Abstract
A long-lived and sequence specific ligand-DNA complex would make possible the modulation of biological processes for extended periods. We have been investigating the threading polyintercalation approach to DNA recognition in which chains of aromatic units thread back and forth repeatedly through the double helix. Here we report the preliminary sequence specificity and detailed kinetic analysis of a structurally characterized threading tetraintercalator. Specific binding on a relatively long DNA strand was observed, strongly favoring a predicted 14-base pair sequence. Kinetic studies revealed a multi-step association process and specificity was found to derive primarily from large differences in dissociation rates. Importantly, the rate-limiting dissociation rate constant of the tetraintercalator complex dissociating from its preferred binding site was extremely slow, corresponding to a 16 day half-life, making it one of the longer non-covalent complex half-lives yet measured, and, to the best of our knowledge, the longest for a DNA binding molecule.
Molecules that bind DNA can modulate transcription, repair, and replication both in vitro and in vivo, so targeting specific sequences of DNA with small molecules has been a field of continued interest for over 30 years. For example, Dervan and co-workers have developed a series of polyamide molecules that bind in dimeric fashion to DNA with programmable specificity via the minor groove,1,2 with a current focus on understanding the rules for delivery and distribution in vivo.3,4 Other modular approaches include triple-helix forming oligonucleotides and peptide-nucleic acids (PNAs).5-8
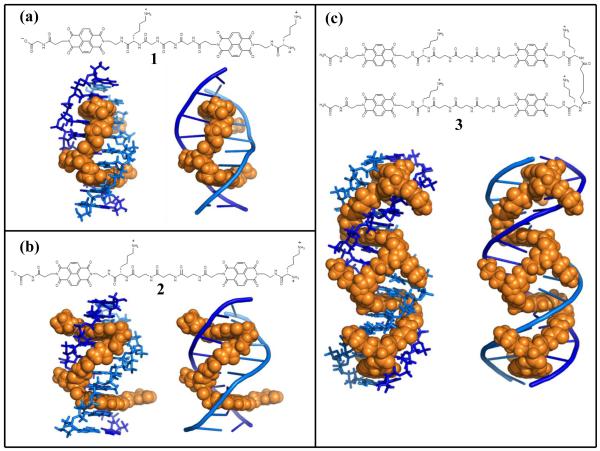
For possible in vivo studies in the laboratory, as well as looking ahead to potential therapeutic applications for addressing long-term chronic diseases, it is of interest to develop DNA binding molecules exhibiting very long-lived complexes, capable of modulating biological processes on timescales relevant to organism lifetimes. With this in mind, we have been investigating threading polyintercalation, a binding mode that places a significant number of functional groups in both DNA grooves. For example, by linking the naphthalene diimide (NDI) units, known threading intercalators,9,10 in a head-to-tail fashion with a variety of flexible peptide linkers, a growing family of unique DNA-binding molecules has been developed.11-17 Threading bis-intercalators 1 and 2 were previously identified through library screening experiments in which the peptide linkers were randomized.12 Structural characterization of the complexes with their respective preferred DNA binding sites revealed that sequence specific recognition was taking place with linkers spanning four base pairs (bp) through the major and minor grooves of DNA for 1 and 2, respectively (Fig. 1a & 1b).12-14 This work complements studies with a variety of other synthetic and naturally occurring mono- and bis- DNA intercalators.10, 18-24
Figure 1. Chemical structures, ligand-DNA complex models, and threading polyintercalation cartoon illustration for NDI polyintercalators.
a, Bisintercalator 1 with a view from the major groove of 1-d(CGGTACCG)2 complex.13b, Bisintercalator 2 with a view from the minor groove of 2-d(CGATAAGC)·d(GCTTATCG) complex.14c, Tetraintercalator 3 with a view of 3-d(GATAAGTACTTATC)2 complex.15 Models and cartoon illustrations were produced by The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.
Tetraintercalator 3 was designed with linkers intended to bind with a minor–major–minor groove topology (Fig. 1c). The two β-alanyl-β-alanyl-β-alanyl-l-lysine (-βAla3-Lys-) minor groove binding linkers were taken from 2. Recall that β-alanine is the biochemical name often used for 3-aminopropanoic acid. Adipic acid with two l-lysine residues attached was chosen to be the central major groove binding linker because modeling indicated this linker would reproduce many of the structural features of the glycyl-glycyl-glycyl-l-lysine linker of 1, while offering the advantage of introducing C2 symmetry into 3, simplifying NMR spectra and thus structural analyses. Consistent with the original design, a detailed NMR structural characterization verified threading polyintercalation, and revealed the linkers to be distributed in the expected minor-major-minor groove topology (Fig. 1c).15
An anticipated consequence of the threading polyintercalator design of 3 is an extremely slow dissociation from its preferred DNA sequence. This expectation is based on the observation that for one of the two internal NDI units to dissociate, the adjacent terminal NDI must dissociate first. For the entire molecule to dissociate, the other two NDI units would have to also dissociate in a coordinated fashion before re-binding occurs. The unlikely nature of this highly concerted set of motions should reduce the dissociation rate significantly. One might expect the association rate to be proportionately slow due to the complicated nature of the bound topology. In addition to kinetics, the overall sequence specificity of the interaction has not previously been addressed with a threading tetraintercalator such as 3.
