Abstract
There have been recent surprising reports that whole genes can evolve de novo from noncoding sequences. This would be extraordinary if the noncoding sequences were random with respect to amino acid identity. However, if the noncoding sequences were previously translated at low rates, with the most strongly deleterious cryptic polypeptides purged by selection, then de novo gene origination would be more plausible. Here we analyze Saccharomyces cerevisiae data on noncoding transcripts found in association with ribosomes. We find many such transcripts. Although their average ribosomal densities are lower than those of protein-coding genes, a significant proportion of noncoding transcripts nevertheless have ribosomal densities comparable to those of coding genes. Most show increased ribosomal association in response to starvation, as has been previously reported for other noncoding sequences such as untranslated regions and introns. In rich media, ribosomal association is correlated with start codons but is not usually consistent and contiguous beyond that, suggesting that translation occurs only at low rates. One transcript contains a 28-codon open reading frame, which we name RDT1, which shows evidence of translation, and may be a new protein-coding gene that originated de novo from noncoding sequence. But the bulk of the ribosomal association cannot be attributed to unannotated protein-coding genes. Our primary finding of extensive ribosome association shows that a necessary precondition for selective purging is met, making de novo gene evolution more plausible. Our analysis is also proof of principle of the utility of ribosomal profiling data for the purpose of gene annotation.
Keywords: ORFan, SUTs, unannotated transcripts, RNA-Seq, evolvability, genetic assimilation
Introduction
Protein-coding sequences found only in a single species, family, or lineage are known as ORFans (Fischer and Eisenberg 1999). Several mechanisms have been proposed for the origin of apparent ORFans (Long et al. 2003; Kaessmann et al. 2009). These include mechanisms by which coding sequences give rise to ORFans, for example, through gene duplication (including via retrotransposition) followed by rapid divergence, through horizontal gene transfer from an uncharacterized source, or through gene fusion/fission. More radically, ORFans also arise de novo from noncoding sequences (Tautz and Domazet-Lošo 2011).
BSC4 in Saccharomyces cerevisiae is a remarkable example of a protein-coding gene that evolved de novo via a series of point mutations in noncoding sequence (Cai et al. 2008). Although at first sight this seems extraordinary, because random polypeptides are unlikely to fold stably (Dobson 1999; Bloom et al. 2007), genome-wide surveys suggest that de novo gene birth from noncoding sequences may not be so rare (Zhou et al. 2008; Tautz and Domazet-Lošo 2011). In addition to BSC4, cases have also been proteomically confirmed in humans (Knowles and McLysaght 2009; Li, Zhang, et al. 2010) and indirectly inferred through fusion constructs for a second open reading frame (ORF) in yeast (Li, Dong, et al. 2010). Cases have been inferred via expression analyses in Drosophila (Chen et al. 2007), Arabidopsis (Donoghue et al. 2011), and rice (Xiao et al. 2009), with protein-coding status yet to be determined for these cases. Other cases have been inferred bioinformatically in Drosophila (Levine et al. 2006; Begun et al. 2007), primates (Tay et al. 2009; Toll-Riera et al. 2009), and Plasmodium vivax (Yang and Huang 2011). On the smaller scale of parts of a gene, the conversion of noncoding sequence to coding can also occur through new coding exons (Kondrashov and Koonin 2003; Sorek 2007; Lin et al. 2009) or incorporation of 3′ untranslated regions (UTRs) (Giacomelli et al. 2007; Vakhrusheva et al. 2011) or 5′ UTRs (Wilder et al. 2009) into coding regions.
Conversion from noncoding to coding seems too unlikely an event to happen in a single evolutionary step. The sequence in question must be transcribed, escape degradation at the nuclear exosome, associate with ribosomes, be translated, and again escape degradation by the proteasome. Finally, it must avoid toxic conformations such as amyloid, for example, in favor of a stable protein fold.
At each stage, molecular errors in the present can provide a preview of mutations in the future (Whitehead et al. 2008; Masel and Trotter 2010; Rajon and Masel 2011). Selection may purge from cryptic sequences those variants whose expression is strongly and unconditionally deleterious, even when the sequences are expressed only at low levels via molecular errors. This purging is predicted to increase evolvability substantially (Masel 2006; Rajon and Masel 2011). At first, this result seems surprising because evolution has no foresight. But whereas it is impossible to know what will be adaptive in the future, it is often possible to rule out what will ‘not’ be adaptive, such as toxic amyloid. The distribution of fitness effects of new mutations is strongly bimodal, with most mutations either being lethal or having a small effect size (Eyre-Walker and Keightley 2007; Fudala and Korona 2009; Wylie and Shakhnovich 2011). If the cryptic lethals are screened out, then whatever is left, by a process of elimination, has a greater chance of being adaptive than random sequences do. This is the cause of increased evolvability. Benign cryptic sequences that persist through a selective filter against low levels of erroneous expression can provide preselected raw material to be co-opted for the evolution of novelty (Masel 2006; Rajon and Masel 2011).
