Abstract
The remarkably coordinated nature of the DNA damage response pathway relies on numerous mechanisms that facilitate the assembly of checkpoint and repair factors at DNA breaks. Post-translational modifications on and around chromatin play critical roles in allowing the timely and sequential assembly of DNA damage responsive elements at the vicinity of DNA breaks. Notably, recent advances in forward genetics and proteomics-based approaches have enabled the identification of novel components within the DNA damage response pathway, providing a more comprehensive picture of the molecular network that assists in the detection and propagation of DNA damage signals.
Linking genome instability and tumorigenesis
Our genome faces constant challenges from genotoxic stress that arises as a result of normal cellular metabolism, including free radicals and DNA replication errors, or from exogenous sources such as ultraviolet (UV) or ionizing radiation (IR). To maintain genome integrity, the cell has evolved the DNA damage response (DDR) pathway, which allows lesion detection, signal propagation, and subsequent activation of a multitude of effector systems. Together, this DDR pathway promotes cell survival by activating cell cycle checkpoint arrest, and allows time for DNA repair prior to resumption of cell cycle progression [1]. In addition, when repair cannot be completed, the cell undergoes senescence or apoptosis, thus eliminating the possibility of passing on damaged or unrepaired genetic material to its progeny. As all cells encounter DNA damage constantly, proper DNA repair must play a major role in the maintenance of genome stability [2]. Indeed, mounting evidence supports a link between genome instability and tumorigenesis [3]; this link is evident from epidemiological studies as well as from human cancer-susceptibility diseases that result from mutations present in various components in the DDR pathway. Moreover, dysregulation of DNA repair pathways not only contributes to tumorigenesis, but also leads to developmental defects and embryonic lethality. The many implications of DNA repair gone awry underscore its importance in cell survival and human health (BOX 1).
Box 1. DNA Repair Pathways.
To maintain genome stability, the mammalian cell has evolved two major DNA repair pathways, namely those that occur through homologous recombination (HR) and non-homologous end joining (NHEJ). Much like the DNA damage-signaling pathway, these repair pathways proceed via a coordinated assembly of repair factors surrounding a DNA lesion. Recent evidence implicates the MRE11–RAD50–NBS1 (MRN) complex as an important and early component in both the HR and NHEJ repair pathways [93-99], supporting the notion that the choice of these repair systems is exquisitely orchestrated at the sites of DNA breaks to protect genome integrity in the cell.
NHEJ describes the ligation of DNA ends with no or limited processing. Because NHEJ does not require template DNA for repair, it is not restricted to any particular cell cycle phase, and is proposed to be the predominant repair pathway of choice in mammalian cells, especially in quiescent cells [100]. However, NHEJ can result in chromosomal fusions and translocations if it is not processed correctly. HR repair, by contrast, is generally viewed as an error-free repair mechanism. This repair pathway requires a homologous template, often a sister chromatid, and hence is believed to be the preferred repair pathway during S and G2 phases of the cell cycle. During HR repair, DNA ends are resected to expose ssDNAs that are rapidly coated by the ssDNA-binding protein RPA. This repair pathway is then initiated by the displacement of RPA from ssDNA, which results in the formation of the RAD51-nucleoprotein filament. Through the concerted action of a number of accessory factors, the RAD51-nucleoprotein filament catalyses strand invasion and homology search [101]. A number of pathways involving various helicases and resolvases have been proposed to dictate the outcome of the HR repair pathway. Nevertheless, mechanistic details regarding the resolution of the repair intermediates awaits further studies.
Recent studies have established the fundamental framework of our current view of DNA damage responses. Roles of post-translational modifications in DNA damage signal transduction and effector activation have been consolidated. Large scale RNA interference (RNAi) screens and proteomics-based approaches have provided an extensive list of damage-responsive proteins and damage-regulated protein modifications [4]. With these in hand, the ultimate goal is to delineate the molecular pathways in the DDR network. Deciphering the composition of these multi-layered pathways should aid not only in the development of clinical interventions, but also in the identification of novel biomarkers for early detection or even cancer predisposition. One particular focus in the field is to understand the hierarchical nature of the mammalian DNA damage-signaling pathway, and how checkpoint and repair proteins form foci structures at sites of DNA damage. In this regard, the canonical signaling pathway involving the phosphorylation of the histone variant H2A.X is pivotal for the concentration of numerous factors at the damage-modified chromatin.
Whereas early studies focused on protein phosphorylation, more recent studies have begun to emphasize the roles of ubiquitylation in the propagation of DNA damage signals and DNA repair [5-8]. In this review, we will discuss the important roles of post-translational modifications in DNA damage responses, with a focus on how they serve as stimuli-inducible switches to promote proper assembly of DNA repair factors at damage-modified chromatin in mammalian cells.
Ionizing Radiation-Induced Foci
DNA lesions, in particular those generated by IR, result in the most lethal type of DNA damage, DNA double-strand breaks (DSBs). Perhaps because of its clinical relevance, IR has been routinely used as a source of DNA damage for the study of damage signaling, checkpoint control and DNA repair pathways. Accordingly, a number of DNA damage responsive proteins that participate in checkpoint control and DNA repair readily accumulate into foci structures upon ionizing radiation. IRIF, the acronym for Ionizing Radiation-Induced Foci, is used to refer to cytologically observable focus formation and describes a general phenomenon of many DDR factors.
Studies related to how proteins concentrate at DNA breaks stemmed from the seminal finding that the repair protein RAD51 forms discrete foci upon DNA damage [9]. Subsequent identification and characterization of damage-induced phosphorylation of the histone variant H2AX proposed that phosphorylated H2AX (γH2AX) marks sites of DNA breaks [10]. As many DNA damage repair proteins co-localize with γH2AX at these IRIF, it was speculated that repair proteins are recruited to the IRIF to facilitate DNA repair. With this as the backdrop, a hierarchical pathway in which many DNA damage-responsive proteins are sequentially assembled at the vicinity of DNA breaks is now beginning to emerge.
