Abstract
Although unnatural amino acids (Uaas) have been genetically encoded in bacterial, fungal and mammalian cells using orthogonal tRNA/aminoacyl-tRNA synthetase pairs, applications of this method to a wider range of specialized cell types, such as stem cells, still face challenges. While relatively straightforward in stem cells, transient expression lacks sufficient temporal resolution to afford reasonable levels of Uaa incorporation and to allow for the study of the longer term differentiation process of stem cells. Moreover, Uaa incorporation may perturb differentiation. Here we describe a lentiviral-based gene delivery method to stably incorporate Uaas into proteins expressed in neural stem cells, specifically HCN-A94 cells. The transduced cells differentiated into neural progenies in the same manner as the wild type cells. By genetically incorporating a fluorescent Uaa into a voltage-dependent membrane lipid phosphatase, we show that this Uaa optically reports the conformational change of the voltage-sensitive domain in response to membrane depolarization. The method described here should be generally applicable to other stem cells and membrane proteins.
Keywords: Neural stem cells, unnatural amino acids, voltage sensing, fluorescence imaging
INTRODUCTION
Unnatural amino acids (Uaas) have been genetically incorporated into proteins in bacterial, fungal, and mammalian cells.[1, 2] This process requires an orthogonal tRNA/aminoacyl-tRNA synthetase pair that is specific for the Uaa, decodes a unique codon such as the amber stop codon UAG, does not crosstalk with endogenous tRNA/synthetase pairs of the host, and functionally couples with the host translation machinery. In bacteria and yeast, these orthogonal tRNA/synthetase pairs are encoded on plasmids that can be stably maintained in parallel with the host chromosome(s).[1, 3, 4] In mammalian cells, genes for the orthogonal tRNA/synthetase are most often transiently transfected into cells.[5–7] By changing the side chain and even the α-amino group of Uaas, a variety of functional groups with non-biologically-based chemical and physical properties have been site-specifically introduced into proteins in living cells. These genetically encoded Uaas possessing new chemical modalities are increasingly exploited to study protein structure and function, to identify protein interactions, to regulate protein activities, and to generate new protein functions.[2]
Stem cells have the unique property of self-renewal and the potential to differentiate into distinct cell lineages with the proper cues; thus they hold great potential for the treatment of various diseases.[8, 9] Signaling networks integrate cell-intrinsic factors with cell-extrinsic signals from stem cell niches to modulate the self-renewal and pluripotency of stem cells. For fundamental studies of stem cell biology and for translational therapeutic applications, it is essential to understand the temporal and spatial patterns underlying these networks at the molecular level. The ability to genetically incorporate Uaas in stem cell proteins would accelerate our understanding of these regulatory networks by affording greater chemical precision when interrogating protein function in stem cells.
In addition, stem cells can be directed into terminally differentiated non-dividing cells such as neurons. In contrast, mature cells lack cell division capabilities and are traditionally isolated from living tissues, making it difficult or expensive to acquire such cells in large amounts. It is also difficult to deliver exogenous genes into mature cells stably and with high efficiency. Therefore, stem cell lines stably incorporating Uaas are an attractive source of mature cell lines capable of genetically incorporating Uaas, thereby obviating the difficulties associated with gene delivery and procurement by traditional routes. However promising, the current method for genetically incorporating Uaas in mammalian cells is not suitable for stem cells given their low transient transfection efficiency.[5–7] More importantly, the differentiation of stem cells takes days to weeks to complete, while the transiently transfected orthogonal tRNA/synthetase pair is lost early on during this long-term process. Moreover, the differentiation of stem cells is accompanied by epigenetic changes, which can suppress and/or activate transient gene expression.[10] For these reasons, it is necessary to stably integrate the orthogonal tRNA/synthetase genes into the chromosome of stem cells in order to fully exploit the Uaas for the study the entire differentiation program.
Here we present a general method to genetically incorporate Uaas in the neural stem cell line, HCN-A94, and demonstrate that a genetically encoded fluorescent Uaa optically reports the membrane depolarization of neurons differentiated from HCN-A94 cells. In brief, lentiviral vectors deliver genes for the orthogonal tRNA/synthetase pair and the target protein efficientlyand stably into HCN-A94 cells. The resultant cell line incorporates Uaas into a green fluorescent protein reporter throughout the differentiation process. Using this strategy, we also designed and deployed a fluorescent Uaa into the voltage-sensitive domain (VSD) of Ciona intestinalis voltage-sensitive phosphatase (CiVSP).[11] Upon differentiation, the fluorescent Uaa in CiVSP provided position-dependent fluorescence changes upon membrane depolarization.