Herein, footprinting studies verified sequence specific binding by 3 in the context of a relatively long piece of DNA that contained a large number of other possible binding sites. Additionally, the detailed kinetic analysis of 3 binding to oligonucleotides with and without its preferred DNA sequence were measured, revealing a multi-step association as well as a sequence specific and remarkably slow dissociation process. We report the slowest dissociation half-life of any non-covalent interaction with DNA, which could ultimately lead to prolonged disruption of transcription or other biological processes.
Results
DNase I footprinting
DNase I footprinting was used to investigate the ability of 3 to bind its 14 bp preferred site within the context of a 467 bp DNA fragment. A clear footprint for 3 binding selectively to sequence A with high specificity was observed at 63 nM after 48 hr (Fig. 2). Non-specific binding was not seen, even at 500 nM, verifying the highly specific and predictable nature of DNA binding by 3. It is assumed that 3 is binding to sequence A with the same threading topology observed in the NMR structural analysis reported previously for this sequence, although there is formally no direct experimental evidence to support this assumption on long pieces of DNA.15 Quantitative footprinting of binding thermodynamics was precluded by the extensive time needed to approach equilibrium at concentrations near our estimated KD value25 and the limited dynamic range of the footprinting method at the low concentrations of labeled DNA required.
Figure 2. DNase I footprinting of 3 bound to a 467 bp DNA segment.
Lane 1 is a control lane without DNase I or 3. Lanes 2 and 4 contain DNA digested with DNase I for 7 min and 4 min, respectively. Lane 3 is an adenine-specific sequencing lane. In lanes 5 – 9, the DNA fragment was incubated with increasing concentrations of 3 (31, 63, 125, 250, 500 nm), respectively, for 48 hr followed by digestion with DNase I for 7 min. The preferred sequence 5′-GATAAGTACTTATC-3′ (1) is highlighted with a bracket.
Dissociation kinetics
A 32P labeled oligonucleotide containing the 14 bp tetraintercalator binding site (oligo A, Fig. 3a) was incubated with a stoichiometric amount of 3, followed by separation using native polyacrylamide gel electrophoresis. The 3 - oligo A complex exhibited retarded migration on the gel. A control oligonucleotide (oligo B, Fig. 3a) containing a randomly chosen sequence was also incubated with 3, but did not produce an observable gel shift.
Figure 3. Oligonucleotide duplex sequences used in these studies and representative dissociation gel mobility-shifts from oligo A and stopped-flow dissociation from oligo B.
a, Duplex sequences abbreviated as oligo A-C as referenced above, with the target sequence underlined for each duplex. b, 3 dissociating from oligo A after 20 days and 34 days of incubation, showing the prolonged dissociation from the preferred binding site. C refers to the control lane which contains oligo A in the absence of 3 in 10 mM piperazine-N,N’-bis(2-ethanesulfonic acid) (PIPES) (pH = 7.0), 1 mM ethylenediaminetetraacetic acid(EDTA), and 100 mM NaCl. All other incubations were run as triplicate measurements in 10 mM PIPES (pH = 7.0), 1 mM EDTA with varying [Na+] as labeled above the respective lanes. Complexed and unbound oligo A bands are as labeled. c, Representative stopped-flow dissociation trace of 3-oligo B complex. The equilibrated complex was mixed with equal volumes of 4% sodium dodecyl sulfate (SDS) in 10 mM PIPES (pH = 7.0), 1 mM EDTA, and 100 mM NaCl monitiored at the visible absorbance maximum of 386 nm, showing the rapid dissociation from a control sequence. The data was fit using a double-exponential equation.
Gel shift assays were used to determine the dissociation rate constant for 3 from oligo A containing the preferred binding site. In these experiments a 100-fold excess of unlabeled oligo A was added to a solution containing the 32P-labeled oligo A occupied by a stoichiometric amount of bound 3 (Fig. 3b). We assume that this technique measures the rate-limiting dissociation rate constant as 3 leaves its binding site. Gel shifts could be analyzed up to 34 days before loss of radioactivity reduced the signal-to-noise ratio and thus the dynamic range of the assay below useful levels. The data was fit to a standard monoexponential decay equation (Fig. S1, Supplementary Information) revealing a dissociation rate constant (kd) of 5.0 ± 0.5 × 10−7 s−1 at 100 mM NaCl, which corresponds to a half-life of 16 days (Table 1). The observed kd increased with increasing NaCl concentrations over the range 25 mM to 250 mM, which is not surprising given the four l-lysine residues within 3 that presumably make electrostatic contacts with the phosphodiester backbone of DNA. A plot of log kd as a function of −log [Na+] was linear with a slope of 0.59 (Fig. S2, Supplementary Information).
Table 1.