Here we focus on the evolutionary stage just before a noncoding sequence is co-opted as a new protein. The likely raw material for such co-option consists of transcripts of unknown function that escape exonucleolytic degradation (stable unannotated transcripts or SUTs; Jacquier 2009) and associate with ribosomes. The occasional accidental translation of these transcripts, at low levels, could be enough to select against ORFs encoding toxic peptides. This preselection would enrich the raw material for those peptides most likely to be benign and so increase the likelihood of de novo gene birth. Because de novo gene birth is a real phenomenon in need of explanation, we predict ample preselected raw material. In other words, we predict that there are many noncoding transcripts associated with ribosomes at high enough levels to be consistent with substantial selection, purging from cryptic sequences those variants whose translation would be strongly deleterious.
Ingolia et al. (2009) profiled the positions of all complete ribosomes bound to RNA, providing a snapshot of translation. Ingolia et al. (2009) then analyzed patterns of ribosomal binding within annotated protein-coding transcripts. Here we reanalyze the ribosomal profiling data, focusing on ribosomes bound to SUTs. An earlier case study looked at three SUTs and found that one of them, NMR026W, was associated with ribosomes (Thompson and Parker 2007). It was unclear whether this SUT was highly unusual or reasonably typical. Here we address this question on a genome-wide basis and find that ribosomal binding to SUTs not only occurs but is also, in agreement with our hypothesis, quite common.
We find that most ribosomal binding of SUTs exhibits a strikingly different pattern from binding to coding sequences. However, we find one clear exception, demonstrating a new example, only 28 amino acids long, where an ORF in S. cerevisiae with evidence of translation appears to have evolved recently. We call this transcript RDT1 for ribosomally detected transcript.
Materials and Methods
The set of S. cerevisiae transcripts not containing annotated genes was downloaded from http://snyderlab.stanford.edu/Naga2008sup/novel_annotations.track. Transcript information for all annotated ORFs was obtained from table S4 in the supporting online material of Nagalakshmi et al. (2008); only those transcripts with well-defined UTRs were used in our analysis (4,419/6,604). Ribosome footprints and corresponding transcriptomes described by Ingolia et al. (2009) were obtained from the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) (GEO accession: GSE13750). These accessions include mappings of footprints to the yeast genome available from Saccharomyces Genome Database (SGD, http://www.yeastgenome.org/) on 22 June 2008. Only footprints that mapped uniquely to a single location in the genome without mismatches (a little more than 60% of the total) were used in our analysis. This yields a false discovery rate of essentially zero (Wang et al. 2009).
Genome sequences for the orthologous intergenic region between SPB1 and KAR4 orthologs in other fungal species were obtained from SGD and aligned using MUSCLE (Edgar 2004) followed by manual alignment. The SPB1 and KAR4 orthologs were used to anchor the alignment. Subsequent alignment was then performed progressively inward until converging on the region containing RDT1. Because the orthologous regions in S. kudriavzevii and S. bayanus could not be identified using the alignment, we searched the orthologous intergenic sequence for a highly divergent ORF. We did this using nucleotide position information for the entire orthologous intergenic region between SPB1 and KAR4.
The serial analysis of gene expression (SAGE) data set was obtained from Affymetrix Yeast S98 arrays and provided by the lab of Allan Jacobson (He et al. 2003) and the GEO (http://www.ncbi.nlm.nih.gov/geo/) (accession number: GSE2579) (Wyers et al. 2005).
AUGCAI was calculated using the method described in Miyasaka (1999). The transfer RNA (tRNA) copy numbers for S. cerevisiae, S. paradoxus, and S. mikatae were obtained from Scannell et al. (2011) and used to calculate tRNA adaptation index (tAI) using the codonR software (http://people.cryst.bbk.ac.uk/∼fdosr01/tAI/).
Results
For each of the two biological replicates of the two experimental conditions (rich and starved) described by Ingolia et al. (2009), we mapped ribosome footprints onto each of the 487 novel transcribed regions (SUTs) described by Nagalakshmi et al. (2008). Of the 404 SUTs for which Ingolia et al. (2009) found evidence of RNA expression, 217 showed some ribosomal association (at least one mismatch-free hit mapping uniquely to that SUT) in at least one of the replicates, in comparison to 4,372 of 4,404 expressed, verified ORF-containing transcripts.
