Abstract
High androgen receptor (AR) level in primary tumour predicts increased prostate cancer-specific mortality. However, the mechanisms that regulate AR function in prostate cancer are poorly known. We report here a new paradigm for the forkhead protein FoxA1 action in androgen signalling. Besides pioneering the AR pathway, FoxA1 depletion elicited extensive redistribution of AR-binding sites (ARBs) on LNCaP-1F5 cell chromatin that was commensurate with changes in androgen-dependent gene expression signature. We identified three distinct classes of ARBs and androgen-responsive genes: (i) independent of FoxA1, (ii) pioneered by FoxA1 and (iii) masked by FoxA1 and functional upon FoxA1 depletion. FoxA1 depletion also reprogrammed AR binding in VCaP cells, and glucocorticoid receptor binding and glucocorticoid-dependent signalling in LNCaP-1F5 cells. Importantly, FoxA1 protein level in primary prostate tumour had significant association to disease outcome; high FoxA1 level was associated with poor prognosis, whereas low FoxA1 level, even in the presence of high AR expression, predicted good prognosis. The role of FoxA1 in androgen signalling and prostate cancer is distinctly different from that in oestrogen signalling and breast cancer.
Keywords: androgen receptor, chromatin marks, glucocorticoid receptor, pioneer factor, transcription programme
Introduction
The androgen receptor (AR) mediates male sex steroid-dependent regulation of cell growth, differentiation and homeostasis (Heinlein and Chang, 2002). This receptor also plays an important role in both androgen-dependent and castration-resistant prostate cancer (Wang et al, 2009). In the nucleus, AR binds to cognate DNA response elements to mediate cell- and tissue-specific regulation of target gene expression (Heinlein and Chang, 2002; Heemers and Tindall, 2007). To form a productive transcription complex on chromatin and to bring about diverse biological actions of androgens, AR needs to communicate with coregulatory proteins (coactivators and corepressors) and collaborating transcription factors (Shang et al, 2002; Kang et al, 2004; Wang et al, 2005). Recent genome-wide studies have shown that nuclear receptors, such as oestrogen receptor (ER), glucocorticoid receptor (GR) and AR, bind to chromatin in vivo far away from transcription start sites of their target genes, which implies that distal enhancers are the primary receptor loading sites and suggests that the receptors utilize a distal regulatory mode of transcriptional control (Carroll et al, 2005; John et al, 2008; Lin et al, 2009; Cheung and Kraus, 2010). However, the underlying mechanisms that guide the receptors to their distal chromatin sites to ensure that regulation of only the intended genes occurs are still elusive.
FoxA1/HNF-3α, a winged-helix transcription factor, is a member of the forkhead family, and it plays a critical role in the growth and differentiation of variety of organs, such as prostate, breast, lung and bladder (Lee et al, 2005; Friedman and Kaestner, 2006; Kaestner, 2010). In mouse prostate development, FoxA1 is required in both epithelial cell differentiation and ductal morphogenesis and patterning (Gao et al, 2005). FoxA1 has been reported to be involved in AR-mediated transcriptional regulation of prostate genes, such as rat probasin and human prostate-specific antigen (PSA) genes (Gao et al, 2003; Mirosevich et al, 2006). FoxA1-binding sites are found in close proximity of AR-binding sites (ARBs) in regulatory regions of these genes, and AR and FoxA1 have been reported to interact through their DNA-binding domains (Gao et al, 2003; Lee et al, 2008).
FoxA proteins can behave as pioneer factors that engage chromatin before other transcription factors (Kaestner, 2010), and FoxA proteins are able to bind to nucleosomal DNA (Belikov et al, 2009; Sekiya et al, 2009). FoxA1 is a pioneer factor in the ERα-mediated transcriptional programme (Carroll et al, 2005; Eeckhoute et al, 2006; Lupien et al, 2008), and it also influences a subset of AR target genes (Gao et al, 2003; Lupien et al, 2008; Jia et al, 2009). On the other hand, FoxA1 is capable of creating a compact chromatin structure through recruitment of corepressor complexes, such as the Groucho family of proteins (Sekiya and Zaret, 2007), and a large proportion of FoxA1-binding sites are located outside the active chromatin regions (Eeckhoute et al, 2009). In this work, we have used LNCaP-1F5 cells, derivatives of LNCaP cells engineered to express the rat GR (Cleutjens et al, 1997) and chromatin immunoprecipitation (ChIP)-sequencing to delineate the genome-wide binding sites of AR and FoxA1—the AR and FoxA1 cistromes—in parental LNCaP-1F5 cells and in cells depleted of FoxA1 protein, in order to assess the role of FoxA1 in AR binding to chromatin and androgen-dependent transcription programme. To delineate the generality of these results, similar experiments on AR binding were conducted in VCaP cells, and on GR binding and glucocorticoid signalling in LNCaP-1F5 cells. Importantly, we also examined the predictive role that FoxA1 protein expression plays in prostate cancer progression, and our results show that, unlike in breast cancer (Badve et al, 2007; Albergaria et al, 2009), high FoxA1 protein level in primary prostate cancer specimens is associated with poor prognosis of the disease, whereas low FoxA1 level predicts good disease outcome, even in the presence of high AR expression.
Results
AR and FoxA1 cistromes in LNCaP-1F5 cells
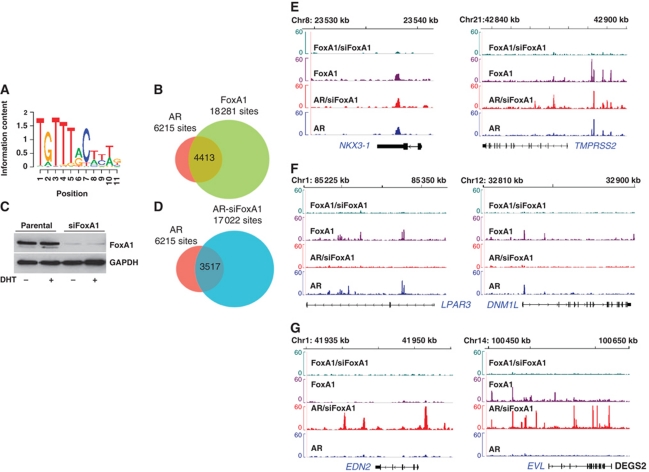
Genome-wide distribution of ARBs on LNCaP-1F5 cell chromatin was analysed by using ChIP-seq after a 2-h exposure to 100 nM 5α-dihydrotestosterone (DHT). The 2-h time point was selected on the basis of previous results on AR loading onto LNCaP cell chromatin (Kang et al, 2004; Wang et al, 2005; Thompson et al, 2006), showing that AR binding peaks by 2 h, and it stays relatively stable at least for the ensuing 12 h. Dose-response experiments indicated that 100 nM DHT was a saturating concentration and that a half-maximal AR loading onto chromatin was achieved by 1–3 nM DHT, as determined by using PSA and TMPRSS2 enhancers as the binding regions (Supplementary Figure S1). By using the MACS algorithm (Zhang et al, 2008), a total of 8419 high-confidence ARBs (false discovery rate (FDR) <2%) were found under these conditions (Sahu et al, in preparation).
Motif over-representation analysis of the AR cistrome revealed that the FoxA1 motif was the most over-represented cis-element (ratio=2.62, P<10−244) (Figure 1A; Supplementary Table S1). This result together with the importance of FoxA1 in ERα signalling prompted us to assess genome-wide FoxA1-binding sites and their relation to the ARBs. FoxA1 ChIP-seq on LNCaP-1F5 chromatin identified initially 23 420 FoxA1-binding sites (FDR <2%). The peaks called from MACS were used to construct a transcription factor association strength (TFAS) for each gene (Ouyang et al, 2009). This analysis identified 6215 ARBs (Supplementary Table S2) and 18 281 FoxA1-binding sites (Supplementary Table S3) that were mapped to the nearest RefSeq gene on the basis of peak intensity and the proximity to the gene within a ±100-kb window. Comparison of AR and FoxA1 cistromes revealed that a high proportion of ARBs (∼71%) overlapped with FoxA1-binding sites (Figure 1B), with the median distance between the respective binding peaks being 43 nt (range, 0–653 nt; Supplementary Figure S2), suggesting a global role of FoxA1 in androgen signalling and providing credence to the pioneering role of FoxA1 in AR binding to chromatin (Wang et al, 2007; Lupien et al, 2008). With regard to the FoxA1 cistrome, however, only 24% of FoxA1-binding sites overlapped with ARBs. Examples of overlapping AR- and FoxA1-binding sites for some well-known AR target genes (KLK3, KLK2, DNM1L and TMPRSS2) are depicted in Supplementary Figure S3.
Figure 1.