Cumulative data for the dissociation of 3 from both oligo A and oligo B at varying [Na+]. Dissociation rate constants of 3 from oligo A calculated from the monoexponential fits to the gel shifts. Error levels estimated at ± 17%. For oligo B, A1 and A2 are the relative amplitudes for the double exponential according to kd,app = A1k1+A2k2. The estimates of the rate constants fit a double exponential equation with a standard error less than 1%. In order to obtain the faster rates and corresponding amplitudes, the fits to the traces obtained at 10 s intervals were coerced to the slower rates and their amplitudes obtained from the 200 s traces.
| Oligo A | Oligo B | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| [Na+] (mM) | kd × 10−7 (s−1) | t1/2 (days) | k1 (s−1) | A1 (%) | k2 (s−1) | A2 (%) | kd (s−1) |
| 25 | 2.3 | 32 | - | - | - | - | - |
| 50 | 3.3 | 23 | 0.27 | 30 | 0.012 | 70 | 0.088 |
| 100 | 5.0 | 16 | 0.29 | 34 | 0.019 | 66 | 0.11 |
| 150 | 5.7 | 13 | 0.38 | 35 | 0.031 | 65 | 0.15 |
| 200 | 9.3 | 8 | 0.39 | 40 | 0.038 | 60 | 0.18 |
| 300 | - | - | 0.48 | 39 | 0.055 | 61 | 0.22 |
Nonspecific dissociation kinetics
In order to quantify the sequence specific behavior of 3, the dissociation rate constant from a randomly chosen sequence, oligo B, was also determined. As mentioned previously, gel electrophoresis could not be used to monitor the dissociation because a gel shift could not be detected for the 3-oligo B complex, presumably due to rapid dissociation. Instead, the dissociation of 3 from oligo B was monitored using stopped-flow methods in the presence of 2% sodium dodecyl sulfate (SDS) monitored for 200 seconds (Fig. 3c).18,26 The absorbance trace was fit to a double exponential function, suggesting a two-step mechanism, with the observed first-order dissociation rate constants of the fast and slow steps, k1 and k2, determined to be 0.29 s−1 and 0.019 s−1, respectively, at 100 mM NaCl. Note that SDS has been shown to enhance the dissociation rate for cationic intercalators that bind DNA, so values reported here should be considered upper-limit estimates.27 Using the slower dissociation step for comparison, these results demonstrate that the dissociation of 3 from oligo A containing the preferred binding site is roughly 4 × 104 times slower compared to oligo B, verifying that the differences in kinetic off-rates is largely, if not exclusively, responsible for sequence selectivity. Salt dependence of the dissociation rate was also examined for the 3-oligo B complex (Table 1), and the log kd versus −log [Na+] plot was linear with a slope of 0.54 (Fig. S3, Supplementary Information).
Association rate measurements
To study the association rates of 3, a multi-pronged approach was pursued: 1D 1H NMR, gel mobility-shift assay, and stopped-flow UV-visible spectroscopy.
1D 1H NMR studies were performed using the same 14 bp binding site oligonucleotide (oligo C, Fig. 3a) used previously,15 at a final concentration of 60 μM mixed directly in an NMR tube with a stoichiometric amount of 3. After a total of 5.6 minutes to mix and scan the sample, analysis of the spectra revealed that all signals were identical to a fully equilibrated sample (Fig. 4).15 In other words, 3 was fully intercalated in the oligo C binding site with the linker in the expected minor-major-minor groove topology after 5.6 min. The 3-oligo C NMR sample was subsequently monitored at different time points over a 24 hr period (Fig. S4, Supplementary Information) with no observable change.
Figure 4. 1D 1H NMR kinetic titrations of 3 binding to oligo C.
All of the NMR peaks are labeled corresponding to the single letter designation for the DNA nucleobase as it would be numbered from 5′to 3′ for oligo C. a, 1D 1H NMR spectra of 3 titration into d(GATAAGTACTTATC)2 completed by Lee et. al15 with the spectra shown for 0 and 1.0 equivalents of 3 added, respectively, showing the emergence of a new set of distinct imino proton peaks. b, 1D 1H NMR spectra of oligo C with both 0 and 1.0 equivalents of 3 added, respectively. The 1:1 association kinetic run with 1.0 eq of 3 was produced after 5.6 min at a final concentration of 60 μM 3:oligo C in 30 mM phosphate buffer (pH = 7.0), with the distinct complexed imino proton peaks indicating structural verification for complete association of the 3-oligo C complex.
As a qualitative approach to monitoring association, the gel shift assay was used with stoichiometric amounts of 3 and oligo A in the concentration range of 250 nM to 2,500 nM. These experiments were used to gain a rough estimate of an association rate constant to be compared with, and inform, the more accurate stopped-flow spectroscopic measurements described below. Using the integrated rate equation for stoichiometric binding28 (Equation (1), Supplementary Information) yielded an estimated association rate constant of 1 ± 0.5 × 104 M−1 s−1. The advantage of the gel shift approach is that we already know that non-specific association does not produce a gel shift with 3. Thus, the estimated association rate constant measured in this way is expected to represent sequence specific binding.