MATERIALS AND METHODS
Cell Culture and Adult Neural Stem Cell Differentiation
HeLa and HEK293T were cultured with Dulbecco’s modified Eagle’s medium (DMEM, Mediatech, Manassas, VA, USA) supplemented with 10% FBS (Mediatech). HCN-A94 derived from dentate gyrus of adult rat brain was cultured as previously reported.[12] Briefly, DMEM/F12 (high glucose) medium containing 1 mM L-glutamine (Irvine Scientific, Santa Ana, CA, USA), 1% N2 supplement (Gibco) and 20 ng/mL FGF-2 was used. FGF-2 was freshly added to the medium before usage. The growth medium was changed every 2 days. When cell confluences reached 90%, TrypLE (0.05%, Invitrogen) was applied to cells at room temperature (25 °C) for 2 min. TrypLE was carefully aspirized and cells were dislodged by gently slapping the culture dishes. DMEM/F12 medium without FGF-2 was used to rinse the dishes and re-suspend the cells. Cells suspensions were centrifuged at 1,200 g for 2 min. The cell pellet was re-suspended in DMEM/F12 medium containing FGF-2 and plated onto plates pre-treated with poly-L-ornithine and freshly coated with Laminin.
The HCN-A94 cells were differentiated into the neural lineages by adding 1 μM retinoic acid and 5 μM forskolin and withdrawing FGF-2. The differentiated cells were fed every 2–3 days. The whole process lasted for 8 days after the initiation of differentiation.
Transfection and Virus Preparation
Polyethylenimine (Polysciences, PA, USA) was used to transfect HEK293T cells. For the lentivirus packaging, 8×106 cells were plated on a 150-mm plate the day before the transfection. DNA for the LV vectors (12.2 μg), MDL (8.1 μg), VSVG (4.1 μg) and REV (3.1 μg) were evenlymixed and dissolved in 1 mL Optimal MEM (Invitrogen). Polyethylenimine (110 μL) was added subsequently. The mixture was vortexed gently. After 5 minof incubation at room temperature, the transfectant was evenly dropped onto the cells. After 5 hr of post transfection, the medium was exchanged for fresh medium. Lentiviruses were collected 48 hr post transfection. For the preparation of concentrated lentiviruses, the above procedure was scaled up to 10 plates.
Virus-containing media were centrifuged at 500 g for 2 min to remove cell debris and then filtered through 0.22 μm filter (Millipore). The filtered virus-containing media were ultracentrifuged at 19,400 rpm for 2 hr (4 °C). The virus-containing white pellet was re-suspended with phosphate-buffered saline (PBS containing Ca2+ and Mg2+, Mediatech) and transferred into new ultracentrifuge tubes for a second spin (19,400 rpm, 2 hr at 4 °C). The final pellet was dissolved in PBS (containing Ca2+ and Mg2+) by gentle vortexing. The concentrated viruses were stored at −80 °C in aliquots.
Lentiviral vector stocks were normalized by HIV-1 p24 antigen content. The p24 antigen content of vector particles was quantified with a commercial HIV-1 p24 enzyme-linked immunosorbent assay kit (PerkinElmer, Boston, USA). For our vector preparations, a p24 concentration of 106–107 pg of p24 per milliliter is routinely obtained. Functional titers were estimated around 109 transducing units (TU)/mL by comparing with the positive control of EGFP expression, which was measured by fluorescence-activated cell-sorting (FACSCalibur, Becton Dickinson) analysis with limiting dilution in HEK293T cells.
Sensing Current and Q-V Curve
Sensing currents were measured on HEK293T cells 48 hr after transient transfection with plasmids expressing VSD-mKate and mutants. Currents were recorded at 25 °C with whole cell recording[13] using MultiClamp700B amplifier (Molecular Devices, Sunnyvale, California). Currents were filtered at 1 kHz and sampled at 10 kHz with Digidata 1440 (MDS Analytical Technologies), which was controlled using pClamp10 software (MDS AnalyticalTechnologies). The components of extracellular solution were: 135 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 5 mM HEPES, 10 mM glucose, 20 mM sucrose, pH 7.4 with NaOH. The intracellular solution contained 145 mM CsCl, 5 mM NaCl, 5.46 mM MgCl2, 10 mM HEPES, 5 mM EGTA, pH7.3 with CsOH. Glass pipettes (2–5 Mohm) were pulled from borosilicate glass using P-97 (Sutter Instrument, CA). For Q-V curve measurement, cell membrane was held stead at −60 mV and a series of pulses with voltage ranging from −120 mV to 80 mV in 10-mV increments were applied sequentially. Offline analysis was applied to subtract the leak components during the recording. The charge (Q) was calculated as the time integral of the sensing currents by excluding the capacitive transient, which charged the cell membrane and was estimated by a single exponential fit to the current traces at the hyperpolarizing steps. Data analysis and curve fitting were performed with Origin 7 (OriginLab) and pClampfit10.
