OSCs. Values were normalized to GFP control knockdowns via rp49. The right chart indicates the efficiencies of the respective knockdowns (n=3; error bars indicate s.d.). (D) The left chart shows changes in steady-state levels of the HeT-A and blood transposons upon knockdown of CG11133, CG31755 or both together in germline cells (MTD-GAL4>shRNA). Values were normalized to ‘no-hairpin’ control flies via rp49. The right chart indicates the efficiencies of the respective knockdowns (n=3; error bars indicate s.d.). (E) Immunofluorescence staining for Piwi in wild-type egg chambers (left) compared with egg chambers where the indicated genes have been knocked down via shRNAs in the germline only. For the CG31755 and CG11133 single knockdowns, most egg chambers displayed a wild-type Piwi pattern (top), while 3–5% displayed the phenotypes of the lower panels. The phenotype for the double knockdown was fully penetrant.
To further characterize the fly Tdrd12 family, we analysed expression and localization of GFP-tagged Yb, CG11133 and CG31755 in ovaries (all proteins were expressed under their respective endogenous control regions). While CG31755 was expressed at comparable levels in follicle and germline cells, Yb was expressed specifically in follicle cells and CG11133 predominantly in germline cells (Figure 8B). Remarkably, the subcellular localizations of the three proteins were highly reminiscent of the Armitage or Vreteno localizations. Double labelling experiments confirmed the co-localization of Armitage with Yb and CG31755 in follicle cells (Figure 8B). Also for CG11133, though present only at very low levels, localization to Armitage foci in follicle cells was observed (not shown). In germline cells, CG31755 localized to peri-nuclear clouds that were also positive for Armitage, while CG11133 was enriched in nuage, where Armitage was also present (Figures 7A and 8B).
Given the follicle cell expression and Yb-body localization of CG31755 and CG11133, we investigated their somatic piRNA pathway involvement in OSCs by knocking them down, individually or in combination with RNAi. Depletion of CG31755, but not CG11133 resulted in a strong increase of mdg1 transposon levels, similar to what has been observed for zuc, armi or Yb knockdowns (Figure 8C; Saito et al, 2010). Double depletion of CG31755 and CG11133 led to an additional, but small increase in mdg1 transcript levels, suggesting a minor role for CG11133 in the somatic piRNA pathway.
We performed a similar analysis in germline cells. The initial survey of TUDOR proteins already indicated that CG11133 was critical for transposon silencing in the germline, while CG31755 was not (Figure 2D and E). We analysed silencing of the HeT-A and blood elements in ovaries depleted for CG31755 and/or CG11133 via the shRNA system (Figure 8D). While knockdown of CG31755 had no significant impact on HeT-A/blood silencing, a nearly 10-fold increase in blood levels was measured in CG11133 knockdown ovaries (Figure 8D). Strikingly, double depletion of CG31755 and CG11133 de-silenced HeT-A and blood to levels comparable to verified piRNA pathway mutants (Figures 2D and 8D). The observed silencing defects correlated with defects in Piwi levels and localization (Figure 8E). In the single knockdowns, most egg chambers displayed wild-type Piwi levels and localization. In about 3–5% of egg chambers, however, we observed reduced Piwi levels or cytoplasmic Piwi localization (representative images in Figure 8E). Strikingly, double depletion of CG11133 and CG31755 resulted in an almost complete loss of germline Piwi (Figure 8E). Based on these observations and the correlation between defects in nuclear Piwi accumulation and defects in primary piRNA biogenesis (Olivieri et al, 2010; Saito et al, 2010), we conclude that CG11133 and CG31755 function together in primary piRNA biogenesis in germline cells. We therefore named CG11133 ‘Brother of Yb’ and CG31755 ‘Sister of Yb’.
Discussion
The set of TUDOR domain-containing proteins in Drosophila
Whereas Tudor, the founding member of TUDOR domain-containing proteins was genetically identified >25 years ago as a grandchild-less gene in Drosophila (Boswell and Mahowald, 1985), the link between TUDOR domains and the piRNA pathway has only recently emerged: On the one side, PIWI proteins have been shown to contain sDMA residues, docking sites for TUDOR domains. In selected cases, sDMA-dependent interactions between TUDOR domains and PIWI proteins could indeed be shown (Nishida et al, 2009; Vagin et al, 2009; Kirino et al, 2010; Huang et al, 2011b). On the other hand, 10 out of the 17 identified proteins in Drosophila that contain an extended TUDOR domain have been directly linked to the piRNA pathway (including the genes identified in this study). We emphasize that our RNAi-based assays could only score those genes, which are individually required for the ovarian piRNA pathway and which perform essential roles in it. Figure 1A suggests that several TUDOR domain-containing genes could act redundantly (in particular yu and papi, CG9925 and CG9684, Tejas and CG8920, CG11133 and CG31755) or specifically in testes (CG15042, CG15930). Double knockdowns of these gene pairs might identify their possible involvements in the pathway.