AR and FoxA1 cistromes in LNCaP-1F5 cells and redistribution of the AR cistrome by FoxA1 depletion. (A) Consensus cis-element for FoxA1 was the most enriched DNA motif in the parental AR cistrome of LNCaP-1F5 cells. (B) Overlap between AR- and FoxA1-binding sites (FDR <2%) in LNCaP-1F5 cells. (C) Depletion of FoxA1 in LNCaP-1F5 cells treated for 72 h with siRNA specific for FoxA1 mRNA (siFoxA1) or control siRNA (parental). (D) Overlap of ARBs (FDR <2%) in parental (AR) and FoxA1-depleted (AR-siFoxA1) LNCaP-1F5 cells. (E) Class I ARBs are independent of FoxA1, despite potential colocalization of AR- and FoxA1-binding sites. The binding sites are shown for NKX3-1 and TMPRSS2 in parental and siFoxA1 cells. (F) Class II ARBs require FoxA1 as a pioneer factor. For LPAR3 and DNM1L, ARBs are present only in parental cells. (G) Class III ARBs involve FoxA1 as a repressor; AR binding occurs only in siFoxA1 cells. There are two ARB subtypes: (i) FoxA1 prevents AR recruitment to the same locus where FoxA1 is bound (EVL) and (ii) new ARBs appear at sites not previously occupied by FoxA1 (EDN2).
Dual role of FoxA1 in AR binding to chromatin
To examine the genome-wide role of FoxA1 in androgen signalling in more detail, we depleted FoxA1 in LNCaP-1F5 cells using FoxA1 mRNA-specific siRNA. FoxA1 mRNA and protein levels decreased by ∼80% upon the 72-h siRNA exposure (Figure 1C; Supplementary Figure S4A), at which time point the cells—treated with control or specific siRNAs—were exposed to 100 nM DHT for 2 h, followed by AR- and FoxA1-binding site identification with ChIP-seq. As expected on the basis of the data shown in Figure 1C, FoxA1 depletion did not result in a complete loss of FoxA1-binding sites; there were 4076 residual FoxA1 sites in the FoxA1-depleted cells (∼17% of that in parental cells) (Supplementary Table S4). Intriguingly, FoxA1 depletion resulted in a marked increase (over 2.5-fold) in the number of ARBs, and the AR cistrome in FoxA1-depleted cells comprises 17 022 ARBs (FDR <2%) as opposed to 6215 ARBs in parental cells (Figure 1D; Supplementary Table S5). Comparison of the AR cistromes in parental and FoxA1-depleted cells (siFoxA1 cells) indicated that 57% of the original ARBs (3517 sites) in parental cells were unchanged in the depleted cells, whereas 43% of the parental cell ARBs (2698 sites) were lost upon FoxA1 depletion. Importantly, 13 505 completely new ARBs were found in siFoxA1 cells, that is, more than twice that of parental cells (Figure 1D). Only a small proportion (<10%) of these new ARBs are found in data sets published thus far on ARBs in different prostate cancer cell lines.
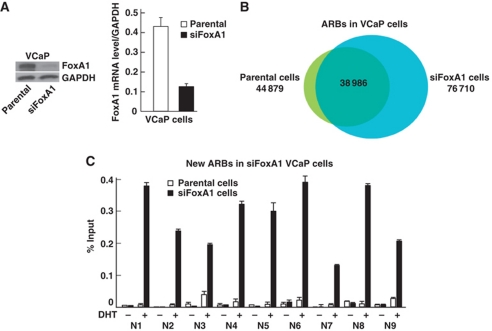
A number of ChIP-seq sites were validated by ChIP–qPCR for the three classes of ARBs (Supplementary Figure S5). The ChIP–qPCR data agreed very well with ChIP-seq results. Similar to LNCaP-1F5 cells, FoxA1 depletion resulted in a marked redistribution of ARBs in another prostate cancer cell line, the VCaP line, which contains a much higher number of ARBs than that in LNCaP-1F5 cells (Figure 2A and B). In VCaP cells, close to 32 000 new ARBs were found in siFoxA1 cells, and around 6000 ARBs were lost upon FoxA1 depletion. ChIP–qPCR validation for a number of FoxA1-independent and FoxA1-pioneered ARBs in VCaP cells is shown in Supplementary Figure S6. The ARBs in parental LNCaP-1F5 cells exhibited 87% overlap with those in parental VCaP cells. In siFoxA1 cells, the three ARB categories in LNCaP-1F5 cells overlapped with those in VCaP cells as follows: FoxA1-independent ARBs, 91%; FoxA1-pioneered ARBs, 10%; and FoxA1-depletion dependent new ARBs, 31%. The new ARBs in siFoxA1 VCaP cells validated in Figure 2C are also present in siFoxA1 LNCaP-1F5 cells.
Figure 2.
FoxA1 depletion in VCaP cells. (A) FoxA1 mRNA and protein levels. VCaP cells were treated for 72 h with siRNA specific for FoxA1 mRNA (siFoxA1) or control siRNA (parental). (B) Overlap of ARBs (FDR <2%) in parental and FoxA1-depleted VCaP cells. (C) Directed ChIP validation of new ARBs in parental (white bars) and FoxA1-depleted (solid bars) VCaP cells. The cells were exposed to 100 nM DHT (+) or vehicle (−) for 2 h prior to ChIP assays. Mean+s.e.m. values for duplicate samples are shown.
FoxA1 defines three classes of ARBs and AR-regulated transcription programmes
FoxA1 depletion results defined three distinct classes of ARBs: (i) the sites that are independent of FoxA1, and FoxA1 depletion does not perturb them (two examples are shown in Figure 1E), (ii) the sites that require FoxA1 as a pioneer factor to recruit AR onto chromatin and that disappear upon FoxA1 depletion (Figure 1F) and (iii) the sites that are masked by FoxA1 and become available for AR binding upon FoxA1 depletion (Figure 1G). This last, previously unrecognized class includes two subgroups; first, FoxA1 prevents AR binding to the same loci where FoxA1 is bound, and second, FoxA1 functions from distance, in that new ARBs appear in siFoxA1 cells at loci not previously occupied by FoxA1 in parental cells. These results indicate that ARBs in prostate cancer cells are remarkably fluid in nature and that their location is highly dependent on the presence (or concentration) of a DNA-binding transcription factor—FoxA1 in this case. Of note, all the above changes in the distribution of ARBs occurred without consistent changes in cellular AR protein content in LNCaP-1F5 or VCaP cells (Supplementary Figure S4B).
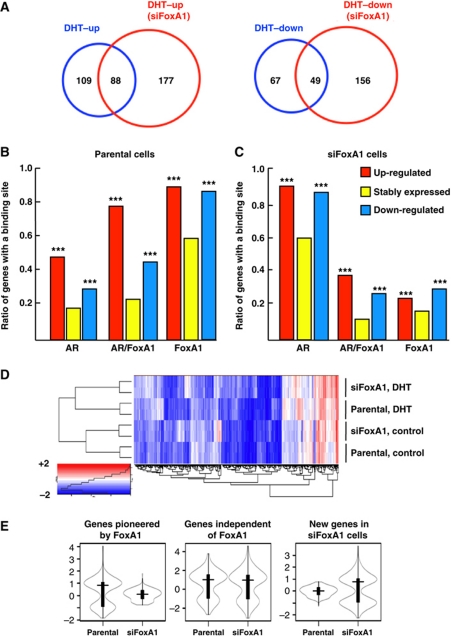
To relate the localization of ARBs to androgen-specific transcription programmes, we profiled gene expression in parental and siFoxA1 cells before and after a 24-h exposure to 100 nM DHT and used a cutoff of ⩾1.7-fold change. Selection of the time interval (24 h) and the DHT concentration (100 nM) was based on the results shown in Supplementary Figure S7A–C. These results showed that maximal responses were achieved in each case at 24 h after exposure to 100 nM DHT. Of note, in the case of several FoxA1-independent genes (PSA and TMPRSS2 ARBs in Supplementary Figure S1, and PSA and NFKBIA mRNA levels in Supplementary Figure S7A), FoxA1 depletion resulted in increased sensitivity to lower DHT concentrations, and full FoxA1 independency was achieved only at higher androgen concentrations. The overall gene expression profiles were commensurate with the three classes of ARBs (cf., Figure 1), in that FoxA1 clearly defined three distinct AR-regulated transcription programmes, as studied in LNCaP-1F5 cells (Figure 3A). Only 45% of genes up-regulated and 42% of genes down-regulated by DHT in parental LNCaP-1F5 cells were androgen dependent in siFoxA1 cells, whereas 55% of the up-regulated and 58% of the down-regulated genes in parental cells lost their androgen dependency upon FoxA1 depletion. Importantly, FoxA1 depletion created new sets of androgen-dependent genes that were not regulated by DHT in parental cells. Similar to ARBs, the total number of androgen-responsive genes was significantly higher in siFoxA1 cells than parental cells.
Figure 3.