Phosphorylation-dependent events target many DNA damage responsive proteins to IRIF
Numerous studies have addressed how genotoxic stress triggers the DDR. It is now established that the ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3 related (ATR) kinases are the major regulators of the cellular response to DNA damage. By regulating the phosphorylation status of more than 700 proteins [11], these kinases orchestrate many steps along the DDR pathway. The ATM/ATR-dependent phosphorylation of H2AX S139 constitutes one of the initial signals upon DNA lesion detection. H2AX phosphorylation is believed to be essential for the sustained accumulation of various checkpoint and DNA repair factors at DNA breaks. Mouse models lacking H2AX are growth retarded, immunologically compromised and display increased tumor incidence [12-13]. These phenotypes are associated with genome instability and repair defects, and interestingly, parallel the IRIF defects observed in the absence of checkpoint and repair proteins including tumor suppressors BRCA1 (breast cancer 1, early onset) and 53BP1 (tumor protein p53 binding protein 1). Since then, a growing list of damage responsive proteins that harbor conserved phospho-protein binding motifs, namely BRCA1 C-terminal (BRCT) and forkhead-associated (FHA) domains [14-16], have been identified, further illustrating the importance of protein phosphorylation in the regulation of the DDR.
The DNA damage signaling pathway began to emerge with the identification of Mediator of DNA damage Checkpoint 1 (MDC1), a BRCT domain-containing protein, as a molecular adaptor that regulates checkpoint functions in mammalian cells [17-20] (Figure 1). MDC1 is recruited to IRIF in a BRCT-dependent manner in vivo, and MDC1 BRCT domains directly bind γH2AX in vitro [21]. Given that H2AX phosphorylation represents one of the earliest damage-induced signals, these data suggest that MDC1 is the major, and at least one of the first, proteins to localize to the sites of DNA breaks in a γH2AX-dependent pathway. Moreover, MDC1 is required for the subsequent localization of many DNA damage repair proteins, including BRCA1 and 53BP1, to IRIF. Although MDC1 has established roles in controlling the assembly of multiple repair factors at DNA breaks and in amplifying the DNA damage signal, mechanistic insights into this process were revealed only by recent findings.
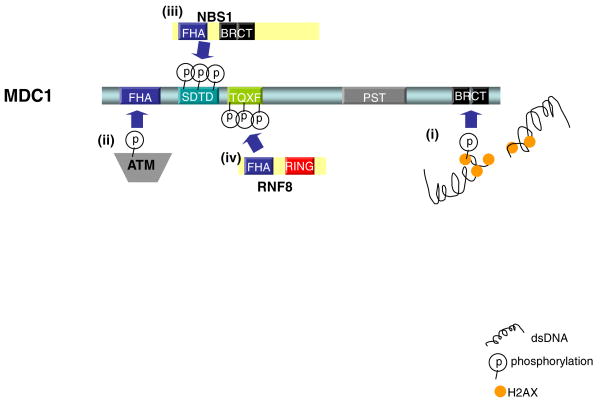
Figure 1. MDC1 is the molecular platform for the assembly of checkpoint and repair proteins at the vicinity of DNA damage sites.
Schematic illustration of MDC1 domain organization and their respective roles in the assembly of various components in the DNA damage response pathway. i) Upon DNA damage, the histone variant H2AX (orange) surrounding DNA damage sites is phosphorylated by the ATM, DNA-PK and ATR kinases, which allows the direct engagement of MDC through its phospho-protein binding BRCT domain (black). ii) At damaged chromatin, the MDC1 FHA domain (blue) recruits phosphorylated ATM (gray) molecules and allows amplification of ATM signaling. iii) The MDC1 tandem SDTD repeats (aqua) are phosphorylated by casein kinase 2, which allows accumulation of the MRN complex through its interaction with NBS1. iv) Phosphorylation of the tandem TQXF repeats (lime green) on MDC1 mediates its interaction with RNF8 through its FHA domain, which initiates the ubiquitin-dependent signaling pathway involving RNF8, RNF168 and UBC13.
The MDC1-NBS1 connection
NBS1 (named for Nijmegen breakage syndrome; also called Nibrin) is a component of the MRE11–RAD50–NBS1 (MRN) complex that is thought to play a role in detecting DNA breaks. Its role in DNA damage responses is supported by the observations that NBS1 patients are immunodeficient, display increased sensitivity to radiation therapy, and are cancer-prone. Like BRCA1, NBS1 harbors a phospho-peptide binding FHA-BRCT domain that is pivotal for its localization at IRIF [22-23]. Recent studies indicate that, distinct from the RAD50–MRE11-mediated interaction with chromatin [24-25], NBS1 concentration at DSBs requires its association with MDC1 [26-29]. Casein kinase 2 phosphorylates a tandem array of conserved acidic SDTD repeats within MDC1, which serve as binding sites for the NBS1 FHA-BRCT domain. Through this interaction, MDC1 facilitates further accumulation of the MRN complex at the damage modified-chromatin. The importance of this sustained localization of the MRN complex is evident as the impairment of the MDC1–NBS1 interaction compromises DNA damage-responsive intra-S phase checkpoint activation and cell survival [26-29].