Fluorescence Microscopy
An Olympus inverted microscope (IX81) equipped with an EM-CCD from Hamamatsu Photonics (Japan) was used for imaging. The Plan-Apochromat 63x objective with N.A. 1.40 was used for the time-lapse imaging of DanAla fluorescence. For DanAla imaging, the excitation filter was 357/44 nm; the emission filter was 535/40 nm; and the dichroic mirror was 409 nm long pass. For mKate imaging, the excitation filter was 580/20 nm, the emission filter was 675/130 nm, and the dichroic mirror was 600 nm long pass. Slidebook5 was used for image acquisition and data analysis. For high potassium shock, the extracellular K+ concentration was raised to 150 mM through perfusion by using an ALA pressure system (Farmingdale, NY, USA). Cells were imaged continuously in the DanAla fluorescence channel during the depolarization process. DanAla fluorescence intensity at the cell membrane was determined from the images and plotted in time sequence. Cells in the same imaging field without DanAla incorporation into VSP-mKate were used as controls to monitor any background fluorescence change due to nonspecific adsoption of DanAla.
RESULTS
Development of lentiviral vectors for genetic incorporation of Uaas in HCN-A94 cells
Neurogenesis in mammals occurs in the sub-ventricular and sub-granular zones of the adult brain and proceeds throughout the lifetime of the organism.[14] Newly differentiated neurons from adult stem/progenitor cells functionally integrate into these regions of the brain and participate in brain circuits controlling measurable organismal activities. The adult rat hippocampal neural stem cell line HCN-A94 can be transplanted in vivo and undergoes differentiation into neurons.[12] Furthermore, formation of observable synaptic connections and the measurement of membrane discharge indicate that these neurons are functional.
We employed lentiviral vectors to deliver and stably integrate genes encoding orthogonal tRNA/synthetase pairs and proteins of interest in HCN-A94 stem cells affording long term expression of transgenes. One challenge facing the incorporation of Uaas in these and many eukaryotic cells is the efficient expression ofa functional orthogonal tRNA, the latter often lacking the conserved A-box and B-box sequences required for eukaryotic polymerase III (Pol III) recognition and tRNA transcription.[2] We previously demonstrated that type-3 or post-transcriptionally cleavable Pol III promoters, such as the H1, U6 and SNR52 promoters, can be used to drive the expression of orthogonal prokaryotic tRNAs in mammalian cells[7] and yeast,[4] regardless of whether the tRNA possesses the A-box or B-box sequences. Here, we specifically adopt the use of the H1 promoter for tRNA expression in HCN-A94 cells. The tyrosyl amber suppressor tRNA ( ) and the tyrosyl-tRNA synthetase (TyrRS) from E. coli behave CUA in an orthogonal manner in mammalian cells[7] and were used here in HCN-A94 cells. The gene for Aequorea victoria green fluorescent protein (GFP) with a UAG stop codon at a permissive site (Tyr182)[7] was used to quantify amino acid incorporation by the orthogonal pair in response to the presence of its UAG codon.