The rates of association between 3 and either oligo A or oligo B were also analyzed using stopped-flow methods. Hypochromism accompanies intercalation, so the time-dependent decrease at the visible absorbance maximum (386 nm) of 3 was monitored following the mixing of equal volumes of solutions containing 3 or DNA. Absorbance traces were collected for fixed concentrations of 3 mixed with increasing DNA concentrations such that the final ratios after mixing ranged from 1:1 to 1:8. An initial rapid decrease in absorbance was observed, reflecting a fast association of 3 with DNA (Fig. 5a).
Figure 5. Stopped-flow association traces of 3 from oligo B with proposed models for association of 3 to both oligo A and oligo B.
a, Representative stopped-flow absorbance traces of the association of 3 (2 μM) with oligo B at four different DNA concentrations (2, 4, 8, 16 μM) in 10 mM PIPES buffer (pH = 7.0), 1 mM EDTA, 100 mM NaCl. The solid lines represent the best global fit to a three-step consecutive mechanism using KinTek Explorer Software. A similar absorbance trace was obtained for the association of 3 to oligo A (Fig. S5, Supplementary Information). b, Rate constants were derived from global fitting the concentration dependence of the data series to a three step model. Both models assume irreversible steps toward complex formation. The data for the association of 3 with oligo B best fit the top model, while the data for the association of 3 with oligo A best fit the bottom model.
Single or two-step models did not fit the data satisfactorily, therefore kinetic traces obtained at each DNA concentration were fit globally29 to a minimal three-step irreversible mechanism (Fig. 5b). The model assumes an initial fast interaction followed by slower consecutive rearrangement steps, where [3-DNA]I and [3-DNA]II represent two different intermediate complexes.30 Although this model describes the kinetic behavior for both DNA sequences, the fit is best for the association of 3 with oligo B. In the case of 3 binding to oligo A, an additional step was required in order to better account for the fast phase, particularly at lower DNA concentration, which is a key component of the data (Fig. 5b). The additional step fits the kinetic data but the parameters are not sufficiently well constrained to uniquely define the rate constants for the two isomerization steps. Nonetheless, globally fitting data according to both mechanisms yielded well-defined second order association rate constants for the two DNA sequences, which were 3.4 ± 0.1 × 106 M−1s−1 and 2.5 ± 0.1 × 106 M−1s−1 (k1) for 3 binding to oligo A and oligo B, respectively. Note that an essentially irreversible rearrangement step is consistent with the remarkably slow off-rate of 3 from the target sequence. The rate constants shown in Fig. 5 are listed as irreversible steps only because there was no information to define the reverse rate constants, and should be considered as only net reaction rate estimates. This model can be reconciled with the apparent second order rate constant of 1 ± 0.5 × 104 M−1s−1 estimated by gel shift assays by allowing the initial binding steps to be readily reversible leading up to the final, largely irreversible step.
Discussion
Previous work with 3 had established the ability to bind the sequence 5′-GATAAGTACTTATC-3′ in a 1:1 stoichiometry with the linkers bound in a minor-major-minor groove topology (Fig. 1c).15 The DNase I footprinting results presented here have verified that 3 can select and bind this 14 bp sequence in the context of nearly 500 bp (Fig. 2). While no structural verification has been attempted, it is reasonable to assume a similar threading topology on a long piece of DNA in which the linkers alternate in the minor-major-minor groove topology as with the oligonucleotide of the same sequence.15
The observed sequence specificity could be the consequence of differences in association rates, dissociation rates or a combination of the two. Based on a 4 × 104-fold slower dissociation rate constant observed for 3 binding its preferred sequence compared to a randomly chosen sequence, it is safe to say that specificity is based largely on differences in the dissociation rates. This conclusion was reinforced by stopped-flow association data that revealed very similar association rate constants for the preferred versus control sequences.
The threading polyintercalation topology is intuitively expected to manifest an extremely slow dissociation due to the seemingly unlikely molecular rearrangements required for full dissociation. In addition, threading intercalator monomers display relatively slow dissociation compared to non-threading monointercalators.10,11 Indeed, using 100 mM NaCl as benchmark conditions, the observed kd = 5.0 ± 0.5 × 10−7 s−1 for 3 dissociating from its preferred binding sequence corresponds to a dissociation half-life of t1/2 = 16 days. We envision that multiple individual steps are required for full dissociation, so these values are best thought of as rate-limiting out of several individual rate constants.
The complex topology of a bound threading polyintercalator also raises questions about the detailed mechanism of association. One might predict a rapid and nonspecific association between positively-charged 3 and negatively-charged DNA driven largely by electrostatic attraction. Following that, a large number of distinct states could be envisioned as the molecule samples various topologies and sequences before ending up in the final threading polyintercalation binding mode. The stopped-flow measurements confirmed at least a three-step process, consistent with expectation. Perhaps surprisingly, there were significant similarities between the association rate profiles and rate constants for 3 binding to its preferred versus a random sequence, although detailed curve fitting indicated there may be some subtle mechanistic differences between the two.