BRCT-domain containing proteins in the DNA damage response
As the prototype of BRCT-domain containing protein, BRCA1 has been reported to interact with a number of phospho-proteins, which in turn allows specific regulation of several cellular processes, including checkpoint control, DNA repair and transcriptional regulation [30-31]. A growing body of evidence suggests that its tumor suppressor function can be attributed primarily to its role in the maintenance of genome stability, partly owing to its stable association with numerous factors involved in the DDR pathway. BRCA1 constitutively heterodimerizes with BRCA1 associated RING domain 1 (BARD1) [32], and forms multiple macromolecular complexes selectively via its BRCT domain, which binds CtBP interacting protein (CtIP), BRCA1 associated C-terminal helicase (BACH1), ATRIP (ATR interacting protein) and Coiled-coil domain-containing protein 98 (CCDC98; also called ABRAXAS) in a phosphorylation-dependent manner [33-39]. Notably, BRCA1 concentrates at DSBs through its BRCT-dependent interaction with CCDC98, which in turn is recruited to DSBs via its binding to RAP80. Recent work from several laboratories revealed that this macromolecular complex contains additional components, namely Mediator of RAP80 interaction and targeting 40 kDa [MERIT40; also called New component of the BRCA1 A complex (NBA1)], BRCA1/BRCA2-containing complex subunit 36 (BRCC36) and BRCA1/BRCA2-containing complex subunit 45 (BRCC45; also called Brain and reproductive organ expressed; BRE) [40-41]. Complex formation stabilizes this five-protein assembly at DNA breaks, promotes BRCA1 accumulation within IRIF, and enforces the BRCA1-dependent DNA damage response.
Microcephalin 1 (MCPH1; also called BRIT) encodes another BRCT domain-containing damage-responsive protein. MCPH1 mutations are implicated in the autosomal recessive disease primary microcephaly, a neurodevelopmental defect resulting in microcephaly and mental retardation [42]. Recent evidence also points to a role for MCPH1 in damage-induced checkpoint control, likely via regulation of the BRCA1-CHK1 pathway [43-45]. This finding raises the possibility that MCPH1 acts early in the DDR pathway. MCPH1 IRIF can be readily observed, and in vitro binding assays [46] suggest that MCPH1 might be directly recruited to DSBs through BRCT domain-dependent γH2AX binding. This finding implies that MCPH1 and MDC1 can be independently recruited by γH2AX; indeed, this idea is supported by the observation that MCPH1 IRIF can be observed in Mdc1-/- cells and that MCPH1 overexpression partially inhibits MDC1 IRIF formation [46]. However, because earlier data suggested that MCPH1 is required for MDC1, phospho-ATM and NBS1 IRIF formation [47], the mechanism(s) by which MCPH1 is recruited to and functions at sites of DNA breaks remain obscure.
Regulatory Ubiquitylation directs IRIF assembly
Although the MDC1–NBS1 interaction can explain MDC1-dependent NBS1 recruitment to IRIF, the mechanism by which MDC1 promotes retention of RAP80–CCDC98–BRCA1 and 53BP1 at the damage-modified chromatin remained murky until the recent identification of a novel ubiquitin-dependent signaling cascade. A role for ubiquitin has long been implicated in the DDR. Cytological observations revealed extensive co-localization of conjugated ubiquitin (detectable by a monoclonal antibody FK2) and γH2AX [48-49], suggesting that certain ubiquitylated protein(s) concentrate at DSBs. Moreover, RAP80 IRIF formation requires its Ubiquitin-Interacting Motif (UIM) [37, 50-51], which selectively binds Lys63- and Lys6 polyubiquitin chains. These findings support the notion that ubiquitylated proteins at sites of DNA breaks might serve as docking sites for RAP80 chromatin loading. The molecular bases linking these observations were revealed when the E3 ubiquitin ligase ring finger protein 8 (RNF8) was discovered to play a role in the DNA damage signaling pathway [5]. RNF8 harbors an FHA domain, and through its phosphorylation-dependent interaction with MDC1, concentrates at γH2AX-marked DSBs. Moreover, in concert with the E2 ubiquitin conjugating enzyme UBC13, RNF8 promotes IRIF formation of conjugated ubiquitin and catalyses histone ubiquitylation at sites of DNA breaks. Most importantly, DSB-associated RNF8 is required for sustained recruitment of RAP80–CCDC98–BRCA1 and 53BP1 to sites of DNA damage. As UBC13 catalyses the formation of non-canonical Lys63-linked polyubiquitin chains, it is generally accepted that RNF8–UBC13-mediated histone ubiquitylation recruits RAP80 to sites of DNA damage.
Intriguingly, an early study reported defective conjugated ubiquitin, BRCA1 and 53BP1 IRIF formation in cells derived from a patient with Riddle syndrome [52], a disease characterized by radiosensitivity, immunodeficiency, dysmorphic features and learning difficulties. Employing siRNA library screen using 53BP1 IRIF as a readout, subsequent studies identified RNF168, which encodes an E3 ubiquitin ligase, as the gene mutated in the Riddle patient [53-54]. Accordingly, RNF168 expression complemented the Riddle cells and restored conjugated ubiquitin, BRCA1 and 53BP1 IRIF formation. RNF168 harbors two motifs that interact with ubiquitin, which promote binding to ubiquitylated histones and support its focal accumulation at DSBs. In concert with UBC13, RNF168 amplifies RNF8-dependent substrate ubiquitylation, and in turn facilitates BRCA1 and 53BP1 IRIF formation. Consistent with previous reports using over-expressed ubiquitin [48, 50], the use of antibodies that reacted specifically with Lys63-linked ubiquitin conjugates revealed damage-induced and RNF168-dependent foci formation, suggesting that RNF8 and RNF168-catalysed ubiquitylation events at DSBs do not target their substrates for proteasomal degradation. Instead these non-canonical ubiquitin chains might serve as molecular platforms for the assembly of the downstream checkpoint and DNA repair factors at the vicinity of DNA breaks. In this regard, although ubiquitylation of H2A-type histones have been proposed to facilitate assembly of downstream checkpoint and repair factors at DSBs, whether endogenous histones are modified by Lys63-linked ubiquitin chains remains to be clarified. Moreover, it remains unknown whether RNF168 might mediate ubiquitylation of other yet-to-be identified substrates.