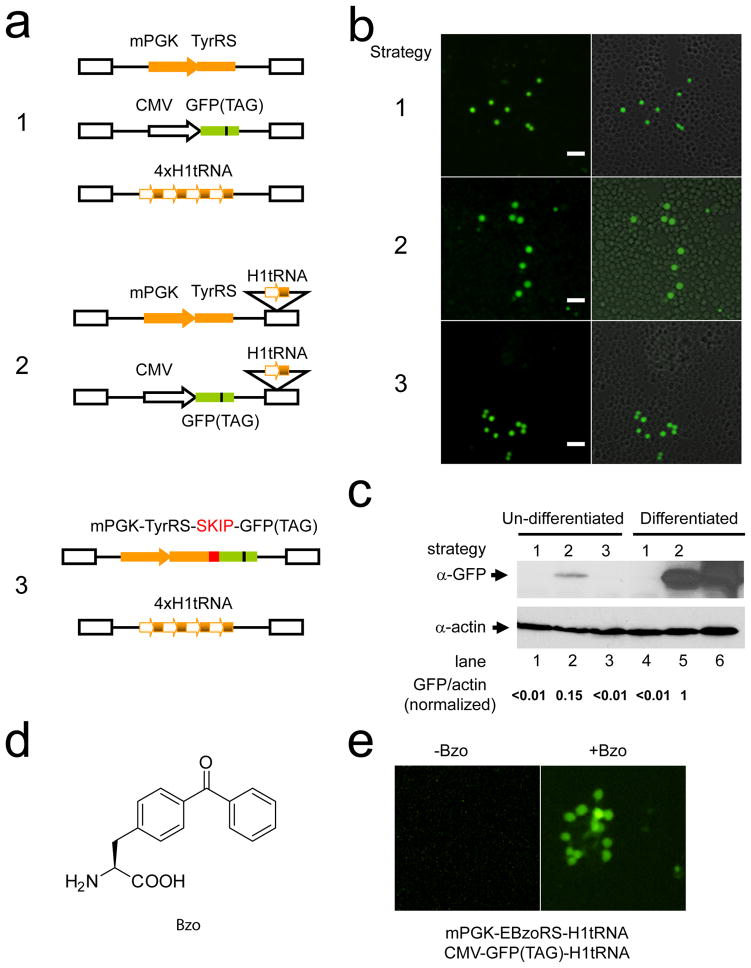
Three strategies were employed to express the , TyrRS and GFP(TAG) genes (Fig. 1a). In strategy 1, the TyrRS and the GFP(TAG) reporter were expressed using two separate lentiviral vectors. The H1 promoter driven expression cassette ( ) was inserted at the regular coding region of the lentiviral vector and repeated 4-times in tandem to increase tRNA expression levels. In strategy 2, the cassette was inserted into the 3′-LTR instead of the regular coding region of the lentiviral vector. During reverse transcription, the LTR is duplicated, and after integration the target cell receives two transcription units of the cassette from each lentival vector. The TyrRS and GFP(TAG) were expressed separately using two lentiviral vectors. Therefore, compared with strategy 1, the number of lentiviruses was reduced from 3 to 2 without decreasing the effective number of cassettes. In strategy 3, the cassette was expressed as 4 tandem repeats in the coding region, as in strategy 1, but the TyrRS and GFP(TAG) genes were combined in one lentiviral vector using the SKIP linker;[15] the latter linker enables two proteins to be expressed from one open reading frame.
Figure 1.
Lentiviral strategy for the genetic incorporation of unnatural amino acids (Uaas) into proteins in adult rat neural stem cells (HCN-A94 cells). (a) Diagram depicting three strategies for expressing the orthogonal tRNA/synthetase pair and the reporter gene GFP(TAG). AUAG amber stop codon was introduced at a permissive site, Tyr182, in the GFP gene. Strategy 1: mPGK promoter for TyrRS, CMV promoter for GFP(TAG), and H1 promoter for tRNA expression in three separate lentiviral vectors. The H1-tRNA cassette was repeated 4-times in tandem in the coding region. Strategy 2: Promoters for TyrRS and GFP(TAG) were identical to those used in Strategy 1; a single copy of H1-tRNA was inserted into the unique Nhe I site of the 3′LTR of lentiviral vectors expressing TyrRS and GFP(TAG). Strategy 3: the mPGK promoter was used for driving the transcription of TyrRS and GFP(TAG), which were connected by the SKIP peptide linker for equivalent translation. Four repeats of the H1-tRNA cassette were encoded in the other lentiviral vector. (b) Incorporation of Tyr at the TAG position in GFP by using the orthogonal E. coli pair in HCN-A94 cells. GFP fluorescence images are shown to the left and overlaid with DIC images to the right. Scale bar, 20 μm. (c) Western blot analysis of GFP expression in HCN-A94 cells before and after differentiation using different lentiviral strategies. Differentiation was initiated 24 hr after lentiviral infection. All cell samples were grown for 7 days post infection and then lysed for Western analyses. Quantification of blot intensity (GFP/actin) is indicated at the bottom. Lane 6 is wild type GFP expressed by a lentiviral vector as a positive control. (d) Structure of the Uaa p-benzoylphenylalanine (Bzo). (e) Incorporation of the Uaa Bzo into GFP in HCN-A94 cells. Bzo-specific and GFP(TAG) reporter genes were delivered by using strategy 2 into HCN-A94 cells. GFP fluorescence was detected in cells only when Bzo was added in the growth media.
Lentiviruses were prepared from the above vectors and used to co-infect HCN-A94 cells. As shown in Fig. 1b, green fluorescence was detected in cells 72 hours post infection, indicating that the orthogonal and GFP(TAG) genes were functionally expressed and incorporated Tyr into GFP using any of the three strategies. As a control, infection of HCN-A94 cells with any single lentivirus alone did not yield green fluorescence. Western blot detection of GFP expression showed that strategy 2 yielded the largest amount of GFP protein (Fig. 1c, lanes 1–3), suggesting that it is the most efficient of the 3 strategies tested.