Unfortunately, no detailed mechanistic interpretation is possible based on the stopped-flow data alone because spectrophotometric monitoring cannot be used to gain an unambiguous structural understanding of the process. For example, it is possible that the observed decreases in NDI absorbance are the result of a step in which the NDI units are simply buried in a DNA groove and not yet intercalated. In addition, some mechanistic features could be very hard to detect as distinct steps because one could imagine several different bound topologies with just two NDI units intercalated, all of which would have very similar overall absorbance values.
Lack of structural information in the stopped-flow experiment was addressed by using 1D 1H NMR studies and qualitative gel shift measurements. A detailed analysis of the entire spectrum and especially the imino proton signals verified that within the limits of NMR detection (greater than 95%), after the 5.6 min mixing time, 3 was fully bound to its preferred site in the expected threading polyintercalator topology as previously assigned (Fig. 4).15 Unfortunately, because the complex was fully formed before the first scan was acquired, no detailed rate information could be obtained. On the other hand, the gel shift association experiments were used to provide an estimate of the association rate constant for 3 binding to its preferred sequence in a manner that survives gel electrophoresis. Recall that the gel shift was only observed for 3 binding its preferred sequence, not a randomly chosen sequence, so this measurement is assumed to verify binding to the oligonucleotide in a sequence specific fashion. The approximate association rate constant obtained in this way was 1 ± 0.5 × 104 M−1 s−1, which is consistent with full binding under the conditions of the NMR experiment, but is still considerably slower than the slower of the two rate constants determined by stopped-flow. It is likely that the stopped-flow assay reports the initial, and readily reversible association steps, which are followed by a largely irreversible step to form the tight complex observed by NMR and the gel shift assay.
It is interesting to compare the dissociation rate constant measured here to those for other reported long-lived complexes. To the best of our knowledge, a kd = 5.0 ± 0.5 × 10−7 s−1, corresponding to a 16-day half-life, compares favorably to all reported dissociation rate constants for molecules bound to DNA. In fact, we know of only a handful of reported slower dissociation rate constants, including that for the synthetic multivalent tris-vancomycin-tris-D-Ala complex (t1/2 ≈ 200 days),31 a few highly engineered antibodies (t1/2 ≈ 5 – 26 days),32-34 and the avidin-biotin system (t1/2 ≈ 89 – 107 days).35,36
Given the lower estimate for the apparent second order rate constant governing the formation of the specific complex of 1 ± 0.5 × 104 M−1s−1 and the slow dissociation rate, we estimate a net dissocation constant (KD) of 50 pM for 3 binding its preferred site on oligo A. Note that binding to a preferred site on a long piece of DNA will almost certainly be complicated by additional mechanistic steps not seen with the relatively short oligonucleotides used here. We are currently embarking on a comprehensive mechanistic study intended to uncover the rates and individual steps involved when a threading polyintercalator such as 3 “scans” long segments of DNA, locates, and then fully intercalates into a preferred site. The results of these analyses will be reported in due course.
In summary, a roughly 4 × 104-fold preference for a specific 14 bp sequence was determined for 3 relative to a randomly chosen sequence. This specificity was almost entirely due to differences in dissociation rate constants. Importantly, a 16-day half-life at 100 mM NaCl is the slowest we know of for DNA binding molecules, and is presumably sufficiently slow to disrupt biological processes such as transcription, repair, and replication for prolonged periods. Noting that adding between 4 and 5 kcal/mol of binding energy to the dissociation rate constant reaches a bound half-life of 90 years, we will be exploiting the modular nature of our threading polyintercalators to make longer molecules with expected bound half-lives relevant to mammalian lifetimes.
Methods
Tetraintercalator (3) synthesis
The Fmoc-Lys-NDI-(β-Ala)3-Lys-NDI-Gly-resin was prepared as previously described,14,37 and thereafter following the tetraintercalator synthesis procedure of Lee et al.15 1H NMR spectra were in agreement with the previously published data,15 and HRMS-ESI predicted for C128H150N30O34[M+2H]+2: 1325.5466, found 1325.8, predicted for C128H151N30O34[M+3H]+3: 884.0336, found 884.5, and predicted for C128H152N30O34[M+4H]+4: 663.681, found 663.6. Tetraintercalator concentration was determined by the absorbance at 386 nm using an extinction coefficient of 51,300 M−1 cm−1.