The notion that ubiquitin chains at DSBs are important for concentration of DNA repair proteins was further substantiated when RAD18 was shown to function in the DDR pathway downstream of RNF8 [55]. RAD18 plays an established role in post-replicative repair (PRR) through regulating proliferating cellular nuclear antigen (PCNA) ubiquitylation [56-57]. Interestingly, distinct from its role in PRR, IR-induced focal accumulation of RAD18 requires its zinc finger, which interacts with ubiquitin chains in vitro. This finding indicates that like RAP80, sustained RAD18 recruitment might be mediated by DSB-associated ubiquitin chains [55].
The importance of the RNF8-RNF168 axis in licensing the assembly of checkpoint and DNA repair factors at DSBs is further illustrated by its requirement for recruitment of other damage responsive factors to IRIF. Recent work indicates that similar to 53BP1, Pax transactivation activation domain-interacting protein (PTIP) IRIF formation requires RNF8 and UBC13 [58-59]. PTIP harbors multiple tandem BRCT repeats, and is thought to promote ATM signaling in response to genotoxic stress through its ability to interact with 53BP1 [60-61]. Significantly, PTIP regulates the recruitment of its associated factor PA1, and proper localization of PTIP and PA1 is required for checkpoint activation in response to irradiation [58]. Although PTIP is a stable component of the Set1-like histone methyltransferase (HMT) complex [62], because depletion of other components of the HMT complex did not affect PTIP IRIF formation, it was proposed that PTIP might participate in the cellular response to DNA damage distinct from its role in transcriptional regulation. This idea was supported by the observation that a portion of PTIP migrates independently of other HMT components in gel filtration experiments [58].
Neither PTIP nor 53BP1 are known to bind ubiquitin, nor do they harbor any recognizable ubiquitin binding motifs; therefore the manner by which this ubiquitin-dependent signaling pathway regulates their recruitment to IRIF remains unresolved. Given the bulky nature of ubiquitin, it is possible that ubiquitylation events at the vicinity of DSBs might open up local chromatin thereby enabling PTIP and 53BP1 accumulation. Of note, 53BP1, in part via its Tudor domain, can interact with methylated histone H3 and H4 [63-64]. As these core histone molecules are constitutively methylated in vivo, DNA damage might expose these methylated histones to facilitate 53BP1 recruitment to IRIF. PTIP foci formation, by contrast, relies on its phospho-protein binding BRCT domain [15], pointing to a role for additional layers of mediators in coordinating its proper localization at DSBs. Further work will be needed to address these possibilities. Nevertheless, the identification of E3 ubiquitin ligases required for the assembly of multiple repair factors highlights the explicit integration between protein phosphorylation and protein ubiquitylation in the cellular response to DNA damage (Figure 2).
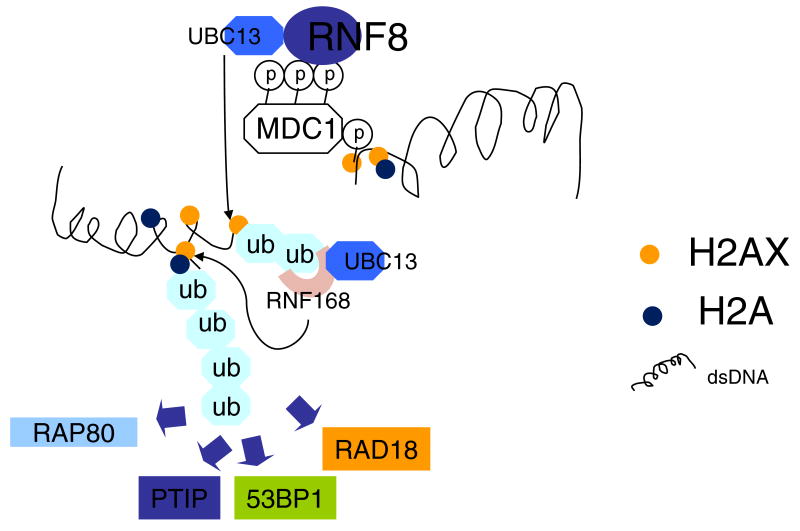
Figure 2. Regulatory ubiquitylation-dependent signaling cascade involving RNF8 and RNF168.
Pictorial representation of the RNF8 and RNF168-mediated substrate ubiquitylation at DSBs to facilitate accumulation of various DNA damage checkpoint and repair proteins. DNA damage triggers the phosphorylation of the histone variant H2AX (orange) at the vicinity of DNA breaks, which in turn directly recruits MDC1 through its phospho-protein binding BRCT domain. Phosphorylated MDC1 then allows accumulation of the E3 ubiquitin ligase RNF8 (dark blue), which in concert with the E2 ubiquitin conjugating enzyme UBC13 (blue), mediates H2A-type histone ubiquitylation. RNF168 (pink) recognizes the RNF8-mediated ubiquitylated histone, and with UBC13, further amplifies the local ubiquitin environment, which is required for focal accumulation of downstream checkpoint and repair factors, including RAP80 (blue), 53BP1 (green), RAD18 (orange) and PTIP (dark blue). Whether histone H2A (dark blue) is poly-ubiquitylated by K63-linked ubiquitin chains remains to be determined. In addition, exactly how RNF168–UBC13-dependent substrate ubiquitylation mediates the recruitment of the downstream DNA damage responsive elements remains unclear. Ubiquitin: ub (light blue).