What could be the role of this surprisingly large set of proteins in the piRNA pathway? The protein domain cartoon of the Drosophila TUDOR family (Figure 1A) suggests that numerous effector domains such as helicase domains, RNA-binding domains, zinc-finger domains, etc. are targeted to PIWI proteins via modular sDMA–TUDOR interactions. Several reports indicate that the sDMA–TUDOR affinity is influenced by the peptide sequence harbouring the sDMA residue (Nishida et al, 2009; Liu et al, 2010a, 2010b; Huang et al, 2011b). This might allow for highly specific and ordered interactions between methylated target proteins and subsets of TUDOR proteins. As many TUDOR proteins contain multiple TUDOR domains, a second scenario is that these multi-domain proteins act as scaffolds (Nishida et al, 2009; Huang et al, 2011b) to bring different effector proteins such as Aubergine and Ago3 into close physical proximity, a likely prerequisite for the efficient functioning of the ping-pong cycle.
It is, however, critical to mention that many of the identified TUDOR domains in Drosophila (similar results emerge in vertebrates) carry mutations in aromatic cage residues, indicating that they lost the ability to interact with sDMA residues. For example, mouse Tdrd12 as well as the three Drosophila counterparts Yb, CG11133 and CG31755 lack nearly all aromatic cage residues. Nevertheless, a genetically identified Yb allele that carries a mutation in the ultra-conserved Arginine residue in β4 of the extended TUDOR domain is a strong loss of function allele (Szakmary et al, 2009). Certain TUDOR domains might thus have evolved to bind alternative residues or post-translational modifications. This might also explain the puzzling observation that loss of the sDMA generating enzyme Csul (PRMT5) results in a surprisingly mild phenotype compared with loss of individual TUDOR domain-containing proteins such as Spindle-E, Krimper or Vreteno (Anne et al, 2007).
The role of Vreteno in primary piRNA biogenesis
Four lines of evidence place Vreteno into the process of primary piRNA biogenesis. First, Vreteno is an essential factor for the piRNA pathway in somatic follicle cells, which has been shown to consist only of the primary branch. Second, piRNA profiles from vreteno mutant ovaries strongly resemble those from armitage and zucchini mutant ovaries with primary piRNA populations collapsing, while ping-pong signatures are not affected. Third, Vreteno physically interacts with Armitage, Yb and Piwi, three factors of the primary piRNA pathway. Finally, Vreteno localizes to Yb bodies, the presumed sites of primary piRNA biogenesis in follicle cells.
Our data provide less insight at the mechanistic level. What is the precise role of the TUDOR domains in Vreteno? Do they interact with sDMA residues in PIWI proteins? What are the functions of the MYND domain and the N-terminal RRM domain? Combinations of biochemical and genetic rescue experiments will be crucial to answer these important questions.
An intriguing observation of our studies in germline cells is that Vreteno is required for the nuage localization of Aubergine and Ago3. Nevertheless, secondary piRNA biogenesis via ping-pong was functional. In this, vreteno mutants differ significantly from armitage mutants. Whether Vreteno merely tethers Aubergine and/or Ago3 to nuage (potentially via sDMA–TUDOR interactions) or whether Vreteno is also important for primary piRNA biogenesis feeding into Aubergine will be an important question for the future. Co-localization experiments in germline cells indicate that Vreteno foci are in very close proximity to Aubergine and especially AGO3 foci (Supplementary Figure S8). sDMA sites in these proteins have been mapped (Nishida et al, 2009) and it will be important to determine whether these peptides mediate physical interactions with Vreteno.