DNase I footprinting
A double-stranded oligonucleotide containing the tetraintercalator binding site was inserted between the SfiI restriction enzyme sites of the pMoPac16 vector.38 The sequence of the construct was confirmed prior to use. The 467 bp DNA fragment was amplified by PCR using a primer which was 5′-32P end-labeled. The PCR product was purified on a 5% native gel, and the bands were excised and eluted in TE buffer (10 mM Tris-Cl, 1 mM EDTA, pH = 7.0). The labeled fragment was incubated for 48 h with 3 at the designated concentration followed by DNase I digestion in 20 mM phosphate buffer (pH = 7.0) and 2 mM MgCl2 for 7 min. A detailed protocol has been described elsewhere.38 To determine the footprinting sequence, the digestion was compared with an adenine specific sequencing lane.40
Gel mobility-shift assays to monitor association and dissociation
The top strand of oligo A was labeled with 32P at the 5′ end, annealed to its compliment strand, and gel purified. After excising the band from the gel, the duplex oligonucleotide was ethanol precipitated and resuspended in ddiH2O. The DNA concentration was quantified with a micro-volume spectrophotometer (Nanodrop ND100). For association experiments, a stoichiometric amount of 3 was added to a solution containing both radiolabeled (50 nM) and unlabeled oligo A such that the final DNA concentration ranged from 250 to 2500 nM in 10 mM PIPES buffer (pH = 7.0), 1 mM EDTA, and 100 mM NaCl in a final volume of 12 μL. Aliquots taken at representative times from each DNA concentration were mixed with CT DNA to a final concentration of 250 μM bp to prevent further association. Samples were then mixed with loading buffer (0.25% Bromophenol Blue; 0.25% Xylene Cyanol; 40% sucrose), and loaded on an 8% non-denaturing gel. The gel was dried, visualized digitally, and the amount of bound and unbound radiolabeled oligo A was quantified using ImageQuant 4.1 imaging software. For dissociation experiments, radiolabeled oligo A (1 μM) was incubated with 3 (1 μM) in 10 mM PIPES buffer (pH = 7.0), 1 mM EDTA, and NaCl concentrations ranging from 25 to 200 mM. Complete association was first verified by gel electrophoresis before conducting the time point dissociation experiments. Once confirmed, a 100-fold excess of non-labeled oligo A was added to the 3-oligo A complex. Aliquots taken at various time points were separated by gel electrophoresis and the bands were quantified as above.
Stopped-flow kinetics
Kinetic data was acquired using a KinTek stopped-flow instrument equipped with 2.5-cm path length optical cell. The absorbance change at 386 nm was monitored for the association and dissociation of 3 with oligo A or oligo B. Kinetic experiments of DNA association were conducted at constant 3 concentration and varying DNA oligonucleotide concentration. DNA oligos (ranging from 4 to 32 μM) in buffer containing 10 mM PIPES (pH = 7.0), 1 mM EDTA, and 100 mM NaCl were mixed with equal volumes of 4 μM 3 in the same buffer. No absorbance change occurred when 3 was mixed with buffer alone. For the dissociation experiments, pre-incubated 3-oligo B complexes in 10 mM PIPES (pH=7.0), 1 mM EDTA, and the designated NaCl concentration (50-300 mM) were rapidly mixed with a 4% SDS solution in the same buffer. Data collected at each timescale consisted of one thousand data points. Global data fitting of the averaged kinetic traces was done using the KinTek Explorer software (KinTek Corp, Austin, TX).
1D 1H NMR sample preparation and spectroscopy
Solutions were prepared in 90% H2O / 10% D2O containing 30 mM phosphate buffer (pH = 7.0) and 100 mM NaCl. One solution contained oligo C (120 μM), concentration verified by nanodrop spectroscopy (ε = 143,400 L mol−1 cm −1, MW = 8,523.6 g mol−1) and the other solution contained 3 (120 μM), as verified by UV-vis spectroscopy (ε = 51,300 M−1 cm−1).11 Control spectra were obtained for oligo C at 60 μM final concentration. The kinetic association was monitored by mixing a stoichiometric amount of 3 as prepared above directly into an NMR tube containing oligo C as prepared above, to a final 1:1 concentration of 60 μM. Experiments were performed using a Varian DirectDrive 600 MHz spectrometer. 1D NMR spectra in 90% H2O / 10% D2O were taken at 27°C using a jump-return solvent suppression method. All spectra were processed using VNMRJ (Varian, Inc.).
Supplementary Material
Acknowledgements
This work was supported by the Robert A. Welch Foundation (grant F1188 to B.L.I, F1604 to K.A.J. and departmental grant AF-0005 to M.Z.F) and the National Institutes of Health (grant GM-069647 to B.L.I). Acknowledgement is made to the Donors of the American Chemical Society Petroleum Research Fund for partial support of this research (M.Z.F). M.Z.F was also supported by the Southwestern University faculty sabbatical program. We thank Steven Sorey for his help with the 1H NMR spectra.
Footnotes
Author contributions
G.G.H., M.Z.F. and A.R.S performed the experiments. G.G.H, M.Z.F., A.R.S., K.A.J., and B.L.I. designed the experiments and analyzed the data. G.G.H., M.Z.F., and B.L.I co-wrote the paper.
Competing Financial Interests
Kenneth A Johnson is President of KinTek Corporation, which provided the KinTek stopped-flow instrument and the KinTek Explorer data fitting software.