The MRN complex in DSB end processing and checkpoint activation
The MRN complex is instrumental in the detection of DNA breaks; it also plays a critical role in ATM activation and chromatin tethering upon genotoxic stress. Recent evidence reveals that the MRN complex, together with CtIP, promotes DNA end resection and enables RPA assimilation onto single-stranded DNA (ssDNA) [65-68]. The three-subunit Replication protein A (RPA) is the canonical eukaryotic single-strand DNA binding protein that stabilizes ssDNA structures, which are crucial for DNA transactions including DNA replication, repair and recombination. At the DSB, the RPA-coated ssDNA recruits ATR–ATRIP to chromatin [69], an event which is critical for the activation of ATR-Chk1 pathway. Although CtIP interacts directly with MRN, it remains unknown whether CtIP accumulation requires the MRN complex.
Much like the trimeric RPA complex, the newly identified human single-stranded DNA binding protein (hSSB1) protein is thought to play a role in sequestering ssDNA for checkpoint activation and DNA repair [70]. Interestingly, hSSB1 (and its homolog hSSB2) forms a trimeric complex with integrator complex subunit 3 (INTS3) and C9ORF80, which accumulates at DSBs [71-72]. Although it remains unclear how RPA and hSSB1 (and hSSB2) differ in functional specificity, genetics studies have revealed differences in the factors that govern their concentration at DSBs. Whereas RPA accumulates at DSBs specifically during S/G2 cell cycle phases, the hSSB complexes are recruited to IRIF independent of cell cycle phases. In addition, although cytological observations revealed distinct localization of damage-induced hSSB1 and RPA IRIF, it remains unknown whether these single-strand DNA binding proteins exhibit preferential binding to selective DNA structures. As hSSB1 retention at DSBs does not require CtIP, it is possible that nucleation of hSSB1 (and hSSB2) onto ssDNA does not require extensive DNA end resection [71].
Recent reports indicate that, in addition to directing damage-induced RPA accumulation, the MRN complex regulates foci formation of the mammalian RECQ homologue, RECQ5, at DNA breaks [73]. RECQ5 is one of the five members of the human RECQ family of helicases, and it has been implicated in the resolution of deleterious DNA structures that arise during DNA replication [74]. Accordingly, MRN complex components were identified by mass spectrometry in RECQ5 immunoprecipitates, and RECQ5 accumulation at damage-modified chromatin was abrogated in MRE11-depleted and ATLD-1 cells, the latter of which expresses the human ataxia-telangiectasia like disorder (A-TLD)-derived MRE11 mutant allele. These observations suggest that the direct interaction with MRN is important for proper RECQ5 localization in response to DNA damage. Moreover, RECQ5 focal accumulation requires neither MDC1 nor CtIP, indicating that MRN might directly target and synergize with RECQ5 in DNA repair. A complete understanding of exactly how RECQ5 and MRN act together awaits further experimentation.
The PALB2-BRCA2-RAD51 Pathway in DNA repair
Although the DNA damage-signaling cascade involving γH2AX plays a major role in the assembly of a cohort of damage response proteins at DSBs to coordinate checkpoint control with DNA repair, recruitment of the homologous recombination-based repair protein RAD51 has long been known to function independently of H2AX phosphorylation status. Previous studies established that BRCA2 dictates RAD51 assimilation onto ssDNA to initiate the early steps of homologous recombination [75-76]. A subsequent study by Livingston and colleagues identified Partner and Localiser of BRCA2 (PALB2) in controlling the DSB-associated accumulation of BRCA2 and RAD51 [77], indicating that PALB2 acts upstream of BRCA2 and RAD51 and plays an instrumental role in coordinating homology-based DNA repair.
Unlike BRCA2, BRCA1 functions not only in DNA repair, but also in checkpoint control and DNA decatenation. Although previous studies suggested that BRCA1 modulates RAD51 focal accumulation at DSBs [78-79], and that homology-based DNA repair is compromised in BRCA1 depleted and mutant cells, the molecular link for BRCA1 and DNA repair was only revealed recently when PALB2 was identified as an integral component for the assembly of the BRCA1–BRCA2 complex [80-82]. Three independent studies now report that BRCA1 associates with BRCA2 through coiled-coil domain-dependent interactions with PALB2. Employing a mutant GFP gene conversion assay as a model to assay DNA repair [83], the BRCA1–PALB2 interaction was found to be important for optimal homology-based DNA repair [80-82]. More importantly, patient missense mutations that affect the PALB2-binding region of BRCA1 specifically disrupted the BRCA1–PALB2 interaction, and led to DNA repair defects. These results solidified the notion that BRCA1 is directly involved in DNA repair, and suggest that impaired homology-based DNA repair is a fundamental cause for the genome instability and cancer predisposition observed among a subset of BRCA1 patients.
Notably, despite the consensus that PALB2 functionally links BRCA1 and BRCA2 in DNA repair [80-82], whether BRCA1 is essential for accumulation of PALB2, BRCA2 and RAD51 at IRIF remains controversial. As BRCA1 directly interacts with PALB2, it is possible that BRCA1 serves as an additional anchor point to stabilize PALB2 retention at DSBs. Thus, in the absence of BRCA1, concentration of the PALB2–BRCA2–RAD51 complex at IRIF might be partially compromised. In support of this idea, PALB2 over-expression in HCC1937 human mammary gland epithelial cancer cells revealed only a mild difference in PALB2 foci forming ability [81]. Moreover, given that deficiencies in any of the components in the H2AX-MDC1-RNF8-RAP80 pathway, which is essential for BRCA1 IRIF formation, do not abolish PALB2-dependent RAD51 foci formation [84], it is likely that BRCA1 plays an accessory role in the DSB-associated concentration of the DNA repair machinery. Exactly how BRCA1 participates in DNA repair requires more in-depth studies, especially as various BRCA1 BRCT domain-associated DNA repair factors, including CtIP and BACH1, are present in PALB immunoprecipitates. The appealing hypothesis, which remains be tested, is that BRCA1 functions as a master regulator to coordinate multiple steps of a homologous recombination reaction.