The three Tdrd12 orthologues are essential primary piRNA pathway factors in flies
The TUDOR domain-containing protein Yb is an essential factor for primary piRNA biogenesis in somatic follicle cells (Olivieri et al, 2010; Qi et al, 2010; Saito et al, 2010). Genetically, this gene is dispensable for the piRNA pathway in germline cells, despite the fact that other primary piRNA biogenesis factors such as Armitage or Zucchini are essential factors in soma and germline. As Yb is not expressed in germline cells, it has been suggested that the sequence-related proteins CG11133 and/or CG31755 fulfil Yb's function in the germline (Olivieri et al, 2010). All three proteins share a similar domain composition, although CG31755 contains a second TUDOR domain and CG11133 and CG31755 harbour a C-terminal CS-like domain (Figure 8A). The single vertebrate orthologue of these three proteins is the uncharacterized Tdrd12 protein (Tdrd12 likely corresponds to the repro23 locus; repro23 mutant mice are male sterile and resemble piRNA pathway mutants; Asano et al, 2009). The molecular role of Tdrd12-family proteins is not known but data from follicle cells suggest that they might be the defining factors of Yb bodies and correspondingly nuage in the germline (Szakmary et al, 2009; Olivieri et al, 2010; Qi et al, 2010; Saito et al, 2010).
Our genetic data indicate that Yb and CG31755 are essential piRNA pathway factors in somatic ovarian cells, while CG11133 and CG31755 function in germline cells. Based on the severe impact on Piwi levels and localization in the germline upon double depletion of CG31755 and CG11133, we suggest that these two proteins are indeed the Yb counterparts in germline cells.
Taken together, our study adds functional data for four novel TUDOR proteins in the Drosophila piRNA pathway. It becomes increasingly clear that this small RNA silencing pathway is by far more complex than the related miRNA and siRNA pathways. The challenge for the future will be to dissect the precise molecular roles for the multitude of genetically identified factors in this fascinating genome defence pathway.
Materials and methods
Drosophila stocks
tj-GAL4 (DGRC stock 104055); tj-Gal4; gypsy-lacZ (Olivieri et al, 2010); MTD-GAL4 (Ni et al, 2011); UAS-Dcr-2; NGT (Bloomington stock 25751); armi[Δ1]/TM3 (Olivieri et al, 2010); fs(1)Yb[72]/FM6 (Swan et al, 2001). zuc[HM27]/CyO and Df(2l)PRL/CyO (Pane et al, 2007); armi-GFP and GFP–Yb (Olivieri et al, 2010); HP36220 (Bloomington stock 22204): P-insertion into the 5′UTR of vreteno (CG4771); vreteno[Δ1] is a strong loss of function mutant obtained by mobilization of the HP36220 element via the Δ2,3 transposase. Molecular analysis of this line revealed an internal deletion of the majority of the P-element without affecting the flanking genomic sequences. We speculate that the enhanced strength of the vret[Δ1] allele compared with the HP36220 allele is due to loss of a cryptic promoter located within the P-element sequence similar to what has been described in Lafave and Sekelsky (2011).
GFP–Vreteno flies carry a genomic rescue construct with an EGFP cassette inserted at the N-terminus of Vreteno (obtained via bacterial recombineering) in the vreteno[Δ1] genetic background.
GFP–CG11133 and GFP–CG31755 flies carry a genomic construct with an EGFP cassette inserted at the respective N-termini (bacterial recombineering).
Lines from the VDRC are listed in Supplementary Table S4.
shRNAi lines were cloned into the Valium-22 vector (Ni et al, 2011) modified with a white selection marker and integrated into the attp2 landing site reported in Markstein et al (2008). Hairpin sequences are listed in Supplementary Table S5.
FRT-based mitotic clones were obtained by crossing armi[Δ1], zuc[HM27], Yb[72], piwi[1] or vret[Δ1] alleles with corresponding FRT insertions to respective hsFlp122; FRT, ubi-GFP flies; clones were induced by heat-shocking freshly enclosed females on 2 consecutive days for 1 h at 37°C; flies were dissected 5 days later.
Antibodies
Antibodies against the N-terminal peptide of Vreteno (MESESSQDDWSAFDP) were raised in rabbits and serum was affinity purified using the same peptide. Other used antibodies were rabbit anti-Armi (peptide sequence in Cook et al, 2004), rabbit anti-Yb (peptide sequence in Szakmary et al, 2009), mouse anti-Piwi (Saito et al, 2006), mouse anti-Armi (Saito et al, 2010), rabbit anti-Piwi, anti-Aub and anti-Ago3 (peptide sequences in Brennecke et al, 2007).