Dual role of FoxA1 in androgen-dependent transcription programme. (A) Venn diagram showing the numbers of androgen-regulated genes in parental and siFoxA1 cells. The cells were exposed to 100 nM DHT for 24 h, after which RNA was isolated for expression profiling by microarray using the cut-off of ⩾1.7-fold change. (B, C) Correlation between androgen-regulated (up- and down-regulated) and androgen-independent (stably expressed) genes and the incidence of binding sites unique to AR, shared by AR and FoxA1, and unique to FoxA1 within a window of ±100 kb of TSSs of the genes in parental cells (B) and in siFoxA1 cells (C). ***Significantly different (P<0.001) from stably expressed genes. (D) Unsupervised hierarchical clustering of androgen-regulated transcripts in parental and siFoxA1 cells before (Control) and after exposure to DHT. Heatmaps of biological duplicate samples are shown. (E) Violin plots (Hintze and Nelson, 1998) summarizing the changes in androgen-dependent transcripts in the three classes of genes defined by FoxA1.
To address the functional significance of AR- and FoxA1-binding sites, occurring either together or alone, we examined their distribution in relation to androgen-regulated genes. To this end, we compared all androgen-dependent and androgen-independent (stably expressed) genes to binding sites unique to AR, shared by AR and FoxA1, and unique to FoxA1 within ±100 kb of the transcription start sites of the genes. AR and FoxA1 sites were significantly more enriched in parental cells among androgen-regulated genes (both up- and down-regulated) than among androgen-independent genes (Figure 3B). Importantly, the majority of androgen-regulated genes in these cells exhibited significant enrichment for binding sites shared by AR and FoxA1 over those unique to AR and, in each instance, the proportion of up-regulated genes exceeded that of down-regulated ones (Figure 3B). By contrast, in FoxA1-depleted cells, the majority of genes that were regulated by androgen exhibited enrichment of binding sites unique to AR, indicating that their regulation was independent of FoxA1 (Figure 3C). In siFoxA1 cells, a minor fraction of the genes was enriched for unique FoxA1 sites or for sites shared by AR and FoxA1, which is likely due to the fact that there were residual FoxA1 sites after siRNA exposure (see above). Thus, FoxA1 depletion relieves a marked repressive feature from chromatin that, in turn, permits emergence of new ARBs that are linked to expression of novel androgen-responsive genes.
FoxA1 depletion altered the transcription programme already in the absence of androgen (Figure 3D), and similar transcript numbers were up-regulated (188) and down-regulated (198) by FoxA1 depletion. Likewise, the androgen-induced transcription programmes were significantly different between parental and siFoxA1 cells (Figure 3D). The differentially expressed genes upon androgen exposure segregated into three classes (Figure 3E). The expression levels of genes unique to parental cells lost their androgen regulation in siFoxA1 cells concomitantly with the loss of ARBs, whereas the genes whose ARBs were FoxA1-independent, maintained androgen responsiveness also in siFoxA1 cells. The new genes that were uniquely regulated in siFoxA1 cells by androgen upon the emergence of new ARBs in their regulatory regions were not regulated by DHT in parental cells.
Even though FoxA1 depletion did not affect androgen-mediated regulation of FoxA1-independent genes, such as PSA, NFKBIA, SPDEF and NDRG1 (Supplementary Figure S8A), and the entire kallikrein cluster on chromosome 19 (Supplementary Figure S9), it influenced their basal expression levels; either up (PSA and NKFBIA) or down (SPDEF and NDRG1). It is currently not know whether these changes in basal expression relate to the changes in androgen sensitivity (see above). Nevertheless, the fold changes in response to androgen were not markedly different in parental and siFoxA1 cells. Basal expression levels of the genes that required FoxA1 pioneering were also altered, but importantly, their androgen regulation vanished upon FoxA1 depletion, as exemplified by LPAR3, LRIG1, EXTL2 and AFF3 genes (Supplementary Figure S8B). The genes that became androgen-regulated in FoxA1-depleted cells, such as ETS2, EDN2, FOXO1 and CITED1, exhibited changes in their basal expression levels, and showed robust androgen induction only in siFoxA1 cells (Supplementary Figure S8C).
Pathway analysis using the WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt/) pathway maps indicated that the three distinct AR-regulated transcription programmes in LNCaP-1F5 cells involved some shared, but to a great extent dissimilar biological processes. In particular, a large number of pathways regulated by DHT in siFoxA1 cells were not androgen-dependent among the pathways that were either independent of or pioneered by FoxA1 (Supplementary Table S6). Under FoxA1 depletion conditions, the new ARBs shared by LNCaP-1F5 and VCaP cells were linked to genes representing a number of new pathways or gene ontology categories related to hormonal signalling and cell proliferation (Supplementary Tables S7 and S8).
Motif and cis-element analyses
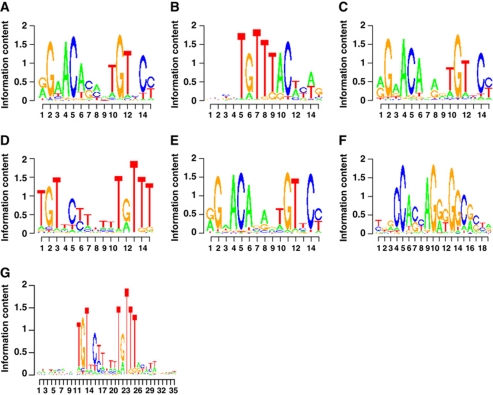
The DNA sequence in itself may act as an allosteric ligand for nuclear receptors, thereby affecting both receptor confirmation and regulatory activity (Meijsing et al, 2009). De novo motif search and motif over-representation analyses identified canonical ARE and FoxA1 motifs as the top-scoring cis-elements in the AR and FoxA1 cistromes of parental LNCaP-1F5 cells, respectively (Figure 4A and B). The ARBs independent of FoxA1 contained a top-scoring ARE very similar to that in parental cells (Figure 4C). Intriguingly, de novo motif search revealed a distinct top-scoring cis-element for the ARBs that were lost upon FoxA1 depletion (Figure 4D). This element may be considered as a tail-to-tail ARE spaced by four nucleotides, or alternatively, as compilation of an ARE half-site and a partial FoxA1 motif (cf., Figure 4A and B). De novo motif search for an extended 35 nt sequence supported the latter possibility (Figure 4G). Importantly, this unique sequence was highly over-represented among the ARBs pioneered by FoxA1 and corresponded 26% of all these sites, but was absent in the two other ARB categories (Supplementary Table S9). The median spacing between AR- and FoxA1-binding sites in the category of lost ARBs was 41 nt (range, 0–454 nt; Supplementary Figure S2). And finally, the new ARBs that appeared in siFoxA1 cells exhibited a canonical ARE as the top-scoring by de novo motif (Figure 4E); these sites were masked in parental cells in a FoxA1-dependent fashion and thus not accessible to AR binding in these cells.
Figure 4.
Top-scoring cis-elements by de novo motif search. (A) Top-scoring cis-element for the ARBs in parental cell AR cistrome. (B) Top-scoring cis-element for the FoxA1-binding sites in parental cell FoxA1 cistrome. (C) Top-scoring cis-element for the FoxA1-independent ARBs. (D) Top-scoring cis-element for ARBs that required FoxA1 pioneering. (E) Top-scoring cis-element for the new ARBs that appeared upon FoxA1 depletion. (F) CTCF as the over-represented cis-element in the unique FoxA1-binding sites in parental cells and the new ARBs in siFoxA1 cells. (G) Top-scoring cis-element for the FoxA1-pioneered ARBs identified by an extended de novo motif search, in which the number of nucleotides in the search was 35 instead of 15 as in (A–F).
Besides the FoxA1 motif, additional cis-elements flanking the ARBs in parental cells included those for forkhead family members, such as FoxA2, and FoxF2, together with E2F1, GATA1 and STAT1 motifs (Supplementary Table S1). Of note, the three ARB categories defined by FoxA1 were enriched for different sets of flanking cis-regulatory elements (Supplementary Table S9). An important cis-element enriched in the unique FoxA1 cistrome, that is, the FoxA1-binding sites that did not overlap with ARBs in parental cells (Figure 1), was that for CTCF (ratio=8.13, P=8.9 × 10−82). This motif (Figure 4F) was also over-represented among the ARBs that appeared upon FoxA1 depletion (ratio=2.8, P=1.32 × 10−22). The new ARBs in siFoxA1 cells were not flanked by FoxA1 motifs; rather, other cis-elements were enriched in the proximity of these sites (Supplementary Table S9). Intriguingly, some transcription factors binding to these latter cis-elements became androgen regulated in siFoxA1 cells, including members of the ETS family and FOXO1 (Supplementary Figure S8). It remains to be established whether they can substitute for FoxA1 in guiding AR binding to the intended genomic sites that subsequently reprogrammes androgen-regulated transcription in FoxA1-depleted cells.