The Fanconi Anemia complex
Fanconi anemia (FA) is a rare genetic cancer-susceptibility syndrome characterized by childhood-onset aplastic anemia, developmental abnormalities, and cellular hypersensitivity to DNA crosslinking agents [85]. To date, 13 different mutated genes have been identified in FA patients, indicating that Fanconi anemia is a highly heterogeneous genetic disorder. Interestingly, three of these genes encode DNA repair factors: PALB2 (FANCN), BRCA2 (FANCD1) and BACH1 (FANCJ) [85-86]. This finding lends support to the idea that FA deficiency and its associated phenotypes result from a compromised cellular capacity for DNA repair. Apart from PALB2 and BRCA2, which have direct roles in chromatin loading of repair protein RAD51, the functions of the other eleven FA proteins remain poorly defined. Emerging evidence suggests that the FA core (FANCA, B, C, E, F, G, L and M) plays a pivotal role in the mono-ubiquitylation and loading of FANCD2 and FANCI to sites of DNA breaks to facilitate repair [87]. This idea stems from the observation that mutations in FA core components compromise FANCD2 and FANCI mono-ubiquitylation and damage-induced focal accumulation, which parallel the cellular hypersensitivity to DNA cross-linking agents. Despite recent advances in FA research, the functional role of FANCD2 and FANCI mono-ubiquitylation remains unknown. Previous evidence proposed that mono-ubiquitylation is required for FANCD2 and FANCI chromatin association, suggesting that an unidentified chromatin factor might facilitate the loading of these monoubiquitylated molecules at DNA damage sites [87]. Moreover, unlike FANCJ, FANCD1 (BRCA2) and FANCN (PALB2), which play crucial roles in homology-based DNA repair [85], suppression of other members of the FA complementation groups results in only mild deficits in gene conversion, suggesting that the FA proteins might be required for DNA repair that involves distinct DNA substrates.
Concluding Remarks
The past few years have witnessed a rapid expansion of our knowledge regarding DDR pathways. A variety of purification strategies have proven invaluable in facilitating the identification of novel components in large damage-responsive protein complexes. Moreover, large-scale RNAi screening strategies coupled with functional readouts facilitate the identification of proteins of interest in specific DDR pathways. IRIF have attracted much attention in the field, and several novel damage-responsive proteins have been identified, in part based on their unique localization to these ionizing radiation induced foci. Interestingly, one recent study reported the use of DNA hybridization and precipitation as a strategic means to capture and identify proteins at specific genomic loci [88]. Whether a similar approach can be adopted to identify additional IRIF-localized DNA repair factors awaits future experimentation.
Although the capacity for IRIF formation has greatly facilitated the identification of novel DNA repair factors and the hierarchical pathways in the DNA damage response, some caution is warranted in the interpretation of protein recruitment and focus formation. Cytologically visible foci reflect the accumulation of hundreds or even thousands of copies of a single protein, and detection is limited by the sensitivity and specificity of antibodies. Failure to observe damage-induced focal accumulation of a protein might reflect impaired protein recruitment, compromised protein stability or affinity for DSBs, or differences in cell cycle-dependent expression and/or localization patterns. Corroborative and complementary assays including chromatin immunoprecipitation are useful tools to further scrutinize protein accumulation at the vicinity of DSBs, especially for evaluating protein recruitment and retention at a specific genomic locus. More importantly, functional assays must be performed in parallel to cytological studies to enable an understanding of the consequences of these interesting subnuclear localizations in the cellular response to DNA damage.
A role for post-translational modifications as molecular switches that direct the assembly of DNA repair factors at damaged chromatin is well-established. The vast numbers of protein modifications and their implicated roles in DNA damage responses suggest that additional levels of regulation are present in various DDR pathways. Indeed, in addition to phosphorylation and ubiquitylation, poly-ADP ribose and Small Ubiquitin-like Modifier (SUMO) conjugates are present at DNA breaks [89-92]. Whether these protein modifications govern the sub-cellular localization of their targets or serve as stimuli-induced recruiting factors deserves careful investigation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28(5):739–45. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Lukas J, Bohr VA, Halazonetis TD. Cellular responses to DNA damage: current state of the field and review of the 52nd Benzon Symposium. DNA Repair (Amst) 2006;5(5):591–601. doi: 10.1016/j.dnarep.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319(5868):1352–5. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 4.Petrini JH. Cell signaling. A touching response to damage. Science. 2007;316(5828):1138–9. doi: 10.1126/science.1143700. [DOI] [PubMed] [Google Scholar]
- 5.Bennett EJ, Harper JW. DNA damage: ubiquitin marks the spot. Nat Struct Mol Biol. 2008;15(1):20–2. doi: 10.1038/nsmb0108-20. [DOI] [PubMed] [Google Scholar]
- 6.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33(3):275–86. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Stewart GS. Solving the RIDDLE of 53BP1 recruitment to sites of damage. Cell Cycle. 2009;8(10):1532–8. doi: 10.4161/cc.8.10.8351. [DOI] [PubMed] [Google Scholar]
- 8.Panier S, Durocher D. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair (Amst) 2009;8(4):436–43. doi: 10.1016/j.dnarep.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Haaf T, et al. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci U S A. 1995;92(6):2298–302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Redon C, et al. Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev. 2002;12(2):162–9. doi: 10.1016/s0959-437x(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 11.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 12.Celeste A, et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114(3):371–83. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bassing CH, et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114(3):359–70. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 14.Yu X, et al. The BRCT domain is a phospho-protein binding domain. Science. 2003;302(5645):639–42. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 15.Manke IA, et al. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302(5645):636–9. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 16.Durocher D, et al. The FHA domain is a modular phosphopeptide recognition motif. Mol Cell. 1999;4(3):387–94. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- 17.Stewart GS, et al. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421(6926):961–6. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 18.Lou Z, et al. MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature. 2003;421(6926):957–61. doi: 10.1038/nature01447. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg M, et al. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421(6926):952–6. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 20.Xu X, Stern DF. NFBD1/KIAA0170 is a chromatin-associated protein involved in DNA damage signaling pathways. J Biol Chem. 2003;278(10):8795–803. doi: 10.1074/jbc.M211392200. [DOI] [PubMed] [Google Scholar]