References
- 1.Dervan PB. Design of sequence-specific DNA-binding molecules. Science. 1986;232:464–471. doi: 10.1126/science.2421408. [DOI] [PubMed] [Google Scholar]
- 2.Dervan PB. Molecular recognition of DNA by small molecules. Bioorg. Med. Chem. 2001;9:2215–2236. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 3.Nickols NG, Dervan PB. Suppression of androgen receptor-mediated gene expression by a sequence-specific DNA-binding polyamide. Proc. Natl. Acad. Sci. U.S.A. 2007;104:10418–10423. doi: 10.1073/pnas.0704217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muzikar KA, Nickols NG, Dervan PB. Repression of DNA-binding dependent glucocorticoid receptor-mediated gene expression. Proc. Natl. Acad. Sci. U.S.A. 2009;106:16598–16603. doi: 10.1073/pnas.0909192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox KR. Targeting DNA with triplexes. Curr. Med. Chem. 2000;7:17–37. doi: 10.2174/0929867003375506. [DOI] [PubMed] [Google Scholar]
- 6.Arya DP. New approaches toward recognition of nucleic acid triple helices. Acc. Chem. Res. 2010;44:134–146. doi: 10.1021/ar100113q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egholm M, Buchardt O, Nielsen PE, Berg RH. Peptide nucleic acids (PNA). Oligonucleotide analogues with an achiral peptide backbone. J. Am. Chem. Soc. 1992;114:1895–1897. [Google Scholar]
- 8.Janowski BA, Hu J, Corey DR. Silencing gene expression by targeting chromosomal DNA with antigene peptide nucleic acids and duplex RNAs. Nat. Protoc. 2006;1:436–443. doi: 10.1038/nprot.2006.64. [DOI] [PubMed] [Google Scholar]
- 9.Yen SF, Gabbay EJ, Wilson WD. Interaction of aromatic imides with deoxyribonucleic acid, spectrophotometric and viscometric studies. Biochemistry. 1982;21:2070–2076. doi: 10.1021/bi00538a014. [DOI] [PubMed] [Google Scholar]
- 10.Tanious FA, Yen SF, Wilson WD. Kinetic and equilibrium analysis of a threading intercalation mode: DNA sequence and ion effects. Biochemistry. 1991;30:1813–1819. doi: 10.1021/bi00221a013. [DOI] [PubMed] [Google Scholar]
- 11.Lokey RS, et al. A new class of polyintercalating molecules. J. Am. Chem. Soc. 1997;119:7202–7210. [Google Scholar]
- 12.Guelev VM, Harting MT, Lokey RS, Iverson BL. Altered sequence specificity identified from a library of DNA-binding small molecules. Chem. Biol. 2000;7:1–8. doi: 10.1016/s1074-5521(00)00007-x. [DOI] [PubMed] [Google Scholar]
- 13.Guelev V, et al. Peptide bis-intercalator binds DNA via threading mode with sequence specific contacts in the major groove. Chem. Biol. 2001;8:415–425. doi: 10.1016/s1074-5521(01)00013-8. [DOI] [PubMed] [Google Scholar]
- 14.Guelev V, Sorey S, Hoffman DW, Iverson BL. Changing DNA grooves – a 1,4,5,8-naphthalene tetracarboxylic diimide bis-intercalator with the linker (β-Ala)3-Lys in the minor groove. J. Am. Chem. Soc. 2002;124:2864–2865. doi: 10.1021/ja016834e. [DOI] [PubMed] [Google Scholar]
- 15.Lee J, Guelev V, Sorey S, Hoffman DW, Iverson BL. NMR structural analysis of a modular threading tetraintercalator bound to DNA. J. Am. Chem. Soc. 2004;126:14036–14042. doi: 10.1021/ja046335o. [DOI] [PubMed] [Google Scholar]
- 16.Chu Y, Sorey S, Hoffman DW, Iverson BL. Structural characterization of a rigidified threading bisintercalator. J. Am. Chem. Soc. 2007;129:1304–1311. doi: 10.1021/ja066480x. [DOI] [PubMed] [Google Scholar]
- 17.Chu Y, Hoffman DW, Iverson BL. A pseudocatenane structure formed between DNA and a cyclic bisintercalator. J. Am. Chem. Soc. 2009;131:3499–3508. doi: 10.1021/ja805676w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaires JB, Dattagupta N, Crothers DM. Kinetics of the daunomycin-DNA interaction. Biochemistry. 1985;24:260–267. doi: 10.1021/bi00323a004. [DOI] [PubMed] [Google Scholar]
- 19.Wilson WD, et al. DNA sequence dependent binding modes of 4′,6-diamidino-2-phenylindole (DAPI) Biochemistry. 1990;29:8452–8461. doi: 10.1021/bi00488a036. [DOI] [PubMed] [Google Scholar]
- 20.Tanious FA, Veal JM, Buczak H, Ratmeyer LS, Wilson WD. DAPI (4′,6-diamidino-2-phenylindole) binds differently to DNA and RNA: minor-groove binding at AT sites and intercalation at AU sites. Biochemistry. 1992;31:3103–3112. doi: 10.1021/bi00127a010. [DOI] [PubMed] [Google Scholar]