X-Gal stainings
Ovaries from 5- to 7-day-old flies were dissected into ice-cold PBS (max 30 min), fixed in 0.5% glutaraldehyde/PBS (RT, 15 min) and washed with PBS. The staining reaction was performed with staining solution (10 mM PBS, 1 mM MgCl2, 150 mM NaCl, 3 mM potassium ferricyanide, 3 mM potassium ferrocyanide, 0.1% Triton, 0.1% X-Gal) at RT overnight.
Immunocytochemistry
IF staining of ovaries and OSCs was as in Olivieri et al (2010). All primary antibodies were used at 1:500.
Cell culture
OSCs were cultured as described (Niki et al, 2006) and transfected with the Cell Line Nucleofector kit V (Amaxa Biosystems), selecting the programme T-029 (K Saito and M Siomi, personal communication).
Immunoprecipitation
A 15-cm dish of OSCs was harvested, cells were extracted twice with 250 μl lysis buffer (20 mM Hepes pH 7.5; 150 mM NaCl; 10% glycerol; 1 mM EDTA; 5 mM MgCl2; 0.5% Triton X-100, complete protease inhibitors (Roche)) and lysates were cleared for 15 min at 16 000 g at 4°C. The supernatant was split and incubated 2 h at 4°C with 20 μg anti-Vreteno, 20 μg anti-Armi antibody cross-linked to 100 μl Invitrogen ProteinG Dynabeads. Empty beads were used in the control reaction. Beads were washed three times for 5 min with wash buffer I (20 mM Hepes pH 7.5; 150 mM NaCl; 10% glycerol; 1 mM EDTA; 5 mM MgCl2; 0.2% Triton X-100), three times for 5 min with wash buffer II (20 mM Hepes pH 7.5; 150 mM NaCl; 10% glycerol; 1 mM EDTA; 5 mM MgCl2; 0.04% Triton X-100) and eluted by boiling in 1 × SDS sample buffer.
Small RNA cloning
Total RNA was isolated from mutant or respective heterozygous ovaries by TRIzol and phenol/chloroform extraction. Small RNA libraries were generated as previously described (Brennecke et al, 2007) and sequenced on an Illumina G2 platform.
Transposon QPCR analysis
cDNA was prepared via random priming of 1 μg total RNA isolated from ovaries of 5- 7-day-old flies. Quantitative PCR was performed using Bio-Rad IQ SYBR Green Super Mix. Each experiment was performed in biological triplicates with technical duplicates. Relative RNA levels were calculated by the 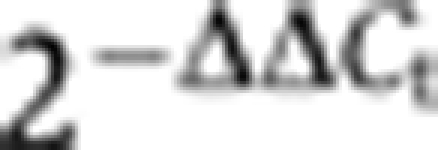 method (Livak and Schmittgen, 2001) normalized to rp49 levels. Fold enrichments were calculated in comparison to respective RNA levels obtained from heterozygote flies or from flies not harbouring a knockdown hairpin. Primers used for QPCR are listed in Supplementary Table S6.
method (Livak and Schmittgen, 2001) normalized to rp49 levels. Fold enrichments were calculated in comparison to respective RNA levels obtained from heterozygote flies or from flies not harbouring a knockdown hairpin. Primers used for QPCR are listed in Supplementary Table S6.
Sterility tests
Ten 3- to 5-day-old female flies were mated with wild-type males overnight. Females were allowed to lay eggs onto apple-juice agar plates for 18 h at 25°C. The number of laid eggs (∼200 per experiment) was counted and the number of hatched eggs was counted 48 h later.
TUDOR domain searches and alignment
A subset of the FlyBase FB2010_01 proteome was generated, which included the longest protein for each protein-coding gene. The obtained set was analysed for the presence of TUDOR-clan similarities as described in the following: Each protein sequence was analysed for its presence in a Pfam24 TUDOR-clan domain definition with the aim to identify known and well-classified TUDOR-clan members. Pfamscan, HMMer2 and HMMer3 were used to query the proteome subset for similarities to models of the Pfam24 TUDOR-clan seed and full alignments. Finally, HHpred 1.5 (with PSI-BLAST homologue collection) was used for even more sensitive searches against SMART, Pfam or PDB-based models that can be associated to the Pfam-TUDOR-clan based on domain similarity. For HMMer hits to be considered, an E-value of ⩽0.01 and a hit length of >10 was required, for HHpred hits the cutoff was set to E⩽0.001 and hit length >10.