- 21.Stucki M, et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123(7):1213–26. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 22.Zhao S, Renthal W, Lee EY. Functional analysis of FHA and BRCT domains of NBS1 in chromatin association and DNA damage responses. Nucleic Acids Res. 2002;30(22):4815–22. doi: 10.1093/nar/gkf612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carney JP, et al. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93(3):477–86. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 24.de Jager M, et al. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell. 2001;8(5):1129–35. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- 25.Hopfner KP, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418(6897):562–6. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 26.Wu L, et al. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc Natl Acad Sci U S A. 2008;105(32):11200–5. doi: 10.1073/pnas.0802885105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008;9(8):795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spycher C, et al. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J Cell Biol. 2008;181(2):227–40. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melander F, et al. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol. 2008;181(2):213–26. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosen EM, Fan S, Ma Y. BRCA1 regulation of transcription. Cancer Lett. 2006;236(2):175–85. doi: 10.1016/j.canlet.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 31.Powell SN, Kachnic LA. Roles of BRCA1 and BRCA2 in homologous recombination, DNA replication fidelity and the cellular response to ionizing radiation. Oncogene. 2003;22(37):5784–91. doi: 10.1038/sj.onc.1206678. [DOI] [PubMed] [Google Scholar]
- 32.Wu LC, et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14(4):430–40. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 33.Greenberg RA, et al. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 2006;20(1):34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venere M, et al. Phosphorylation of ATR-interacting protein on Ser239 mediates an interaction with breast-ovarian cancer susceptibility 1 and checkpoint function. Cancer Res. 2007;67(13):6100–5. doi: 10.1158/0008-5472.CAN-07-0369. [DOI] [PubMed] [Google Scholar]
- 35.Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol. 2004;24(21):9478–86. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim H, Huang J, Chen J. CCDC98 is a BRCA1-BRCT domain-binding protein involved in the DNA damage response. Nat Struct Mol Biol. 2007;14(8):710–5. doi: 10.1038/nsmb1277. [DOI] [PubMed] [Google Scholar]
- 37.Wang B, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316(5828):1194–8. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z, Wu J, Yu X. CCDC98 targets BRCA1 to DNA damage sites. Nat Struct Mol Biol. 2007;14(8):716–20. doi: 10.1038/nsmb1279. [DOI] [PubMed] [Google Scholar]
- 39.Cantor SB, et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105(1):149–60. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 40.Heinrichs A. Higher-order BRCA1 complexity. Nat Rev Mol Cell Biol. 2009;10(5):301. doi: 10.1038/nrm2683. [DOI] [PubMed] [Google Scholar]
- 41.Dong Y, et al. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol Cell. 2003;12(5):1087–99. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- 42.Woods CG, Bond J, Enard W. Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. Am J Hum Genet. 2005;76(5):717–28. doi: 10.1086/429930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alderton GK, et al. Regulation of mitotic entry by microcephalin and its overlap with ATR signalling. Nat Cell Biol. 2006;8(7):725–33. doi: 10.1038/ncb1431. [DOI] [PubMed] [Google Scholar]
- 44.Lin SY, et al. BRIT1/MCPH1 is a DNA damage responsive protein that regulates the Brca1-Chk1 pathway, implicating checkpoint dysfunction in microcephaly. Proc Natl Acad Sci U S A. 2005;102(42):15105–9. doi: 10.1073/pnas.0507722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu X, Lee J, Stern DF. Microcephalin is a DNA damage response protein involved in regulation of CHK1 and BRCA1. J Biol Chem. 2004;279(33):34091–4. doi: 10.1074/jbc.C400139200. [DOI] [PubMed] [Google Scholar]
- 46.Wood JL, et al. MCPH1 functions in an H2AX-dependent but MDC1-independent pathway in response to DNA damage. J Biol Chem. 2007;282(48):35416–23. doi: 10.1074/jbc.M705245200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rai R, et al. BRIT1 regulates early DNA damage response, chromosomal integrity, and cancer. Cancer Cell. 2006;10(2):145–57. doi: 10.1016/j.ccr.2006.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris JR, Solomon E. BRCA1 : BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum Mol Genet. 2004;13(8):807–17. doi: 10.1093/hmg/ddh095. [DOI] [PubMed] [Google Scholar]
- 49.Polanowska J, et al. A conserved pathway to activate BRCA1-dependent ubiquitylation at DNA damage sites. Embo J. 2006;25(10):2178–88. doi: 10.1038/sj.emboj.7601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sobhian B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316(5828):1198–202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316(5828):1202–5. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 52.Stewart GS, et al. RIDDLE immunodeficiency syndrome is linked to defects in 53BP1-mediated DNA damage signaling. Proc Natl Acad Sci U S A. 2007;104(43):16910–5. doi: 10.1073/pnas.0708408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doil C, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136(3):435–46. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 54.Stewart GS, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136(3):420–34. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 55.Huang J, et al. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol. 2009;11(5):592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoege C, et al. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419(6903):135–41. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 57.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. Embo J. 2000;19(13):3388–97. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gong Z, et al. Accumulation of Pax2 transactivation domain interaction protein (PTIP) at sites of DNA breaks via RNF8-dependent pathway is required for cell survival after DNA damage. J Biol Chem. 2009;284(11):7284–93. doi: 10.1074/jbc.M809158200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu J, et al. PTIP Regulates 53BP1 and SMC1 at the DNA Damage Sites. J Biol Chem. 2009;284(27):18078–84. doi: 10.1074/jbc.M109.002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munoz IM, et al. Phospho-epitope binding by the BRCT domains of hPTIP controls multiple aspects of the cellular response to DNA damage. Nucleic Acids Res. 2007;35(16):5312–22. doi: 10.1093/nar/gkm493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jowsey PA, Doherty AJ, Rouse J. Human PTIP facilitates ATM-mediated activation of p53 and promotes cellular resistance to ionizing radiation. J Biol Chem. 2004;279(53):55562–9. doi: 10.1074/jbc.M411021200. [DOI] [PubMed] [Google Scholar]