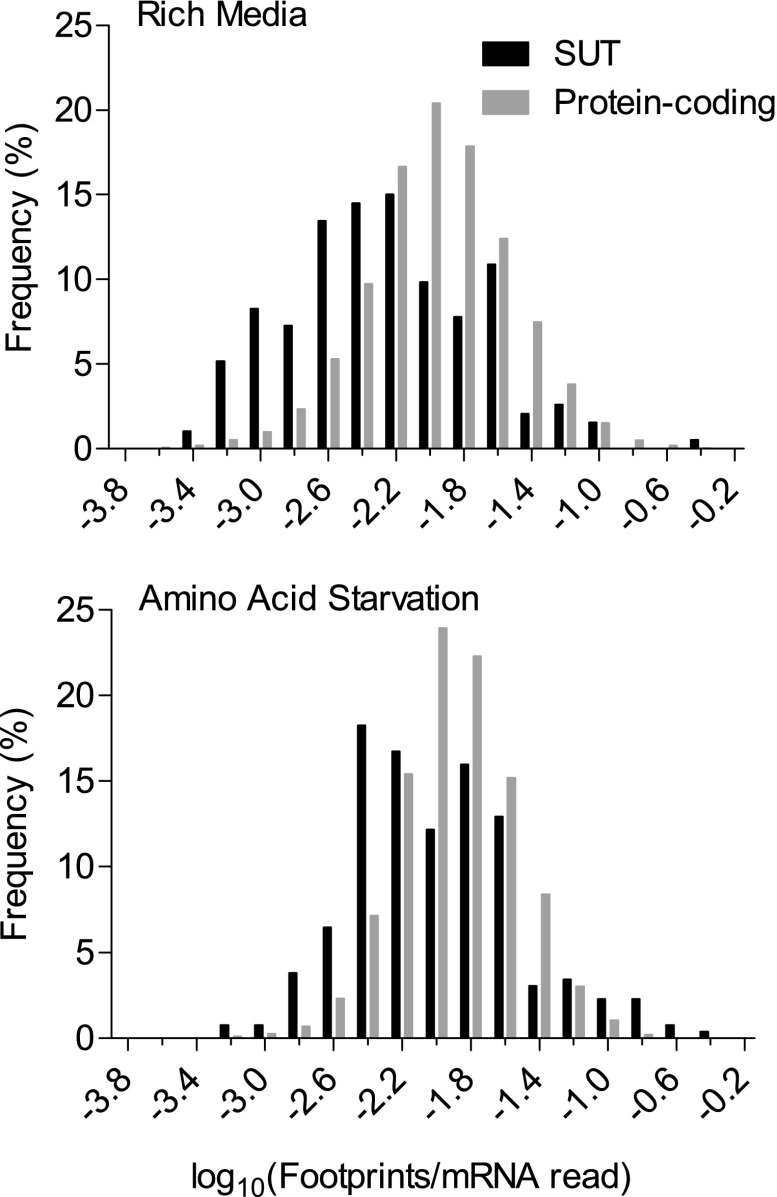
Next we quantified the level of ribosomal association to produce a histogram of average ribosomal density per ribosomally associated transcript (fig. 1). Ribosome association is not uncommon for SUTs and can occur at high frequency relative to messenger RNA (mRNA) concentration, especially but not exclusively in starved conditions (fig. 1). Although SUTs have, on average, lower ribosomal densities than protein-coding genes do (P < 10−17 for each of the four replicates, Welch two-sample t-test with unequal variance), many individual SUTs have high levels of ribosomal association.
FIG. 1.–
Histogram of ribosomal densities for transcripts annotated as protein coding and for other ribosomally associated transcripts (SUTs). Although ribosomal densities are lower for SUTs, there is substantial overlap, with many SUTs associated with ribosomes at levels typical for protein-coding transcripts. SUT–ribosome associations increase under starved conditions, whereas associations with protein-coding transcripts do not. Ribosomal density is calculated as the number of ribosome footprint mappings for a given transcript normalized by the number of mRNA mappings for the same transcript in the same experimental replicate. We pooled the two replicates available for each of the two (rich vs. starved) conditions. Transcripts showing no ribosomal association were excluded: numbers for these are given in the text.
Next we produced traces of ribosomal association as a function of position along each transcript. Each time a footprint mapped to a nucleotide position, we incremented its occupancy by the tag count of the footprint. A typical SUT’s ribosomal trace shows only a single peak (fig. 2A) or several, noncontiguous peaks (fig. 2D; supplementary fig. S1, Supplementary Material online). Ribosomal footprints that map to SUTs are 50% more likely to include an AUG triplet than an alternative NUN triplet in rich media (P < 10−3; contingency table) but are nonspecific with respect to triplet identity in starved conditions (P = 0.06). Note that it is difficult to know for sure whether ribosomal association always leads to translation. In this regard, it must be noted that translation can occasionally initiate even in the absence of an AUG start codon (Ingolia et al. 2009).