FoxA1 translates the H3K4me2 mark in a genome-wide fashion
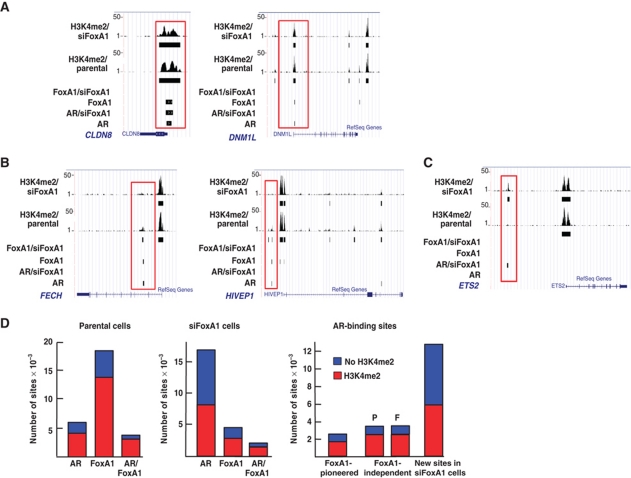
FoxA1-binding sites have been predominantly found at genomic sites that are rich in dimethylated histone H3 lysine 4 (H3K4me2) marks and poor in dimethylated H3 lysine 9 (H3K9me2) marks (Lupien et al, 2008; He et al, 2010). H3K4me2 marks are associated with both enhancers and transcription start sites of genes (Barski et al, 2007; Heintzman et al, 2007). H3K4me2 ChIP-seq in both parental and FoxA1-depleted LNCaP-1F5 cells was conducted to examine whether the genome-wide distribution of H3K4me2 marks was altered by FoxA1 depletion and if yes, whether there was any correlation between these marks and the new ARBs in siFoxA1 cells. Distribution of the majority of H3K4me2-marked sites in parental cells was in agreement with that in a previous report (Lupien et al, 2008); FoxA1 translated this epigenetic mark and guided AR to bind to proper chromatin sites upon androgen exposure, in that ∼70% of the sites shared by AR and FoxA1 possessed H3K4me2 marks. Of note, FoxA1 depletion did not affect the majority of the H3K4me2 marks, as illustrated by the two examples in Figure 5A. CLDN8 does not require FoxA1 to guide receptor binding, and AR can recognize H3K4me2 marks either on its own or through another collaborating factor. DNM1L is a FoxA1-pioneered locus that maintains H3K4me2 marks despite the absence of both FoxA1 and AR binding in siFoxA1 cells. In some instances, however, H3K4me2 marks disappeared upon FoxA1 depletion with a subsequent loss of both AR binding and androgen-dependent gene expression, as illustrated by FECH and HIVEP1 loci in Figure 5B. In addition, FoxA1 depletion could also result in the appearance of new H3K4me2 marks that were occupied by new ARBs in siFoxA1 cells, as shown by the ETS2 locus in Figure 5C.
Figure 5.
H3K4me2 marks in parental and FoxA1-depleted cells. (A) AR-/FoxA1-binding sites and H3K4me2 marks in parental and siFoxA1 cells at CLDN8 and DNM1L loci. H3K4me2 marks are shown by raw tag counts and the solid black bar shows the binding sites after peak calling on the UCSC genome browser. (B) AR-/FoxA1-binding sites and H3K4me2 marks in parental and siFoxA1 cells at FECH and HIVEP1 loci. (C) AR-/FoxA1-binding sites and H3K4me2 marks in parental and siFoxA1 cells at the ETS2 locus. AR recruitment upon FoxA1 depletion occurs concomitantly with the appearance of new H3K4me2 marks. (D) Distribution of AR-/FoxA1-binding sites and H3K4me2 marks in parental cells (left panel), siFoxA1 cells (middle panel), and in the three classes of ARBs defined by FoxA1 (right panel). H3K4me2 marks for FoxA1-independent ARBs are shown separately for parental (P) and FoxA1-depleted cells (F).
As mentioned above, localization of most AR- and FoxA1-binding sites correlated with the presence of H3K4me2 marks in parental LNCaP-F15 cells (Figure 5D). Nevertheless, about one-third of the unique ARBs, one-fourth of FoxA1-binding sites, and one-fifth of shared AR/FoxA1 sites were without the H3K4me2 marks in parental cells (Figure 5D). For the three classes of ARBs defined by FoxA1, about 70% of the sites pioneered by FoxA1 and the sites independent of FoxA1 contained H3K4me2 marks. However, close to 50% of the ARBs in siFoxA1 cells did not have H3K4me2 marks, suggesting that other epigenetic marks and/or other collaborating transcription factors are required to guide AR to recognize these sites in the absence of FoxA1.
Correlation of AR and FoxA1 binding with DNaseI-hypersensitive sites
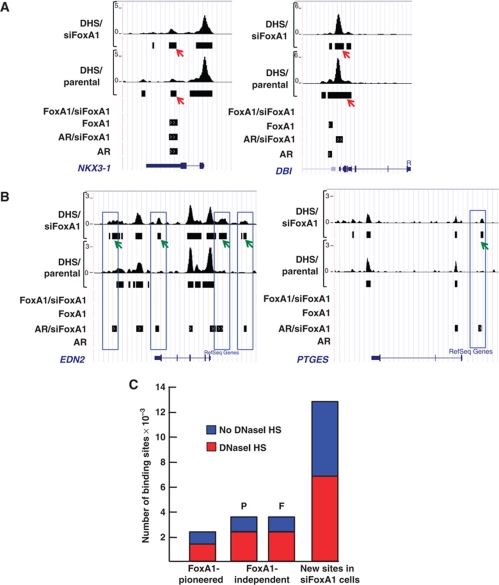
The nuclease accessible sites (DNaseI-hypersensitive (DHS) sites) are good indicators to distinguish between accessible and non-accessible chromatin sites that are known to be highly cell-specific and often dictated by the available compilation of chromatin remodellers and other associated factors (John et al, 2008; Boyle et al, 2008). Genome-wide mapping of DHS sites (Song and Crawford, 2010) was used to examine in LNCaP-1F5 cells whether FoxA1 depletion resulted in an open chromatin conformation that subsequently created new ARBs, or whether chromatin was already in an open conformation prior to FoxA1 depletion. The results showed that most of the AR- and FoxA1-binding sites associate with DHS sites in parental cells and that the majority of these sites (∼70%) were constitutively open already prior to FoxA1 and AR binding to chromatin. Examples of this arrangement are depicted in Figure 6A for NKX3-1 and DBI loci. However, FoxA1 depletion also affected chromatin conformation in a number of instances, as illustrated by the appearance of new DHS sites concomitantly with the appearance of new ARBs, which is exemplified by EDN2 and PTGES loci in Figure 6B. These new ARBs occupying de novo DHS sites comprises 16% of all the ARBs mapped in siFoxA1 cells, and they may be formed through recruitment of chromatin modifying proteins to these loci in an AR-dependent manner.
Figure 6.
Comparison of DHS sites in parental and FoxA1-depleted LNCaP-1F5 cells. (A) DHS sites together with AR- and FoxA1-binding sites in parental and siFoxA1 cells at the NKX3-1 and DBI loci. The constitutively open sites are shown marked by red arrows. (B) DHS sites together with AR- and FoxA1-binding sites in parental and siFoxA1 cells at the EDN2 and PTGES loci. The de novo DHS sites are marked by green arrows. EDN2 and PTGES genes are androgen-regulated only in siFoxA1 cells. (C) Proportion of the DHS sites in the three ARB classes defined by FoxA1. DHS sites for FoxA1-independent ARBs are shown separately for parental (P) and FoxA1-depleted cells (F).
Comparison of genome-wide colocalization of DHS sites with the ARBs in the three categories defined by FoxA1 revealed that approximately the same proportion of the DHS sites—two-thirds—overlapped with ARBs that were either FoxA1-independent or pioneered by FoxA1 (Figure 6C). However, similar to the H3K4me2 marks, only one-half in the new ARBs in siFoxA1 cells overlapped with DHS sites (cf., Figures 5D and 6C).
FoxA1 depletion elicits redistribution of GR-binding sites and reprogrammes glucocorticoid signalling
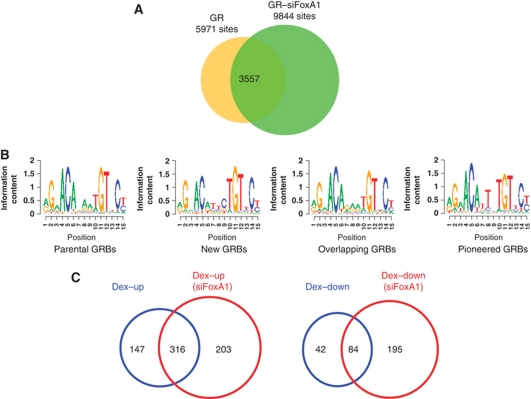
LNCaP-1F5 cells have been engineered to express the rat GR (Cleutjens et al, 1997) and, therefore, provided an opportunity to examine in a single cell line whether reprogramming of androgen signalling by FoxA1 depletion in LNCaP-1F5 or VCaP cells was unique to AR. This issue is of particular significance in view of a recent report showing that FoxA1 depletion abrogates almost all genome-wide ER-binding sites and blunts concomitantly oestrogen-dependent signalling in breast cancer cells (Hurtado et al, 2011). ChIP-seq analyses revealed the presence of 5971 GR-binding sites (GRBs) in parental LNCaP-1F5 cells and 9844 GRBs in siFoxA1 cells after a 2-h dexamethasone (Dex) exposure (Figure 7A). Redistribution of GRBs upon FoxA1 depletion showed tripartite classification very similar to that of ARBs, in that 60% of the GRBs in parental cells were maintained and 40% were lost upon FoxA1 depletion, and a large number of completely new GRBs (6287 sites) emerged in siFoxA1 cells (Figure 7A). When the top-scoring cis-elements were analysed by de novo motif search, similar elements were found in the three subclasses and, unlike the situation with ARBs, the GRBs that were lost upon FoxA1 depletion did not possess a clearly distinct cis-element (Figure 7B). Despite their very similar top-scoring cis-elements, the two receptors did not bind to identical chromosomal loci, as only one-fifth of the new ARBs overlapped with the new GRBs in siFoxA1 cells.