- 62.Cho YW, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282(28):20395–406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huyen Y, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432(7015):406–11. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 64.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127(7):1361–73. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459(7245):460–3. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen L, et al. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283(12):7713–20. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 67.Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284(14):9558–65. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450(7169):509–14. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300(5625):1542–8. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 70.Richard DJ, et al. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature. 2008;453(7195):677–81. doi: 10.1038/nature06883. [DOI] [PubMed] [Google Scholar]
- 71.Huang J, et al. SOSS complexes participate in the maintenance of genomic stability. Mol Cell. 2009;35(3):384–93. doi: 10.1016/j.molcel.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Y, et al. hSSB1 and hSSB2 form similar multi-protein complexes that participate in DNA damage response. J Biol Chem. 2009 doi: 10.1074/jbc.C109.039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng L, et al. MRE11 complex links RECQ5 helicase to sites of DNA damage. Nucleic Acids Res. 2009;37(8):2645–57. doi: 10.1093/nar/gkp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci. 2008;33(12):609–20. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Katagiri T, et al. Multiple possible sites of BRCA2 interacting with DNA repair protein RAD51. Genes Chromosomes Cancer. 1998;21(3):217–22. [PubMed] [Google Scholar]
- 76.Sharan SK, et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386(6627):804–10. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 77.Xia B, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22(6):719–29. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 78.Bhattacharyya A, et al. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275(31):23899–903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 79.Zhang J, et al. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol Cell Biol. 2004;24(2):708–18. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang F, et al. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol Cancer Res. 2009;7(7):1110–8. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci U S A. 2009;106(17):7155–60. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang F, et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19(6):524–9. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pierce AJ, et al. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13(20):2633–8. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sy SM, et al. PALB2 Regulates Recombinational Repair through Chromatin Association and Oligomerization. J Biol Chem. 2009;284(27):18302–10. doi: 10.1074/jbc.M109.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moldovan GL, D'Andrea AD. How the Fanconi Anemia Pathway Guards the Genome. Annu Rev Genet. 2009 doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8(10):735–48. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 87.Alpi AF, Patel KJ. Monoubiquitylation in the Fanconi anemia DNA damage response pathway. DNA Repair (Amst) 2009;8(4):430–5. doi: 10.1016/j.dnarep.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 88.Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136(1):175–86. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.El-Khamisy SF, et al. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31(19):5526–33. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brown ML, et al. Role of poly(ADP-ribosyl)ation in DNA-PKcs- independent V(D)J recombination. Proc Natl Acad Sci U S A. 2002;99(7):4532–7. doi: 10.1073/pnas.072495299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu ZX, et al. PML colocalizes with and stabilizes the DNA damage response protein TopBP1. Mol Cell Biol. 2003;23(12):4247–56. doi: 10.1128/MCB.23.12.4247-4256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Carbone R, et al. PML NBs associate with the hMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene. 2002;21(11):1633–40. doi: 10.1038/sj.onc.1205227. [DOI] [PubMed] [Google Scholar]
- 93.Dimitrova N, de Lange T. Cell cycle dependent role of MRN at dysfunctional telomeres: ATM signaling-dependent induction of NHEJ in G1 and resection-mediated inhibition of NHEJ in G2. Mol Cell Biol. 2009 doi: 10.1128/MCB.00476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dinkelmann M, et al. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol. 2009;16(8):808–13. doi: 10.1038/nsmb.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16(8):814–8. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rass E, et al. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16(8):819–24. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 97.Deng Y, et al. Multiple roles for MRE11 at uncapped telomeres. Nature. 2009;460(7257):914–8. doi: 10.1038/nature08196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mimitou EP, Symington LS. DNA end resection: Many nucleases make light work. DNA Repair (Amst) 2009;8(9):983–95. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Deriano L, et al. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol Cell. 2009;34(1):13–25. doi: 10.1016/j.molcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yano K, et al. Molecular mechanism of protein assembly on DNA double-strand breaks in the non-homologous end-joining pathway. J Radiat Res (Tokyo) 2009;50(2):97–108. doi: 10.1269/jrr.08119. [DOI] [PubMed] [Google Scholar]
- 101.Mimitou EP, Symington LS. Nucleases and helicases take center stage in homologous recombination. Trends Biochem Sci. 2009;34(5):264–72. doi: 10.1016/j.tibs.2009.01.010. [DOI] [PubMed] [Google Scholar]