Bioinformatics analysis
Only sequences matching the Drosophila genome (release 5; excluding Uextra) 100% were retained. Libraries from heterozygous flies were scaled to 1 million repeat-derived 23–30 nt small RNAs. Libraries from mutant flies were normalized to their respective heterozygous counterparts using endogenous siRNAs originating from convergent gene pairs and from long hairpin loci as in Malone et al (2009). For the piRNA cluster profiles, only piRNAs mapping uniquely to the respective region were considered. For the transposon profiles, all piRNAs mapping with up to three mismatches to the Repbase reference sequence (Jurka et al, 2005) were considered. For the calculation of fold changes in piRNA sequences (transposon analysis), only piRNAs mapping antisense to the respective element were considered. This avoids the influence of random degradation products of transposon sense transcripts that are seen for some elements in piRNA pathway mutants. These small RNA populations have a uniform size distribution and lack a clear nucleotide bias as seen for bona fide piRNAs, classifying them as likely degradation products. The ping-pong signatures for individual elements were calculated as described in Malone et al (2009).
Supplementary Material
Acknowledgments
We thank the members of the Brennecke laboratory for helpful discussions and M Sattler for discussions on Tudor proteins. We are grateful to Sara Farina Lopez for fly injections, to Andreas Sommer for help with deep sequencing and to Mathias Madalinski for peptide synthesis and antibody purification. We thank M Siomi for antibodies and OSCs and the Bloomington stock center and the VDRC for fly lines. We thank the IMBA/IMP Bio-optics Department for excellent support. Work in the Brennecke laboratory is supported by an ERC starting grant from the European Union and a START grant from the Austrian Science Foundation. Small RNA libraries are deposited at GEO (accession no. GSE30955, data sets GSM767594 to GSM767601).
Author contributions: DH, DO and JB conceived the experiments; DH, DO and FSG conducted the experiments; MN, DH and JB performed the TUDOR analysis; KMeixner generated the vret[Δ1] allele; KMechtler supervised the mass spectrometry analysis; AS mapped small RNAs to transposons; RS provided the small RNA database; JB performed the computational analysis of small RNA libraries; JB and DH wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anne J, Ollo R, Ephrussi A, Mechler BM (2007) Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development 134: 137–146 [DOI] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ (2008) A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ (2007) Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747 [DOI] [PubMed] [Google Scholar]
- Asano Y, Akiyama K, Tsuji T, Takahashi S, Noguchi J, Kunieda T (2009) Characterization and linkage mapping of an ENU-induced mutant mouse with defective spermatogenesis. Exp Anim 58: 525–532 [DOI] [PubMed] [Google Scholar]
- Boswell RE, Mahowald AP (1985) Tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell 43: 97–104 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ (2008) An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Jin J, James DA, Adams-Cioaba MA, Park JG, Guo Y, Tenaglia E, Xu C, Gish G, Min J, Pawson T (2009) Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc Natl Acad Sci USA 106: 20336–20341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Pane A, Schupbach T (2007) Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol 17: 637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA (2007) Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet 39: 715–720 [DOI] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, Theurkauf WE (2004) The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell 116: 817–829 [DOI] [PubMed] [Google Scholar]
- Cote J, Richard S (2005) Tudor domains bind symmetrical dimethylated arginines. J Biol Chem 280: 28476–28483 [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156 [DOI] [PubMed] [Google Scholar]
- Friberg A, Corsini L, Mourao A, Sattler M (2009) Structure and ligand binding of the extended Tudor domain of D. melanogaster Tudor-SN. J Mol Biol 387: 921–934 [DOI] [PubMed] [Google Scholar]
- Gillespie DE, Berg CA (1995) Homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes Dev 9: 2495–2508 [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA (2006) A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202 [DOI] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP (2008) Early origins and evolution of microRNAs and PIWI-interacting RNAs in animals. Nature 455: 1193–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC (2007) A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, Chen C, Gordon A, Hannon GJ (2010) Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev 24: 2499–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B, Hendrix D, Trang V, Levine M (2008) A simplified miRNA-based gene silencing method for Drosophila melanogaster. Dev Biol 321: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Gao Q, Peng X, Choi SY, Sarma K, Ren H, Morris AJ, Frohman MA (2011a) piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell 20: 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HY, Houwing S, Kaaij LJ, Meppelink A, Redl S, Gauci S, Vos H, Draper BW, Moens CB, Burgering BM, Ladurner P, Krijgsveld J, Berezikov E, Ketting RF (2011b) Tdrd1 acts as a molecular scaffold for Piwi proteins and piRNA targets in zebrafish. EMBO J 30: 3298–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J (2005) Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110: 462–467 [DOI] [PubMed] [Google Scholar]