- 21.Wilson WD, Krishnamoorthy CR, Wang YH, Smith JC. Mechanism of intercalation: ion effects on the equilibrium and kinetic constants for the interaction of propidium and ethidium with DNA. Biopolymers. 1985;24:1941–1961. doi: 10.1002/bip.360241008. [DOI] [PubMed] [Google Scholar]
- 22.Westerlund F, Wilhelmsson LM, Nordén B, Lincoln P. Monitoring the DNA binding kinetics of a binuclear ruthenium complex by energy transfer: evidence for slow shuffling. J. Phys. Chem. B. 2005;109:21140–21144. doi: 10.1021/jp0534838. [DOI] [PubMed] [Google Scholar]
- 23.Westerlund F, Nordell P, Nordén B, Lincoln P. Kinetic characterization of an extremely slow DNA binding equilibrium. J. Phys. Chem. B. 2007;111:9132–9137. doi: 10.1021/jp072126p. [DOI] [PubMed] [Google Scholar]
- 24.Leng F, Priebe W, Chaires JB. Ultratight DNA binding of a new bisintercalating anthracycline antibiotic. Biochemistry. 1998;37:1743–1753. doi: 10.1021/bi9720742. [DOI] [PubMed] [Google Scholar]
- 25.Hampshire AJ, Rusling DA, Broughton-Head VJ, Fox KR. Footprinting: A method for determining sequence selectivity, affinity and kinetics of DNA-binding ligands. Methods. 2007;42:128–140. doi: 10.1016/j.ymeth.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Müller W, Crothers DM. Studies of the binding of actinomycin and related compounds to DNA. J. Mol. Biol. 1968;35:251–290. doi: 10.1016/s0022-2836(68)80024-5. [DOI] [PubMed] [Google Scholar]
- 27.Westerlund F, Wilhelmsson LM, Nordén B, Lincoln P. Micelle-sequestered dissociation of cationic DNA-intercalated drugs: unexpected surfactant-induced rate enhancement. J. Am. Chem. Soc. 2003;125:3773–3779. doi: 10.1021/ja029243c. [DOI] [PubMed] [Google Scholar]
- 28.Anslyn EV, Dougherty DA. Modern Physical Organic Chemistry. University Science Books; 2006. [Google Scholar]
- 29.Johnson KA, Simpson ZB, Blom T. Global Kinetic Explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal. Biochem. 2009;387:20–29. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 30.Bevilacqua PC, Kierzek R, Johnson KA, Turner DH. Dynamics of ribozyme binding of substrate revealed by fluorescence-detected stopped-flow methods. Science. 1992;258:1355–1358. doi: 10.1126/science.1455230. [DOI] [PubMed] [Google Scholar]
- 31.Rao J, Lahiri J, Isaacs L, Weis RM, Whitesides GM. A trivalent system from vancomycin-D-Ala-D-Ala with higher affinity than avidin-biotin. Science. 1998;280:708–711. doi: 10.1126/science.280.5364.708. [DOI] [PubMed] [Google Scholar]
- 32.Boder ET, Midelfort KS, Wittrup KD. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10701–10705. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graff CP, Chester K, Begent R, Wittrup KD. Directed evolution of an anti-carcinoembryonic antigen scFv with a 4-day monovalent dissociation half-time at 37°C. Protein Eng. Des. Sel. 2004;17:293–304. doi: 10.1093/protein/gzh038. [DOI] [PubMed] [Google Scholar]
- 34.Rajpal A, et al. A general method for greatly improving the affinity of antibodies by using combinatorial libraries. Proc. Natl. Ac. Sci. U.S.A. 2005;102:8466–8471. doi: 10.1073/pnas.0503543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Green NM. Avidin: 1-The use of [14C]biotin for kinetic studies and for assay. Biochem. J. 1963;89:585–591. doi: 10.1042/bj0890585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piran U, Riordan WJ. Dissociation rate constant of the biotin-streptavidin complex. J. Immunol. Meth. 1990;133:141–143. doi: 10.1016/0022-1759(90)90328-s. [DOI] [PubMed] [Google Scholar]
- 37.Guelev VM, Cubberley MS, Murr MM, Lokey RS, Iverson BL. Design, synthesis, and characterization of polyintercalating ligands. Methods Enzymol. 2001;340:556–570. doi: 10.1016/s0076-6879(01)40442-3. [DOI] [PubMed] [Google Scholar]
- 38.Hayhurst A, et al. Isolation and expression of recombinant antibody fragments to the biological warfare pathogen Brucella melitensis. J. Immunol. Meth. 2003;276:185–196. doi: 10.1016/s0022-1759(03)00100-5. [DOI] [PubMed] [Google Scholar]
- 39.Trauger JW, Dervan PB. Footprinting methods for analysis of pyrrole-imidazole polyamide/DNA complexes. Methods Enzymol. 2001;340:450–466. doi: 10.1016/s0076-6879(01)40436-8. [DOI] [PubMed] [Google Scholar]
- 40.Iverson BL, Dervan PB. Adenine specific DNA chemical sequencing reaction. Nucleic Acids Res. 1987;15:7823–7830. doi: 10.1093/nar/15.19.7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.