To determine if a Uaa could be incorporated into GFP in HCN-A94 cells, we replaced the TyrRS gene with a mutant synthetase gene, EBzoRS, in the lentivector employed in strategy 2. EBzoRS encodes a synthetase evolved from the E. coli TyrRS to be specific for the Uaa p-benzoylphenylalanine (Bzo, Fig. 1d).[16] HCN-A94 cells were co-infected with the two lentiviruses and grown in the presence and absence of 1 mM of Bzo. After 72 hr, bright green fluorescence was detected in cells with Bzo added to the growth media (Fig. 1e), indicating that Bzo was incorporated into GFP in HCN-A94 cells. Compared to Tyr incorporation by wild-type TyrRS, the incorporation efficiency of Bzo in HCN-A94 cells was 27% (±3%) as determined by flow cytometry.[7]
Uaa incorporation and HCN-A94 cell differentiation
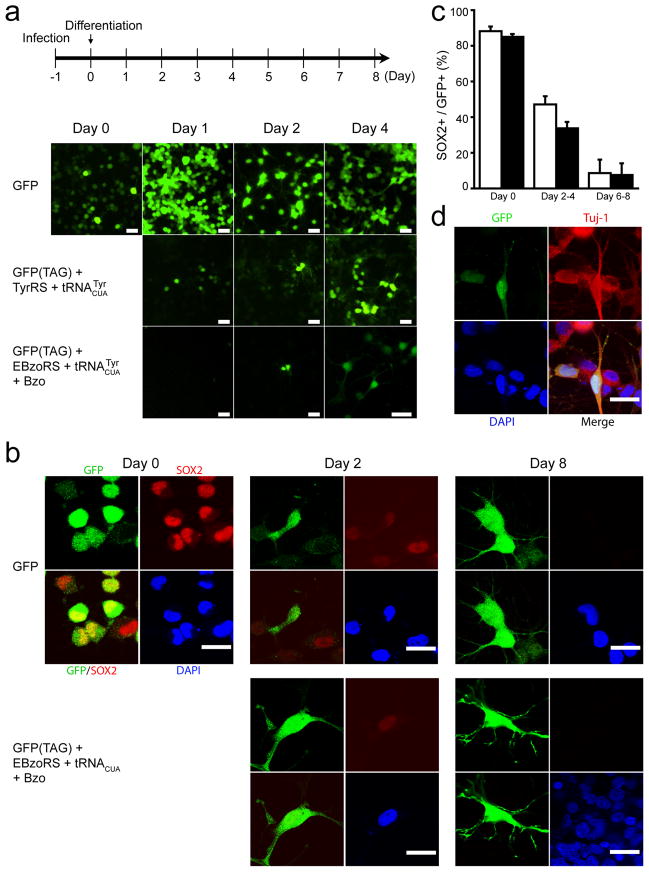
Genetically encoded Uaas were first tested to see if the expression of the orthogonal tRNA/synthetase and the incorporation of the Uaa would negatively affect the differentiation process of HCN-A94 cells. As a positive control, HCN-A94 cells were infected with a lentivirus encoding GFP and GFP expression and neural morphogenesis were monitored during cellular differentiation over 6 days (Fig. 2a). HCN-A94 cells began to express GFP at day 0 post-infection and retained their normal round shape. Differentiation was induced 24 hr post-infection by replacing FGF-2 with retinoic acid and forskolin. Neurites appeared after 48 hr and remained albeit with varying lengths through day 8.
Figure 2.
Tyr and Bzo incorporation does not affect the differentiation program of HCN-A94 cells into neuron-like progenies. (a) Time sequences of lentiviral infection and neural differentiation are shown at the top. Fluorescence images of cells were recorded on different days to monitor the differentiation process. Imaging parameters were the same for all samples, and images are displayed using the same intensity scale. Scale bar, 20 μm. Transgenic genes delivered by lentiviral vectors into HCN-A94 cells are indicated to the left. The orthogonal tRNA/synthetases pairs were delivered using strategy 2 as described. (b) Immunocytochemical analysis of HCN-A94 cells expressing GFP and cells expressing GFP(TAG) together with the Bzo-specific . An antibody against the stem cell-specific marker SOX2 was used. Scale bar, 10 μm. (c) Percentage of SOX2 postive cells relative to GFP positive cells was similar between HCN-A94 cells expressing GFP (white bars) and HCN-A94 cells expressing GFP with Bzo incorporation by the (dark bars). Error bars represent s.d. (d) Immunocytochemistry confirmed the neural differentiation of HCN-A94 cells when Bzo was incorporated into GFP. An antibody against the neural marker Tuj-1 was applied to HCN-A94 cells expressing GFP(TAG) and the in the presence of Bzo on day 6 after differentiation. Scale bar, 10 μm.