Figure 7.
FoxA1 depletion in LNCaP-1F5 cells brings about redistribution of GR binding and commensurate changes in GR-dependent transcription programme. (A) Overlap analysis of GRBs (FDR <2%) in parental (GR) and FoxA1-depleted cells (GR–siFoxA1). (B) Top-scoring cis-elements, as identified by de novo motif search in different subclasses of GRBs. (C) The number of genes up- and down-regulated by Dex (Dex–up and Dex–down, respectively) exposure in parental and FoxA1-depleted (siFoxA1) LNCaP-1F5 cells.
Glucocorticoid-dependent transcription programmes in parental and FoxA1-depleted LNCaP-1F5 cells revealed a pattern similar to that of androgen-mediated programmes, in that redistribution of GRBs in siFoxA1 cells was commensurate with changes in GR-dependent gene expression profiles, in both genes up- and down-regulated by Dex (Figure 7C). Importantly, only a small proportion (∼15%) of androgen-regulated new transcripts in siFoxA1 cells were also regulated by glucocorticoid in the same cells.
Collectively, our results on both androgen and glucocorticoid signalling indicate that binding of their cognate receptors to chromatin is a mobile event and significantly regulated by another DNA-binding transcription factor, FoxA1. As a consequence, the steroid, the receptor and the cis-element are necessary but not sufficient in most instances to guide AR and GR to the appropriate chromosomal locations, in order to initiate the intended hormonal signalling.
FoxA1 expression in prostate cancer and its relationship to disease outcome
Increased nuclear AR protein expression in either diagnostic biopsy and/or radical prostatectomy specimens is associated with a reduced time to prostate cancer-specific mortality (Donovan et al, 2008, 2009). This finding was verified in the patient cohort examined in this work (Supplementary Figure 10A). AR amplification and overexpression are also major features of castration-resistant prostate cancers (Visakorpi et al, 1995; Chen et al, 2004), and retinoblastoma tumour suppressor controls prostate cancer progression through modulation of AR expression (Sharma et al, 2010). These previous results, high expression of FoxA1 mRNA in normal and malignant prostate (Supplementary Figure S10B), and the ability of FoxA1 protein to reprogramme androgen signalling prompted us to investigate FoxA1 protein expression in primary tumours of 350 prostate cancer patients who underwent radical prostatectomy and had clinical follow-up for 11.3–25.0 years (Supplementary Tables S10–S12).
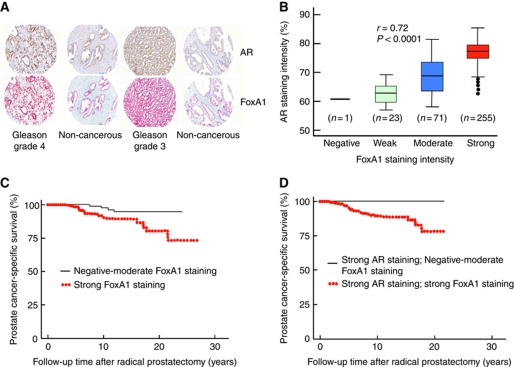
Almost all primary prostate cancer specimens expressed FoxA1 protein, and the antigen staining was predominantly nuclear (Figure 8A). FoxA1 staining intensity exhibited significant positive correlation with that of AR (Figure 8B). When FoxA1 expression was graded according to the staining intensity, 24 samples (6.9%) were negative or weakly staining, 71 samples (20.3%) contained moderate and 255 (72.9%) strong FoxA1 staining. FoxA1 staining intensity was significantly weaker in the adjacent non-cancerous than in the cancerous tissue, as illustrated by the two examples in Figure 8A (P<0.01 for the entire material). Importantly, FoxA1 staining in primary prostate cancer specimens showed a significant relationship to disease outcome (P<0.05), with a strong nuclear FoxA1 staining being associated with poor prognosis, that is, a reduced time to prostate cancer-specific mortality (Figure 8C). The hazard ratio was 2.89 (95% confidence interval 1.02–8.21). There was also a significant positive association of FoxA1 staining intensity and the percentage of FoxA1-stained nuclei to the Gleason grade of the tumours (Supplementary Table S12). Of note, low FoxA1 protein expression was associated with good prognosis, even in the presence of high AR protein expression that, in and of itself (see above), spells poor disease outcome (Figure 8D). This latter finding agrees with our data from cell line experiments, indicating that the level of FoxA1 plays an important role in AR pathway reprogramming.
Figure 8.
FoxA1 protein expression in prostate cancer tissue specimens and prostate cancer-specific survival. (A) Representative FoxA1 and AR staining patterns in prostate cancer and adjacent non-cancerous tissues of two patients. (B) Correlation between FoxA1 and AR staining intensity in the prostate cancer patient cohort (n=350). (C) Disease-specific survival of 350 patients with prostate cancer according to the intensity of FoxA1 staining in the primary tumour. χ2=4.36. P=0.04 (log-rank test). (D) Disease-specific survival of patients with high AR staining classified according to FoxA1 staining in the same specimen; either negative–moderate (n=18) or high (n=222). χ2=2.57. P=0.10 (log-rank test).
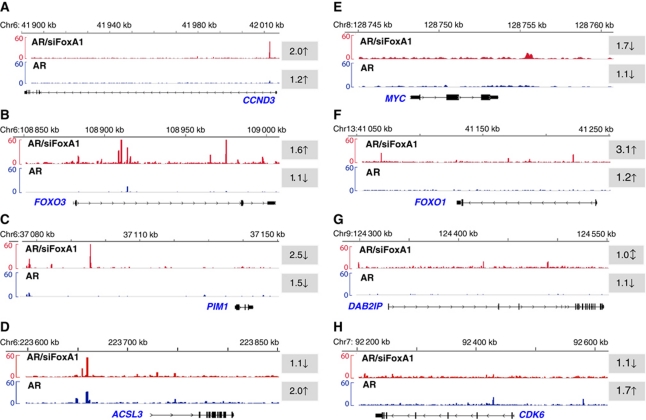
In prostate cancer, high FoxA1 protein expression-dependent signalling appears to maintain a transcription programme with oncogenic potential. This may occur, at least in part, through active AR signalling that is supported by high FoxA1 expression; the support disappears upon FoxA1 depletion. Low FoxA1 expression would, in turn, be connected with reprogrammed AR pathway that involves activation of tumour suppressors or attenuation of AR-dependent oncogenic signalling. Potential examples of this dual role of FoxA1 are shown in Figure 9. In the case of CCND3, FOXO3, PIM1, ACSL3, MYC, FOXO1 and DAB2IP loci (Figure 9A–G), FoxA1 depletion resulted in emergence of new ARBs; as a consequence, androgen regulation of the encoded mRNAs in LNCaP-1F5 cells was altered in a fashion that is consistent with activation of tumour suppression or attenuation of oncogenic potential. In the case of CDK6, loss of AR binding upon FoxA1 depletion blunted androgen-dependent expression of CDK6 mRNA (Figure 9H). Comparison of FOXO1 and FOXO3 nuclear staining in 10 low–moderate FoxA1/high AR spots and 30 high FoxA1/high AR spots on three primary prostate cancer tissue microarray (TMA) slides revealed significant association of low–moderate FoxA1 protein level with stronger FOXO1 and FOXO3 staining intensity (Supplementary Table S13), thus being in concert with the results shown in Figure 9, in that FoxA1 depletion in LNCaP-1F5 cells brings about up-regulation of FOXO1 and FOXO3 mRNA accumulation.
Figure 9.
Examples of prostate cancer-associated genes with altered ARBs and expression profiles upon FoxA1 depletion. Distribution of ARBs in parental (blue) and siFoxA1 (red) cells in eight loci known to encode genes that are involved in prostate cancer development and/or progression. The shaded numbers with arrows (↑, up-regulation; ↓, down-regulation; ↕, no effect) refer to fold induction of the encoded mRNA after a 24-h exposure to 100 nM DHT in LNCaP-1F5 cells, as determined by the microarray experiments.