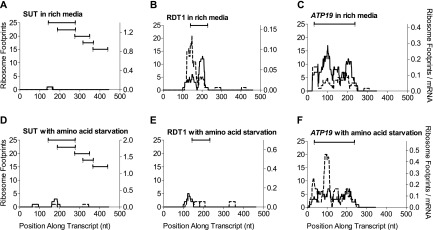
FIG. 2.–
Ribosomal traces for RDT1, compared with a representative SUT and with a verified short protein-coding gene ATP19. We see that the RDT1 traces show a very similar pattern to the protein-coding traces, whereas most other SUTs have dramatically different traces. Solid and dashed lines indicate each of the two replicates for the given condition. Lines are drawn above the positions of ORFs longer than 15 codons: the example of a SUT shown here contains multiple overlapping ORFs. The raw number of footprints per nucleotide is given in the 5′–3′ direction of each transcript. The y axis on the right of each figure is normalized for mRNA concentration; the y axis on the left is not. The otherwise representative SUT and protein-coding gene shown were chosen because of their length similarity to RDT1.
It is possible that some or even many of the SUTs with very high levels of ribosomal association are in fact short unannotated protein-coding genes. We therefore looked for ORFs within SUTs that might be protein-coding sequences. We examined each of the SUT ribosomal association traces manually. We chose to do this manually because we were interested in contiguity in addition to peak occupancy and have no validated a priori quantitative metric for contiguity. Five transcripts have particularly intriguing ribosomal traces, with locations along the transcript having peak occupancy of 10 or more footprints in at least one of the four replicates. For four of these transcripts, ribosomal occupancy did not correspond to an ORF and was much higher in starved conditions (supplementary fig. S2, Supplementary Material online). Increased association under starved conditions is typical for other noncoding sequences such as UTRs and introns (Ingolia et al. 2009).
However, one transcript contained a 28 amino acid ORF whose position corresponded to the region of highest ribosome occupancy relative to all other positions on that transcript (fig. 2B and E). The transcript showed higher ribosomal association in rich media. This transcript had a higher total number of ribosomal hits than any of the 486 other SUTs in both of the rich condition replicates and ranked 19th and 8th on this measure in the two starved condition replicates. The start codon context adaptation index (AUGCAI) is 0.32, which is well within the range of other yeast mRNA (Miyasaka 1999; supplementary fig. S5A, Supplementary Material online). These observations are all consistent with translation as a protein-coding gene, rather than merely occasional accidental translation. However, it should also be noted that the tAI is 0.18, falling only just within the range of other yeast mRNA (dos Reis et al. 2004; supplementary fig. S5B, Supplementary Material online). We named this transcript RDT1, for ribosomally detected transcript. RDT1 is located on the Watson strand of chromosome III between positions 30768 and 31228.
We blasted RDT1 using BlastN on the nt/nr nucleotide database, and the only significant hits (e value < 10−3), other than the same location in S. cerevisiae (i.e., self-hits), were found in the syntenic region in S. paradoxus. We also blasted the ORF sequence using the TBlastX algorithm on the nt/nr database, in case nucleotide divergence had masked amino acid conservation with another species, perhaps one related only through horizontal gene transfer. Again, we found only self-hits.
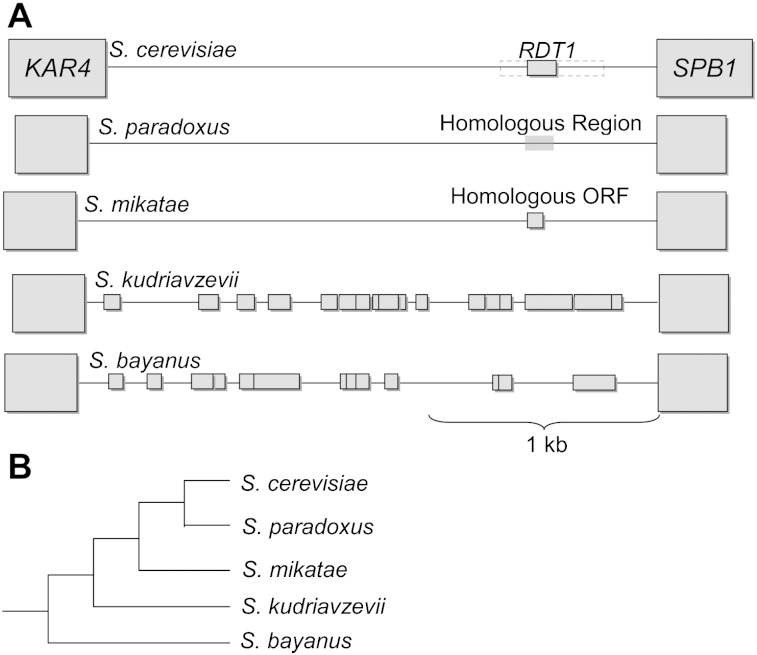
Through the inclusion of adjacent genes, we then forced an alignment of known syntenic sequences of other Saccharomyces species (Byrne and Wolfe 2005; see Materials and Methods). Although nucleotide sequence identity is low, we can confirm sequence homology among S. cerevisiae, S. paradoxus, and S. mikatae (fig. 3); sequences from S. kudriavzevii and S. bayanus were too divergent from these three to be aligned reliably. The start codon is present in the reference sequence of all three species; however, it is followed almost immediately by a stop codon in the S. paradoxus reference sequence. Saccharomyces mikatae does, however, contain a homologous 20 amino acid ORF. We looked in the syntenic region of S. kudriavzevii and S. bayanus for any syntenic ORF too divergent to detect homology but did not find a match (fig. 4).