When HCN-A94 cells were co-infected with two lentiviruses, one encoding the pair and one encoding the GFPgene now containing an amber UAG codon, GFP CUA (TAG), using strategy 2 as described above, green fluorescence appeared between day 0 and day 1 post-infection, indicating that the orthogonal pair led to the incorporation of Tyr into GFP. On day 1, green fluorescent HCN-A94 cells were round in shape, indicative of their un-differentiated state. After induction of differentiation as described for control cells, neurites emerged in green fluorescent cells on day 2 and maintained morphology and green fluorescence for the 6 days of the experiment (Fig. 2a). Relative to control HCN-A94 cells infected with a non-amber codon containing GFP gene, GFP (TAG) cells did not show a delay of differentiation or morphological differences after differentiation. Notably, the differentiation of HCN-A94 cells infected with the pair and the GFP (TAG) lentiviral vectors appeared to promote the incorporation of Tyr into GFP, as the number of green cells and their fluorescence intensity increased during the progression of differentiation (Fig. 2a). To confirm this observation, differentiated cells were analyzed on day 6 for GFP expression, and they indeed showed an 85% increase in GFP expression in comparison to the un-differentiated HCN-A94 cells based upon the GFP band intensity normalized to α-actin on Western blot (lanes 5 and 2, Fig. 1c).
We then used the Uaa, Bzo, for incorporation into GFP in HCN-A94 cells by co-infecting 2 lentiviruses harboring the and GFP(TAG) genes. Green fluorescence began to appear on day 1 post-infection and post-Bzo addition. As seen before in control cells as well as GFP(TAG) cells with Tyr incorporated, long neurites emerged from green fluorescent cells on day 2 and both remained for the duration of the experiment (Fig. 2a).
To monitor the differentiation of HCN-A94 cells, an antibody against the neural stem cell marker SOX2 was used to immunostain HCN-A94 cells expressing GFP or GFP(TAG) together with the (Fig. 2b). SOX2 expression decreased as the differentiation progressed in both HCN-A94 cells expressing GFP and cells expressing GFP with Bzo incorporated (Fig. 2c). To confirm the neural identity of differentiated HCN-A94 cells, an antibody against the neural marker Tuj-1 was used for immunocytochemical analysis on day 6 (Fig. 2d); the GFP-expressing cells with Bzo incorporated were specifically marked by these antibodies. In addition, doublecortin, a microtubule-associated phosphoprotein expressed by neuronal precursor cells and immature neurons, was detected on day 8 after differentiation in both HCN-A94 control cells and HCN-A94 cells expressing GFP(TAG) together with the for Bzo incorporation (Supporting Information Fig. S1). Collectively, these results suggested that the expression of the orthogonal tRNA/synthetase pair and the incorporation of the natural amino acid Tyr or the Uaa Bzo in response to the UAG codon occurred throughout the differentiation process, and their expression did not significantly alter the differentiation process of HCN-A94 cells.
Detection of VSD movement with a fluorescent Uaa in neurons differentiated from HCN-A94 cells
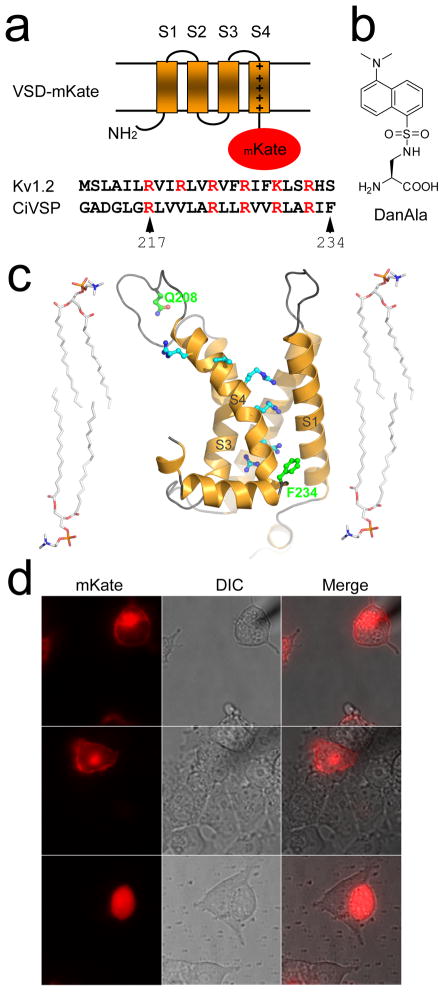
To demonstrate the utility of genetically encoded Uaas in HCN-A94 differentiated cells, we then set out to incorporate a fluorescent Uaa into a VSD and determine if the Uaa would then optically report on the VSD conformational change in response to membrane polarization in differentiated neurons. The Uaa 2-amino-3-(5-(dimethylamino) napththalene-1-sulfonamido) propanoic acid (abbreviated as DanAla, Fig. 3b) was chosen as an environmentally sensitive fluorescent reporter as the dansyl fluorophore is sensitive to environmental polarity;[17] its fluorescence intensity increases when the polarity (solvent accessibility) of the microenvironment decreases. The VSD target protein encompassed the N-terminal domain of CiVSP. The phosphatase activity of CiVSP changes in response to membrane potential through its N-terminal VSD. This N-terminal VSD is highly homologous to that of voltage-gated channels.[11] Importantly, CiVSP represents the first discovered member of voltage-dependent proteins that is not an ion channel. Sequence alignment of the VSDs of ion channels and CiVSP delineates CiVSP’s 4 transmembrane segments S1 through S4 encompassing the VSD. S4 contains the canonical basic residues that sense the membrane potential change (Fig. 3a). Moreover, unlike voltage-gated channels that operate as tetrameric assemblies, CiVSP functions as a monomer,[18] and thus serves as an tractable model to study voltage sensing in S4-based voltage-dependent proteins without potential complications due to heteromeric assembly of oligomers in the membrane.