Discussion
The present work shows several novel features in the AR–chromatin interaction. First, AR binding to chromatin is remarkably fluid and highly dependent on the forkhead protein FoxA1. Second, FoxA1 plays a dual role in regulating the accessibility of AR to chromatin; it serves as a pioneer factor for a subset of sites but importantly, it also creates—perhaps via recruitment of corepressor complexes (Sekiya and Zaret, 2007; Eeckhoute et al, 2009)—an environment on chromatin that precludes AR binding to its cognate cis-elements. These latter sites become accessible to AR binding upon FoxA1 depletion. Third, there is a subset of ARBs that is independent of FoxA1 and may collaborate with or be pioneered by other DNA-binding transcription factors. They are present in parental cells and by this means FoxA1-independent; however, depletion of FoxA1 leads, at least in some instances, to augmented AR binding and increased androgen sensitivity. Fourth, redistribution of ARBs upon FoxA1 depletion correlates remarkably well with changes in androgen-dependent transcription programme, in that FoxA1-dependent loss of an ARB signifies blunted androgen responsiveness, and the appearance of a new ARB upon FoxA1 depletion is linked in many instances to the emergence of androgen responsiveness of a nearby gene. Fifth, the subset of ARBs that require FoxA1 pioneering utilizes a cis-element different from the canonical ARE. Importantly, these novel features in the AR–chromatin interaction are also true for GR binding to chromatin and glucocorticoid-dependent transcription programme upon FoxA1 depletion.
In the case of both AR and GR signalling, it was remarkable that FoxA1 depletion resulted in the appearance of a large number of new ARBs or GRBs which was linked to androgen or glucocorticoid regulation of nearby genes. Intriguingly, FoxA1 regulates ER binding and oestrogen signalling in breast cancer cells (Hurtado et al, 2011) in a fashion very different from that of AR and GR in prostate cancer cells. The fact that FoxA1 can prevent steroid receptor binding to specific chromatin sites, and that this property is reversible, has not been previously recognized in the nuclear receptor field. Of note, we did not knockout FoxA1 protein completely in our experiments; rather, the level was depleted by ∼80%. FoxA1 null mice exhibit neonatal mortality with a complex phenotype, including persistent hypoglycemia, progressive starvation, wasting and hypotriglyceridemia (Shih et al, 1999), and FoxA1 expression is critical for growth and differentiation of variety of organs, such as prostate, breast, lung and bladder (Kaestner, 2010). In addition, it has been reported that there is FoxA1 haploinsufficiency in mammary gland development (Bernardo et al, 2010). In view of these results, we envision that FoxA1 expression level—not only its presence or absence—plays an important regulatory role during development, in order to establish a proper chromatin environment for temporal and tissue- and cell-specific recruitment of steroid receptors, such as AR and GR, which, in turn, activate or attenuate the intended transcription programmes.
There was a considerable overlap between AR and FoxA1 cistromes in parental cells, and over 70% of the ARBs were shared by FoxA1-binding sites. However, FoxA1 pioneered only a subset of ARBs. That the FoxA1-pioneered ARBs possessed a unique cis-element different from canonical ARE-like motifs present in other ARBs suggests that this particular element plays an important role in the establishment of FoxA1-pioneered ARBs, which is supported by the finding that the unique cis-element was not present among the other ARB classes, in either parental or siFoxA1-depleted cells. FoxA1 binding to the unique cis-element may be the requisite initial event, and the adjacent ARE half-site of the element is recognized by the AR, which is stabilized by the interaction of the two proteins (Gao et al, 2003; Wang et al, 2007). We cannot, however, exclude the possibility that AR binds to chromatin at these loci indirectly through FoxA1. Multiple overlapping AR- and FoxA1-binding sites were independent of the pioneering function of FoxA1. Some well-known AR target genes, such as PSA and some other kallikrein cluster genes, NKX3-1 and TMPRSS2 (Supplementary Figures S2 and S5) have ARBs of this category. In these cases, FoxA1 may bind to chromatin via AR or alternatively, chromatin structure has been modified by factors other than FoxA1, and chromatin is therefore available for independent AR and FoxA1 binding. Some ARBs or androgen-regulated genes of this class were not totally FoxA1-independent, in that FoxA1 depletion resulted in increased AR binding and enhanced androgen sensitivity over that in parental LNCaP-1F5 cells. This phenomenon could be due to competition between AR and FoxA1 for binding to their respective cis-elements that overlap at these sites.
The signals that determine whether FoxA1 binding to chromatin leads to its pioneering function or to formation of a compact chromatin structure are currently unknown. Recently, the histone mark H3K4me2 was shown to be instrumental in guiding FoxA1 to correct chromatin sites (Lupien et al, 2008), and our genome-wide results on androgen signalling support this result in most, but not in all cases. On the other hand, FoxA1 can also bind to unique sites on reconstituted chromatin in vitro without any specific histone modifications (Cirillo et al, 2002; Cirillo and Zaret, 2007) or on reconstituted nucleosomes in Xenopus oocytes in vivo (Belikov et al, 2009). Our genome-wide results in parental cells are in agreement with both situations, in that the majority of genomic sites possessed H3K4me2 marks together with AR- and FoxA1-binding sites, but there are also examples of sites in which FoxA1 depletion altered H3K4me2 marks and AR binding.
Three-fourths of FoxA1-binding sites did not overlap with ARBs in parental cells, and these sites along with the new ARBs in siFoxA1 cells were enriched for the CTCF cis-element. The CTCF insulator proteins (Fu et al, 2008; Zhang et al, 2010) appear to be important in recruiting FoxA1 to bind to these loci, and the DNA-bound FoxA1 is subsequently the signal to recruit the Groucho family of proteins to the same loci, in order to generate compact chromatin structure (Cirillo et al, 2002; Sekiya and Zaret, 2007) not available for AR binding. Genome-wide DHS assays showed that most of the ARBs in both parental and FoxA1-depleted cells were located at constitutively open chromatin loci, a result that is in agreement with the data demonstrating that GR binding in a mammary tumour cell line occurs manly to pre-existing DHS sites (John et al, 2008). However, our results also showed that, in about one-sixth of the cases in siFoxA1 cells, the emergence of a new ARB was concomitant with the presence of a new DHS site. Whether or not these particular DHS sites are formed through AR-dependent recruitment of chromatin-remodelling enzymes, such as the Swi/Snf proteins, remains to be established.
ER-binding sites and oestrogen-dependent signalling in breast cancer cells vanish upon FoxA1 depletion (Hurtado et al, 2011), and high FoxA1 expression in breast cancer predicts favourable prognosis of the disease (Badve et al, 2007; Albergaria et al, 2009). These results on ER pathways are in sharp contrast to our data on AR and GR pathways in a prostate cancer cell line, and the role of FoxA1 expression in prostate cancer tissue. Patients with high FoxA1 protein-expressing primary tumours had significantly higher prostate cancer-specific mortality than those with moderate or low FoxA1 levels, suggesting that high FoxA1 protein expression regulates signalling pathways important in prostate cancer progression, maybe in metastasis as well. In the absence of this signalling, or through activation of other events by low FoxA1 protein expression, prostate cancer does not seem able to progress. Consistent with this latter possibility is the up-regulation of FOXO family members, such as FOXO1 and FOXO3, known to act as tumour suppressors, and down-regulation of oncogenes such as PIM1 and c-Myc in siFoxA1 cells (Figure 8; Supplementary Figure S8). FOXO1 mediates PTEN suppression (Ma et al, 2009), inhibits AR signalling in prostate cancer, and loss of FOXO1 by chromosomal deletion (13q14) promotes castration-resistant prostate cancer (Dong et al, 2006). Moreover, prostate cancer patients with FOXO1 protein positive tumours have been shown to have higher cancer-specific survival rates than those with FOXO1 negative tumours (Nakajima et al, 2011). Transcriptional down-regulation of FOXO3 correlates inversely with increasing tumour grade in prostate cancer (Shukla et al, 2009). Down-regulation of PIM1 and c-Myc in siFoxA1 cells should also be a beneficial feature, in that PIM1 promotes c-Myc transcriptional activity and prostate cancer cell tumourigenicity (Kim et al, 2010), and c-Myc is known to repress FOXO3-dependent transcriptional targets (Chandramohan et al, 2008).
Interestingly, a recent risk SNP block analysis (Lu et al, 2011) revealed two annotation sets that were significantly enriched in prostate cancer, one for ARBs and the other one for FoxA1-binding sites, defined in previous ChIP-on-chip studies (Carroll et al, 2005; Wang et al, 2007), adding to the evidence that FoxA1 plays an important role in the pathogenesis of prostate cancer. Further studies are warranted to explore in detail the mechanisms and down-stream events by which FoxA1 modulates androgen signalling and regulates progression of prostate cancer. And finally, prostate-specific targeting of FoxA1 would potentially offer a novel therapeutic modality for prostate cancer.
While this work was under consideration for publication, similar findings on the role of FoxA1 in reprogramming of the AR pathway in a related prostate cancer cell line (LNCaP cells) was published (Wang et al, 2011).