FIG. 3.–
Alignment of sequences homologous to RDT1 in Saccharomyces mikatae, S. paradoxus, and S. cerevisiae. Amino acids are given underneath the center position of each putatively coding codon. Frameshifts mean that not all amino acid positions are homologous. Nucleotides that match in any two species are highlighted. Polymorphism among the S. cerevisiae strains is underlined. S. cer. A corresponds to the most common allele, also found in the SGD reference sequence, S. cer. B is the alternative putatively protein-coding allele (found in DBVPG6040, UWOPS83 787, and UWOPS87 2421), and S. cer. C (found in the Malaysian strains UWOPS05 217, UWOPS05 227, and UWOPS03 461) does not contain a start codon. S. par. H refers to UWOPS91 917.1, a Hawaiian strain of S. paradoxus.
FIG. 4.–
Synteny alignment of region including RDT1. (A) Synteny alignment of the Watson strand in the ∼2.5-kb intergenic region between KAR4 and SPB1 and their orthologs in related species. The position of RDT1 in Saccharomyces cerevisiae is shown, as is the homologous ORF in S. mikatae. All ORFs >15 codons long, in all three reading frames of the Watson strand (including overlapping ORFs), are shown for S. bayanus and S. kudriavzevii. Horizontal position represents distance (in nucleotides) from SPB1. (B) Known phylogenetic relationships of Saccharomyces species used (Rokas et al. 2003).
To study polymorphism in RDT1, we downloaded 39 S. cerevisiae and 36 S. paradoxus strains sequenced by the Saccharomyces Genome Resequencing Project (http://www.sanger.ac.uk/research/projects/genomeinformatics/sgrp.html, 2010 Sep). Thirty three S. cerevisiae strains share the same ORF allele as the S288C reference strain, and three strains (DBVPG6040, UWOPS83 787, and UWOPS87 2421) share a second allele of the same ORF with three nucleotide substitutions leading to two amino acid differences. The remaining three strains (UWOPS05 217, UWOPS05 227, and UWOPS03 461; this is the Malaysian cluster identified by Liti et al. 2009) share these three nucleotide differences and have two more, one of which abolishes the start codon and hence the ORF (fig. 3). This shows that translation of the RDT1 ORF is not essential in S. cerevisiae.
All but one of the S. paradoxus strains clearly lack the ORF. Twenty five strains have a stop codon in the third codon position, whereas 10 strains do not contain a start codon within the plausible length of a homologous transcript (supplementary fig. S3, Supplementary Material online). Assuming that the apparent start codon of the one remaining strain, UWOPS91 917.1, is not merely the result of a sequencing error, this strain has a homologous ORF 46 amino acids long. This strain is highly divergent from other S. paradoxus isolates and was sampled from a native plant in Hawaii (Liti et al. 2009).
The start codon context adaptation index (AUGCAI) described by Miyasaka (1999) was similar in S. cerevisiae RDT1 (0.32), in the homologous ORF in the Hawaiian S. paradoxus strain (0.32), and in the short ORF in S. mikatae (0.35). The tAI values for the Hawaiian S. paradoxus homolog and the S. mikatae homolog are slightly higher at 0.26 and 0.19, respectively, compared with 0.18 and 0.17 in the two S. cerevisiae alleles.