Figure 3.
Incorporation of Uaa DanAla into the voltage-sensitive domain (VSD) of CiVSP. (a) Cartoon depicting the topology of the VSD of the membrane protein CiVSP. The C-terminal phosphatase domain of CiVSP was replaced by a red fluorescent protein mKate. Sequence alignment of the S4 residues of CiVSP and Kv1.2 shows the conserved voltage sensitive basic residues, which are represented by + signs along S4 in the cartoon. (b) Structure of Uaa DanAla. (c) Three-dimensional model of the VSD of CiVSP generated using the rat Shaker potassium channel Kv1.2 as a structural template. Side chains corresponding to the six voltage-sensing basic residues in S4 are highlighted in cyan. Phe234 and Gln208 are highlighted by green coloring. Four lipid molecules are shown to indicate the likely membrane embedded region of the VSD. (d) Fluorescence images of HEK293 cells expressing VSD-mKate (top), VSD-mKate(F234TAG) and with DanAla in the growth media (middle), VSD-mKate(F234TAG) and without DanAla (bottom). In the absence of DanAla, the VSD-mKate(F234TAG) gene expressed a truncated form localizing throughout the cell. When DanAla was added, full-length VSD-mKate(F234TAG) was expressed and correctly localized to the membrane as for the wild type VSD-mKate.
To identify the most likely site to incorporate a Uaa sensitive to S4 conformational changes, we first aligned the amino acid-sequences of the S1 through S4 segments of CiVSP with the rat Shaker potassium channel Kv1.2 using protein threading.[19] On the basis of this alignment, a homology model of the three-dimensional structure of the VSD domain of CiVSP was generated using the refined Kv1.2 structure (PDB entry 3LUT)[20] as a guide (Fig. 3c). The membrane bilayer-embedded region of this VSD model was estimated by comparison against the lipid-interacting regions observed in the structure of a Kv1.2-Kv2.1 chimeric channel (PDB entry 2R9R).[21] First, the R217Q mutation, known to shift the VSD voltage dependency closer to the physiological membrane potential, was introduced into the VSD.[22] Next, the C-terminal phosphatase domain of CiVSP was replaced with a red fluorescent protein mKate to monitor protein expression. Finally, Phe234 of the VSD of CiVSP was chosen for DanAla incorporation using an amber UAG codon. Residue 234 was chosen because it sits after the last basic residue of the canonical voltage sensor segment S4 and near the cytoplasmic side of the membrane bilayer (Fig. 3a, b).
Incorporation of DanAla into VSD-mKate by the orthogonal pair was first tested in HEK293 cells using transient transfection. As a positive control, VSD-mKate without the UAG amber codon was used. Red fluorescence of mKate indicated that most of the fusion protein localized to the membrane bilayer (Fig. 3d, top). When VSD-mKate(F234TAG) was co-expressed with the orthogonal but lacking exogenous DanAla Uaa in the growth medium, red fluorescence was observed throughout the cell without clear membrane localization (Fig. 3d, bottom). Cytoplasmic localization is consistent with the internal initiation of protein translation after the F234TAG codon, resulting in the expression of mKate fused with a translationally truncated VSD lacking any polypeptide segment capable of targeting to the cell membrane. In contrast, when DanAla was added to the growth medium, red fluorescence localized on the cell membrane (Fig. 3d, middle) as observed in the VSD-mKate control consistent with resumption of proper translation of the full-length VSD-mKate fusion protein. The incorporation efficiency of DanAla by the pair was 38(±4)% relative to Leu incorporation by the pair.