Materials and methods
Cell culture
LNCaP-1F5 cell line (Cleutjens et al, 1997) was a kind gift from Dr Jan Trapman (Erasmus Medical Center). The presence of rat GR in LNCaP-1F5 cells does not alter the growth response of these cells to androgen from that of the parental LNCaP cells (Cleutjens et al, 1997). Likewise, the profile of androgen-regulated transcripts is very similar in LNCaP and LNCaP-1F5 cells (Sahu et al, manuscript in preparation).
FoxA1 depletion by RNA interference
Cells were cultured in RPMI-1640 containing 10% charcoal-stripped FBS to 60% confluence and then transfected with control siRNA or siRNA targeting FoxA1 mRNA (ON-TARGETplus™ SMARTpool siRNA, Dharmacon, Thermo Scientific) with Dharmafect-3 transfection reagent according to the manufacturer's instructions. The cells were incubated for 72 h and then exposed to steroid (DHT or Dex) for 2 h (ChIP-seq) or for 24 h (gene expression profiling). The siRNA sequences are shown in Supplementary Table S14.
Chromatin immunoprecipitation
The antibodies used in the present work for ChIP assays were from the following sources: AR (Kang et al, 2004), FoxA1 (ab23738, Abcam), H3K4me2 (07-030, Millipore), CTCF (ab70303, Abcam), GR (Widen et al, 2000), normal rabbit IgG (sc-2027, Santa Cruz) and normal mouse IgG (sc-2025, Santa Cruz). The cells were exposed to DHT (100 nM) or Dex (100 nM) for 2 h prior to ChIP. Cells were fixed in 1% formaldehyde (Merck KGaA) for 10 min at room temperature and washed twice with ice-cold PBS. The cell suspension was centrifuged and the pellet resuspended for lysis in 400 μl RIPA buffer (1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS in 1 × PBS) containing 2 × protease inhibitors (Roche). The chromatin was sonicated to an average fragment size of 100–500 bp using Bioruptor UCD-300-TO (Diagenode), after which the samples were centrifuged at 15 000 r.p.m. for 15 min and the supernatant saved.
ChIP-sequencing
ChIP samples were processed according to Illumina's instructions, and DNA library was sequenced using Illumina Genome Analyzer II at the Biomedicum Functional Genomics core facility. Aligned read numbers are listed in Supplementary Table S15. The peak calling was carried out using the MACS algorithm (Zhang et al, 2008) in which binding sites are determined by comparing the density of reads at a genomic location in the antibody-specific ChIP-seq experiment to a local Poisson background density model fit to background reads that were control IgG ChIP-seq reads. MACS estimates a FDR by swapping the signal and background ChIP-seq data sets and repeating the peak-calling process to give an estimate of statistical significance. Using an FDR of <2%, the peaks called from MACS were used to map the binding site to the nearest RefSeq gene by building a TFAS. TFAS was built by computing the weighted sum of the corresponding ChIP-seq signal strength, where the weights reflect the proximity of the signal to the gene (Ouyang et al, 2009).
Gene expression profiling
LNCaP-1F5 cells were cultured in RPMI-1640 medium supplemented with 10% charcoal-stripped FBS for 4 days prior to siRNA transfection. Seventy-two hours after transfection, the cells (both parental and siFoxA1 cells) were exposed to 100 nM DHT or 100 nM Dex for 24 h. Total RNA was isolated using RNAeasy kit (Qiagen). RNA samples from two biological duplicates were hybridized to Illumina HumanHT-12 v3 Expression BeadChip Kits at the Biomedicum Functional Genomics core facility. The data analysis was performed by using Anduril software (Ovaska et al, 2010) together with ‘R’ (http://www.r-project.org/) and Bioconductor ‘lumi’ package (http://www.bioconductor.org). Raw intensity values were quantile normalized separately for the AR and GR experiments. The mean value of sample replicates was used to calculate differentially expressed genes. Fold changes of ⩽1.7 and ⩾1.7 were set as the cutoff values.
ChIP-seq and gene expression microarray data have been deposited in the Gene Expression Omnibus database with accession number GSE30624.
DNAaseI-hypersensitive site sequencing
DHS sites sequencing (DHS-seq) was performed essentially as described by Song and Crawford (2010). Additional details are described in Supplementary data.
Real-time PCR
Quantitative RT–PCR was performed using SYBR green Mastermix (Roche). cDNA synthesis was carried out from 2 μg total RNA using Transcriptor High Fidelity cDNA synthesis kit (Roche). Primers are listed in Supplementary Table S16.
Immunoblotting
Immunoblot analyses were performed using anti-AR (Kang et al, 2004), anti-FoxA1 (ab23738, Abcam) and anti-GAPDH (sc-47724, Santa Cruz) antibodies.
Prostate cancer patients
The use of the tissue specimens and patient information was approved by the Institutional Review Board of the Helsinki University Central Hospital. The median follow-up time of the 350 patients was 13.3 years (range, 11.33–25.0 years). Patient cohort details are in Supplementary Tables S10–S12.
TMA construction
TMAs were constructed from formalin-fixed paraffin-embedded blocks of 350 patients who underwent prostatectomy at the Helsinki University Central Hospital. To account for tumour heterogeneity, two TMA cores were from the most dominant Gleason grade area and one from the second most dominant Gleason grade area. One core of each patient contained an adjacent non-cancerous area.
Immunohistochemistry
Freshly cut TMA sections were mounted on electrically charged glass slides (SuperFrost® Plus, Menzel-Gläser) that were stained with the Benchmark XT system (Ventana Medical Systems) using a biotin-free multimer-based detection system (ultraView™ Universal Red, Ventana). Antigen staining was carried out using anti-FoxA1 (ab23738, Abcam) or anti-AR antibody (NCL-AR-318, Immuno Diagnostics), and staining intensity was evaluated without prior knowledge of the Gleason grades, as described in detail in Supplementary data. Anti-FOXO1 antibody (HPA001252) was from Sigma-Aldrich and anti-FOXO3 antibody (9467) from Cell Signaling.
Supplementary Material
Acknowledgments
We thank Jaana Vuopio and Susanna Vähäkuopus for help with pulsed field gel electrophoresis. We thank Saija Kotola for excellent technical assistance. This work was supported by the Academy of Finland, Sigrid Jusélius Foundation, Finnish Cancer Foundations, Biocentrum Helsinki, ERANET SysBio+ (Synergy), Helsinki University Central Hospital, European Union (contract No. LSHM-CT-2005-018652) and HBGS, GPBM and FICS Graduate Programmes.
Author contributions: The overall study was conceived and designed by BS and OAJ. The experiments were performed by BS with the help of TM, JL, ML, JK and TV in immunohistochemistry. BS, ML, KO, TM, ML and SH analysed the data. AR, AS, J-PT and SN collected the clinical data and prepared TMAs. BS and OAJ wrote the paper with substantial contributions from OK and SH.
Footnotes
The authors declare that they have no conflict of interest.