- Khurana JS, Theurkauf W (2010) piRNAs, transposon silencing, and Drosophila germline development. J Cell Biol 191: 905–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z (2009) Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 11: 652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Vourekas A, Sayed N, de Lima Alves F, Thomson T, Lasko P, Rappsilber J, Jongens TA, Mourelatos Z (2010) Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA 16: 70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE (2007) Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell 12: 45–55 [DOI] [PubMed] [Google Scholar]
- Lafave MC, Sekelsky J (2011) Transcription initiation from within P elements generates hypomorphic mutations in Drosophila melanogaster. Genetics 188: 749–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Robine N, Martin R, Chung WJ, Niki Y, Berezikov E, Lai EC (2009) Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res 19: 1776–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE (2006) Characterization of the piRNA complex from rat testes. Science 313: 363–367 [DOI] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, Kittler EL, Zapp ML, Klattenhoff C, Schulz N, Theurkauf WE, Weng Z, Zamore PD (2009) Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Kai T (2007) Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci USA 104: 6714–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang JY, Huang Y, Li Z, Gong W, Lehmann R, Xu RM (2010a) Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev 24: 1876–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Chen C, Guo Y, Lam R, Bian C, Xu C, Zhao DY, Jin J, MacKenzie F, Pawson T, Min J (2010b) Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc Natl Acad Sci USA 107: 18398–18403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Qi H, Wang J, Lin H (2011) PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development 138: 1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ (2009) Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ (2009) Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N (2008) Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet 40: 476–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP (2003) The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci 28: 69–74 [DOI] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, Binari R, Booker M, Brennecke J, Perkins LA, Hannon GJ, Perrimon N (2011) A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods 8: 405–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki Y, Yamaguchi T, Mahowald AP (2006) Establishment of stable cell lines of Drosophila germ-line stem cells. Proc Natl Acad Sci USA 103: 16325–16330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Okada TN, Kawamura T, Mituyama T, Kawamura Y, Inagaki S, Huang H, Chen D, Kodama T, Siomi H, Siomi MC (2009) Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J 28: 3820–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J (2010) An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J 29: 3301–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schupbach T (2007) zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell 12: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil VS, Kai T (2010) Repression of retroelements in Drosophila Germline via piRNA pathway by the Tudor domain protein Tejas. Curr Biol 20: 724–730 [DOI] [PubMed] [Google Scholar]
- Qi H, Watanabe T, Ku HY, Liu N, Zhong M, Lin H (2010) The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J Biol Chem 286: 3789–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS (2009) Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol 16: 639–646 [DOI] [PubMed] [Google Scholar]
- Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC (2009) A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr Biol 19: 2066–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC (2009) A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299 [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC (2010) Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev 24: 2493–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC (2006) Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschene G, Bucheton A, Pelisson A (2004) Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics 166: 1313–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenko P, Sprangers R, Stier G, Buhler D, Fischer U, Sattler M (2001) SMN tudor domain structure and its interaction with the Sm proteins. Nat Struct Biol 8: 27–31 [DOI] [PubMed] [Google Scholar]
- Senti KA, Brennecke J (2010) The piRNA pathway: a fly's perspective on the guardian of the genome. Trends Genet 26: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33 (Web Server issue): W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprangers R, Groves MR, Sinning I, Sattler M (2003) High-resolution X-ray and NMR structures of the SMN Tudor domain: conformational variation in the binding site for symmetrically dimethylated arginine residues. J Mol Biol 327: 507–520 [DOI] [PubMed] [Google Scholar]
- Swan A, Hijal S, Hilfiker A, Suter B (2001) Identification of new X-chromosomal genes required for Drosophila oogenesis and novel roles for fs(1)Yb, brainiac and dunce. Genome Res 11: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakmary A, Reedy M, Qi H, Lin H (2009) The Yb protein defines a novel organelle and regulates male germline stem cell self-renewal in Drosophila melanogaster. J Cell Biol 185: 613–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA (2009) Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev 23: 1749–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, Noce T, Nakano T, Nakatsuji N, Lin H, Sasaki H (2011) MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell 20: 364–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.