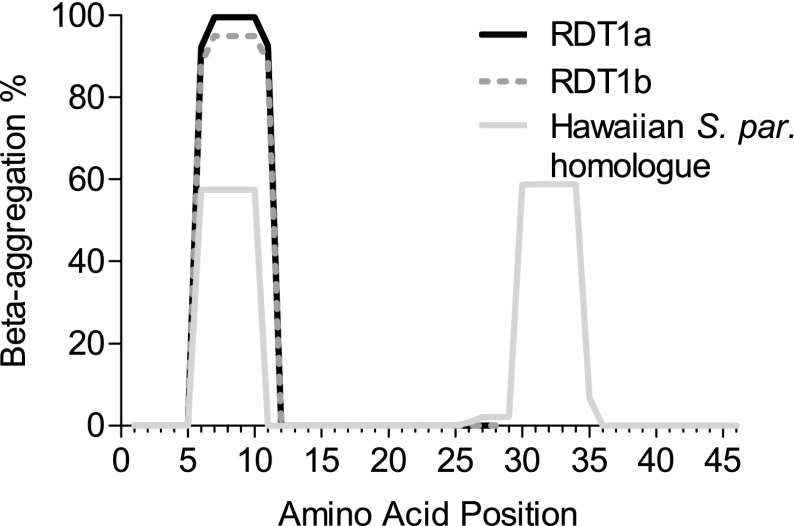
Protein aggregation was predicted for the ORF using TANGO (Fernandez-Escamilla et al. 2004). Surprisingly, an aggregation-prone hexapeptide is strongly predicted for both S. cerevisiae alleles and is weakly predicted in the single ORF-containing S. paradoxus strain (fig. 5). However, TANGO scores apply only to peptides in isolation and not to entire proteins in context, and so this result does not necessarily imply that RDT1 will aggregate. For example, RDT1 might form a homo-oligomer or a complex with other proteins, in which the aggregation-prone segment is sequestered deep within a protein fold. No aggregation propensity was detected for the S. mikatae ORF.
FIG. 5.–
Protein aggregation propensity (%) along the length of the putative protein, as predicted by TANGO. The y axis indicates the estimated likelihood that a peptide would be found in an aggregated structure rather than, for example, as an α-helix or β-sheet (Fernandez-Escamilla et al. 2004). These probabilities are based on thermodynamic stability and the Boltzmann equation and apply to the peptide in isolation from the rest of the protein. Both Saccharomyces cerevisiae RDT1 alleles have a predicted aggregation-prone sequence from amino acids 6–11 inclusive. No aggregation is predicted for S. mikatae, whose amino acids are not homologous in this region due to a frameshift. The single strain of S. paradoxus that contained an ORF shows a weak aggregation propensity, at a position shifted by one amino acid.
Discussion
We do not know whether RDT1 codes for a functional protein: Its translation could be accidental rather than a product of adaptation. It is clearly not essential in S. cerevisiae, as it is absent in Malaysian isolates. Nevertheless, its origin would still be interesting as a possible intermediate along the pathway to de novo gene birth.
There are two scenarios regarding the evolutionary origin of RDT1 as a protein-coding sequence. First, it may have evolved de novo on the branch leading to S. cerevisiae.
Second, RDT1 might already have been present as a protein-coding gene in the common ancestor of S. cerevisiae and S. mikatae. In this scenario, it was then lost in most or all the S. paradoxus lineages and also lost in the Malaysian S. cerevisiae lineage. The question is then whether it originated de novo after divergence with S. bayanus or whether it is evolving so fast that recognizable homology to older lineages is lost, making it appear to be ORFan. With or without recognizable homology in the nucleotide sequence, there is no syntenic ORF in S. bayanus (fig. 4). There is syntenic overlap with a much larger ORF in S. kudriavzevii but no indication whatsoever of homology regardless of how we (manually) align the sequences to attempt to force a homologous match. For this reason, de novo origination is suggested, but not proved, by the homology data.
The short length of RDT1 is also compatible with, but not proof of, its recent de novo origination. Recent de novo origination on the S. cerevisiae branch would be further supported if the homologous 20–amino acid S. mikatae ORF were found not to be transcribed or if its transcript is not ribosomally associated. However, it is difficult to obtain conclusive proof of absence of transcription because transcription may only occur under particular environmental conditions that do not match those assayed in the laboratory. Our finding of comparable codon adaptation indices in S. mikatae is consistent with translation in that species but might just as easily be a simple product of chance or phylogenetic confounding.
ORFs appearing by chance in SUTs are likely to be very short. Even after they have evolved to become functional proteins, they are likely to remain short for substantial periods of evolutionary time. Most classical gene annotation methods exclude short ORFs (Basrai et al. 1997) because they often appear by chance alone and do not code for proteins. This means that proteins recently evolved de novo will be missed due to their short length. Other gene annotation methods rely on evolutionary conservation (Cliften et al. 2003; Kellis et al. 2003); obviously, these methods will also fail to annotate recently evolved de novo protein-coding genes. The best methods to date for finding short protein-coding genes are proteomic (Kim et al. 2009). Our approach represents a novel proteomic method, strongly suggesting that RDT1 is translated. This could be demonstrated more conclusively in the future by artificially expressing RDT1, validating a mass spectrometry protocol to detect it in spiked yeast extracts and then assaying native RDT1 peptide levels in yeast.
We also used our method on an earlier SAGE “noncoding” data set used by Thompson and Parker (2007) (see Materials and Methods for details) and identified multiple protein-coding genes not annotated at the time that the data set was produced (not shown). All these have since been annotated as protein coding. This suggests that ribosomal profiling may be a powerful gene annotation method for taxa less well studied than S. cerevisiae.