References
- Albergaria A, Paredes J, Sousa B, Milanezi F, Carneiro V, Bastos J, Costa S, Vieira D, Lopes N, Lam EW, Lunet N, Schmitt F (2009) Expression of FOXA1 and GATA-3 in breast cancer: the prognostic significance in hormone-receptor negative tumours. Breast Cancer Res 11: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badve S, Turbin D, Thorat MA, Morimiya A, Nielsen TO, Perou CM, Dunn S, Huntsman DG, Nakshatri H (2007) FOXA1 expression in breast cancer – correlation with luminal subtype A and survival. Clin Cancer Res 13: 4415–4421 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Belikov S, Astrand C, Wrange O (2009) FoxA1-binding directs chromatin structure and the functional response of a glucocorticoid receptor-regulated promoter. Mol Cell Biol 29: 5413–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo GM, Lozada KL, Miedler JD, Harburg G, Hewitt SC, Mosley JD, Godwin AK, Korach KS, Visvader JE, Kaestner KH, Abdul-Karim FW, Montano MM, Keri RA (2010) FOXA1 is an essential determinant of ERα expression and mammary gland ductal morphogenesis. Development 137: 2045–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE (2008) High-resolution mapping and characterization of open chromatin across the genome. Cell 132: 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 [DOI] [PubMed] [Google Scholar]
- Chandramohan V, Mineva ND, Burke B, Jeay S, Wu M, Shen J, Yang W, Hann SR, Sonenshein GE (2008) c-Myc represses FOXO3α-mediated transcription of the gene encoding the p27(Kip1) cyclin dependent kinase inhibitor. J Cell Biochem 104: 2091–2106 [DOI] [PubMed] [Google Scholar]
- Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL (2004) Molecular determinants of resistance to antiandrogen therapy. Nat Med 10: 33–39 [DOI] [PubMed] [Google Scholar]
- Cheung E, Kraus WL (2010) Genomic analyses of hormone signaling and gene regulation. Annu Rev Physiol 72: 191–218 [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS (2002) Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 9: 279–289 [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Zaret KS (2007) Specific interactions of the wing domains of FOXA1 transcription factor with DNA. J Mol Biol 366: 720–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleutjens CB, Steketee K, van Eekelen CC, van der Korput JA, Brinkmann AO, Trapman J (1997) Both androgen receptor and glucocorticoid receptor are able to induce prostate-specific antigen expression, but differ in their growth-stimulating properties of LNCaP cells. Endocrinology 138: 5293–5300 [DOI] [PubMed] [Google Scholar]
- Dong XY, Chen C, Sun X, Guo P, Vessella RL, Wang RX, Chung WKL, Zhou W, Dong JT (2006) FOXO1 is a candidate for the 13q14 tumor suppressor gene inhibiting androgen receptor signaling in prostate cancer. Cancer Res 66: 6998–7006 [DOI] [PubMed] [Google Scholar]
- Donovan MJ, Hamann S, Clayton M, Khan FK, Sapir M, Bayer-Zubek V, Fernandez G, Mesa-Tejada M, Teverovskiy M, Reuter VE, Scardino PT, Cordon-Cardo C (2008) Sytems pathology approach for the prediction of prostate cancer progression after radical prostatectomy. J Clin Oncol 26: 3923–3929 [DOI] [PubMed] [Google Scholar]
- Donovan MJ, Osman I, Khan FM, Vengrenyuk Y, Capodieci P, Koscuiszka M, Anad A, Cordon-Cardo C, Costa J, Scher HI (2009) Androgen receptor expression is associated with prostate cancer-specific survival in castrate patients with metastatic disease. BJU Int 105: 462–467 [DOI] [PubMed] [Google Scholar]
- Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M (2006) A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev 20: 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhoute J, Lupien M, Meyer CA, Verzi MP, Shivdasani RA, Liu XS, Brown M (2009) Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Res 19: 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Kaestner KH (2006) The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci 63: 2317–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sinha M, Peterson CL, Weng Z (2008) The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet 4: e1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, Ishii K, Mirosevich J, Kuwajima S, Oppenheimer SR, Roberts RL, Jiang M, Yu X, Shappell SB, Caprioli RM, Stoffel M, Hayward SW, Matusik RJ (2005) Forkhead box A1 regulates prostate ductal morphogenesis and promotes epithelial cell maturation. Development 132: 3431–3443 [DOI] [PubMed] [Google Scholar]
- Gao N, Zhang J, Rao MA, Case TC, Mirosevich J, Wang Y, Jin R, Gupta A, Rennie PS, Matusik RJ (2003) The role of hepatocyte nuclear factor-3 alpha (Forkhead Box A1) and androgen receptor in transcriptional regulation of prostatic genes. Mol Endocrinol 17: 1484–1507 [DOI] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, Mieczkowski P, Lieb JD, Zhao K, Brown M, Liu XS (2010) Nucleosome dynamics define transcriptional enhancers. Nat Genet 42: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemers HV, Tindall DJ (2007) Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr Rev 28: 778–808 [DOI] [PubMed] [Google Scholar]
- Heinlein CA, Chang C (2002) Androgen receptor (AR) coregulators: an overview. Endocr Rev 23: 175–200 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Hintze JL, Nelson RD (1998) Violin plots: a box plot-density trace synergism. Am Stat 52: 181–184 [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Smidt D, Carroll JS (2011) FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 43: 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Landan G, Pomerantz M, Jaschek R, Herman P, Reich D, Yan C, Khalid O, Kantoff P, Oh W, Manak JR, Berman BP, Henderson BE, Frenkel B, Haiman CA, Freedman M, Tanay A, Coetzee GA (2009) Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet 5: e1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, Hager GL (2008) Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell 29: 611–624 [DOI] [PubMed] [Google Scholar]
- Kaestner KH (2010) The FoxA factors in organogeensis and diferentiation. Curr Opin Genet Dev 20: 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Z, Jänne OA, Palvimo JJ (2004) Coregulator recruitment and histone modifications in transcriptional regulation by the androgen receptor. Mol Endocrinol 18: 2633–2648 [DOI] [PubMed] [Google Scholar]
- Kim J, Roh M, Abdulkadir SA (2010) Pim1 promotes human prostate cancer cell tumorigenicity and c-MYC transcriptional activity. BMC Cancer 10: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, Kaestner KH (2005) The initiation of liver development is dependent on Foxa transcription factors. Nature 435: 944–947 [DOI] [PubMed] [Google Scholar]
- Lee HJ, Hwang M, Chattopadhyay S, Choi H-S, Le K (2008) Hepatocyte nuclear factor-3 alpha (HNF-3α) negatively regulates androgen receptor transactivation in prostate cancer cells. Biochem Biophys Res Commun 367: 481–486 [DOI] [PubMed] [Google Scholar]
- Lin B, Wang J, Hong X, Yan X, Hwang D, Cho JH, Yi D, Utleg AG, Fang X, Schones DE, Zhao K, Omenn GS, Hood L (2009) Integrated expression profiling and ChIP-seq analyses of the growth inhibition response program of the androgen receptor. PLoS One 4: e6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Zhang Z, Yu H, Zheng SL, Isaacs WB, Xu J, Sun J (2011) Functional annotation of risk loci identified through genome-wide association studies for prostate cancer. Prostate 71: 955–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M (2008) FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132: 958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Fu W, Li P, Nicosia SV, Jenster G, Zhang X, Bai W (2009) FoxO1 mediates PTEN suppression of androgen receptor N- and C-terminal interactions and coactivator recruitment. Mol Endocrinol 23: 213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR (2009) DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 324: 407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirosevich J, Gao N, Gupta A, Shappell SB, Jove R, Matusik RJ (2006) Expression and role of Foxa proteins in prostate cancer. Prostate 66: 1013–1028 [DOI] [PubMed] [Google Scholar]
- Nakajima Y, Akaogi K, Suzuki T, Osakabe A, Yamaguchi C, Sunuhara N, Ishida J, Kako K, Ogawa S, Fujimura T, Homma Y, Fukamizu A, Murayama A, Kimura K, Inoue S, Yanagisawa J (2011) Estraogen regulates tumor growth through a nonclassical pathway that includes the transcription facotrs ERb and KLF5. Sci Signal 4: ra22. [DOI] [PubMed] [Google Scholar]
- Ouyang Z, Zhou Q, Wong WH (2009) ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc Natl Acad Sci USA 106: 21521–21526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovaska K, Laakso M, Haapa-Paananen S, Louhimo R, Chen P, Aittomäki V, Valo E, Nunez-Fontarnau J, Rantanen V, Karinen S, Nousiainen K, Lahesmaa-Korpinen AM, Miettinen M, Saarinen L, Kohonen P, Wu J, Westermarck J, Hautaniemi S (2010) Large-scale data integration framework provides a comprehensive view on glioblastoma multiforme. Genome Med 2: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS (2009) Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev 23: 804–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Zaret KS (2007) Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell 28: 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Myers M, Brown M. (2002) Formation of the androgen receptor transcription complex. Mol Cell 9: 601–610 [DOI] [PubMed] [Google Scholar]
- Sharma A, Yeow W-S, Ertel A, Coleman I, Clegg N, Thangavel C, Morrissey C, Zhang X, Comstock CES, Witkiewicz AK, Gomella L, Knudsen ES, Nelson PS, Knudsen KE (2010) The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest 120: 4478–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih DO, Navas MA, Kuwajima S, Duncan SA, Stoffel M (1999) Impaired glucose homeostasis and neonatal mortality in hepatocyte nuclear factor 3α-deficient mice. Proc Natl Acad Sci USA 96: 10152–10157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Shukla M, Maclennan GT, Fu P, Gupta S (2009) Deregulation of FOXO3a during prostate cancer progression. Int J Oncol 34: 1613–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Crawford GE (2010) DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc 2010, pdb.prot5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J, Lepikhova T, Teixido-Travesa N, Whitehead MA, Palvimo JJ, Jänne OA (2006) Small carboxyl-terminal domain phosphatase 2 attenuates androgen-dependent transcription. EMBO J 25: 2757–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinänen R, Palmberg C, Palotie A, Tammela T, Isola J, Kallioniemi OP (1995) In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet 9: 401–406 [DOI] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, Glass CK, Rosenfeld MG, Fu X-D (2011) Reprogramming of transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474: 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Carroll JS, Brown M (2005) Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell 19: 631–642 [DOI] [PubMed] [Google Scholar]
- Wang Q, Li W, Liu XS, Carroll JS, Jänne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M (2007) A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell 27: 380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, Wu T, Regan MM, Meyer CA, Carroll JS, Manrai AK, Jänne OA, Balk SP, Mehra R, Han B, Chinnaiyan AM et al. (2009) Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 138: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widen C, Zilliacus J, Gustafsson J-Å, Wikström AC (2000) Glucocorticoid receptor interaction with 14-3-3 and Raf-1, a proposed mechanism for cross-talk of two signal transduction pathways. J Biol Chem 275: 39296–39301 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liang J, Li Y, Xuan C, Wang F, Wang D, Shi L, Zhang D, Shang Y (2010) CCCTC-binding factor acts upstream of FOXA1 and demarcates the genomic response to estrogen. J Biol Chem 285: 28604–28613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.