Note that although our method can detect shorter proteins than many other methods, we still have a detection threshold of minimum protein length. This is because we looked for contiguous ribosomal association, which is more striking for longer ORFs. In addition, because our hits do not have complete codon specificity, bias caused by overlap means that traces have stronger signals in their central region and weaker signals at the edges (see supplementary fig. S4, Supplementary Material online, for an illustration). Very short translated ORFs would have a signal strength corresponding to that found at edges and hence be harder to detect.
We do not yet know whether the peptide encoded by RDT1 has been co-opted for a function or whether it is part of background evolutionary “noise.” But what is really striking is our more general finding of widespread ribosomal binding to SUTs. A high proportion of the noncoding genome is transcribed into SUTs (David et al. 2006). Here we have shown that just over half of all SUTs are transported to the cytoplasm and bind there to ribosomes, especially at AUG codons.
Although we do not know the extent to which this ribosomal association leads to translation, these SUTs, apart from RDT1, do not appear to encode functional protein-coding genes. Given the extraordinarily low false discovery rate associated with RNA-Seq data (Wang et al. 2009), this supports the hypothesis that the high level of ribosome association is due to intrinsically error-prone molecular processes.
This biological noise may ultimately and fortuitously facilitate de novo gene birth (Rajon and Masel 2011). Short ORFs appear frequently by chance and are then likely to be translated by accident, at least at low levels. A low level of expression is ideal for purging strongly deleterious sequences, whereas benign sequences remain effectively neutral (Masel 2006; Rajon and Masel 2011). These low rates of accidental expression leading to preadaptive purging could help provide the raw material for de novo birth of protein-coding genes.
Supplementary Material
Supplementary figures S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals .org/).
Acknowledgments
We thank Yuriko Harigaya, Roy Parker, Etienne Rajon, and Debrah Thompson for helpful discussions and the associate editor and anonymous reviewers for constructive comments on the manuscript. This work was supported by the National Institutes of Health (R01GM076041, R25GM072733) and by the Undergraduate Biology Research Program at the University of Arizona. J.M. is a Pew Scholar in the Biomedical Sciences.
References
- Basrai MA, Hieter P, Boeke JD. Small open reading frames: beautiful needles in the haystack. Genome Res. 1997;7:768–771. doi: 10.1101/gr.7.8.768. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Lindfors HA, Kern AD, Jones CD. Evidence for de novo evolution of testis-expressed genes in the Drosophila yakuba/Drosophila erecta clade. Genetics. 2007;176:1131–1137. doi: 10.1534/genetics.106.069245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JD, Raval A, Wilke CO. Thermodynamics of neutral protein evolution. Genetics. 2007;175:255–266. doi: 10.1534/genetics.106.061754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH. The yeast gene order browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Zhao R, Jiang H, Wang W. De novo origination of a new protein-coding gene in Saccharomyces cerevisiae. Genetics. 2008;179:487–496. doi: 10.1534/genetics.107.084491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S-T, Cheng H-C, Barbash DA, Yang H-P. Evolution of hydra, a recently evolved testis-expressed gene with nine alternative first exons in Drosophila melanogaster. PLoS Genet. 2007;3:e107. doi: 10.1371/journal.pgen.0030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften P, et al. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- David L, et al. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci U S A. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM. Protein misfolding, evolution and disease. Trends Biochem Sci. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- Donoghue M, Keshavaiah C, Swamidatta S, Spillane C. Evolutionary origins of Brassicaceae specific genes in Arabidopsis thaliana. BMC Evol Biol. 2011;11:47. doi: 10.1186/1471-2148-11-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat Rev Genet. 2007;8:610–618. doi: 10.1038/nrg2146. [DOI] [PubMed] [Google Scholar]
- Fernandez-Escamilla A-M, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- Fischer D, Eisenberg D. Finding families for genomic ORFans. Bioinformatics. 1999;15:759–762. doi: 10.1093/bioinformatics/15.9.759. [DOI] [PubMed] [Google Scholar]
- Fudala A, Korona R. Low frequency of mutations with strongly deleterious but nonlethal fitness effects. Evolution. 2009;63:2164–2171. doi: 10.1111/j.1558-5646.2009.00713.x. [DOI] [PubMed] [Google Scholar]
- Giacomelli MG, Hancock AS, Masel J. The conversion of 3′ UTRs into coding regions. Mol Biol Evol. 2007;24:457–464. doi: 10.1093/molbev/msl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, et al. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell. 2003;12:1439–1452. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kim W, et al. Proteomic detection of non-annotated protein-coding genes in Pseudomonas fluorescens Pf0-1. PLoS One. 2009;4:e8455. doi: 10.1371/journal.pone.0008455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles DG, McLysaght A. Recent de novo origin of human protein-coding genes. Genome Res. 2009;19:1752–1759. doi: 10.1101/gr.095026.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA, Koonin EV. Evolution of alternative splicing: deletions, insertions and origin of functional parts of proteins from intron sequences. Trends Genet. 2003;19:115–119. doi: 10.1016/S0168-9525(02)00029-X. [DOI] [PubMed] [Google Scholar]
- Levine MT, Jones CD, Kern AD, Lindfors HA, Begun DJ. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc Natl Acad Sci U S A. 2006;103:9935–9939. doi: 10.1073/pnas.0509809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C-Y, Zhang Y, et al. A human-specific de novo protein-coding gene associated with human brain functions. PLoS Comput Biol. 2010;6:e1000734. doi: 10.1371/journal.pcbi.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Dong Y, et al. A de novo originated gene depresses budding yeast mating pathway and is repressed by the protein encoded by its antisense strand. Cell Res. 2010;20:408–420. doi: 10.1038/cr.2010.31. [DOI] [PubMed] [Google Scholar]
- Lin L, et al. Large-scale analysis of exonized mammalian-wide interspersed repeats in primate genomes. Hum Mol Genet. 2009;18:2204–2214. doi: 10.1093/hmg/ddp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betrán E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- Masel J. Cryptic genetic variation is enriched for potential adaptations. Genetics. 2006;172:1985–1991. doi: 10.1534/genetics.105.051649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel J, Trotter MV. Robustness and evolvability. Trends Genet. 2010;26:406–414. doi: 10.1016/j.tig.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasaka H. The positive relationship between codon usage bias and translation initiation AUG context in Saccharomyces cerevisiae. Yeast. 1999;15:633–637. doi: 10.1002/(SICI)1097-0061(19990615)15:8<633::AID-YEA407>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajon E, Masel J. Evolution of molecular error rates and the consequences for evolvability. Proc Natl Acad Sci U S A. 2011;108:1082–1087. doi: 10.1073/pnas.1012918108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Williams BL, King N, Carroll SB. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- Saccharomyces Genome Resequencing Project [Internet]. 2008. Cambridge (England): Wellcome Trust Sanger Institute. [updated 2008 Sep 13; cited 2009 May 20]. Available from: http://www.sanger.ac.uk/research/projects/genomeinformatics/sgrp.html. [Google Scholar]
- Scannell DR, et al. The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for the Saccharomyces sensu stricto genus. G3 (Bethesda) 2011;1:11. doi: 10.1534/g3.111.000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R. The birth of new exons: mechanisms and evolutionary consequences. RNA. 2007;13:1603–1608. doi: 10.1261/rna.682507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Domazet-Lošo T. The evolutionary origin of orphan genes. Nat Rev Genet. 2011;12:692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- Tay S-K, Blythe J, Lipovich L. Global discovery of primate-specific genes in the human genome. Proc Natl Acad Sci U S A. 2009;106:12019–12024. doi: 10.1073/pnas.0904569106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Parker R. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:92–101. doi: 10.1128/MCB.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toll-Riera M, et al. Origin of primate orphan genes: a comparative genomics approach. Mol Biol Evol. 2009;26:603–612. doi: 10.1093/molbev/msn281. [DOI] [PubMed] [Google Scholar]
- Vakhrusheva A, Kazanov M, Mironov A, Bazykin G. Evolution of prokaryotic genes by shift of stop codons. J Mol Evol. 2011;72:138–146. doi: 10.1007/s00239-010-9408-1. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead DJ, Wilke CO, Vernazobres D, Bornberg-Bauer E. The look-ahead effect of phenotypic mutations. Biol Direct. 2008;3:18. doi: 10.1186/1745-6150-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder JA, Hewett EK, Gansner ME. Molecular evolution of GYPC: evidence for recent structural innovation and positive selection in humans. Mol Biol Evol. 2009;26:2679–2687. doi: 10.1093/molbev/msp183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Wylie CS, Shakhnovich EI. A biophysical protein folding model accounts for most mutational fitness effects in viruses. Proc Natl Acad Sci U S A. 2011;108:9916–9921. doi: 10.1073/pnas.1017572108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, et al. A rice gene of de novo origin negatively regulates pathogen-induced defense response. PLoS One. 2009;4:e4603. doi: 10.1371/journal.pone.0004603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Huang J. De novo origin of new genes with introns in Plasmodium vivax. FEBS Lett. 2011;585:641–644. doi: 10.1016/j.febslet.2011.01.017. [DOI] [PubMed] [Google Scholar]
- Zhou Q, et al. On the origin of new genes in Drosophila. Genome Res. 2008;18:1446–1455. doi: 10.1101/gr.076588.108. [DOI] [PMC free article] [PubMed] [Google Scholar]