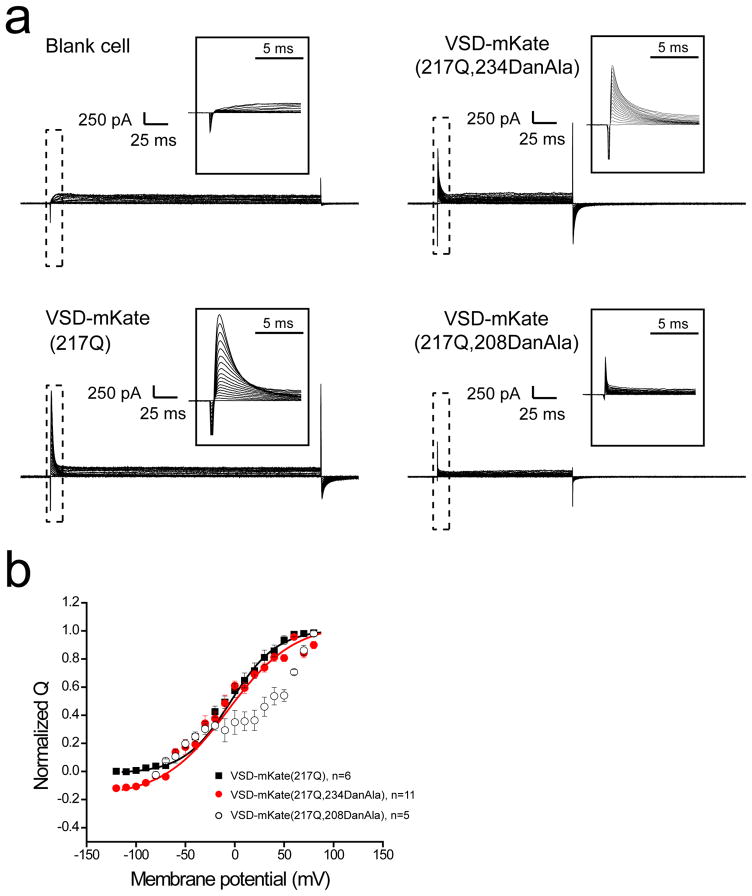
Whole-cell patch clamping of transfected cells was applied to record voltage-dependent sensing current, a characteristic of functional VSDs.[23] VSD-mKate expressing cells exhibited sensing currents similar to CiVSP (Fig. 4a), indicating that the VSD functions normally in this artificial fusion protein.[23, 24] Cells expressing the VSD-mKate(F234TAG) and pair also exhibited sensing currents but only in the presence of exogenous addition of the DanAla Uaa, confirming DanAla incorporation into a functional VSD. The sensing current traces were integrated to calculate the transported charge at different membrane potentials (the Q-V curve, Fig. 4b). There was no significant Q-V shift between the VSD-mKate and VSD-mKate(F234DanAla) mutant, indicating that the incorporation of DanAla at position 234 did not alter the VSD response to membrane potential changes.
Figure 4.
(a) Sensing current traces recorded from HEK293T cells expressing VSD-mKate and its DanAla mutants. The membrane was clamped at −60 mV and a series of voltage pulses with voltages ranging from −60 mV to 110 mV in 10-mV increments were applied sequentially over a period of 2 seconds for each pulse. Online compensation for cell capacitance was applied during recording of these traces. The inserts highlight the time scale in expanded format. (b) Sensing charge as a function of membrane potential (Q-V curve) for VSD-mKate and its DanAla-containing mutants. Solid lines are single Boltzmann fits.
A second site was selected for DanAla incorporation, namely Gln208 of CiVSP. Gln208 resides on the S3–S4 loop at the opposite end of S4 relative to Phe234 (Fig. 3c). Similarly, sensing currents were recorded from HEK293 cells expressing VSD-mKate(Q208TAG) and the pair only when DanAla was added to the growth medium, indicating that again, the VSD containing DanAla at position 208 was functional (Fig. 4a). However, the Q-V curve of the Q208DanAla mutant exhibited hysteresis (Fig. 4b). Although the curve in the negative membrane potential range was similar to the wild-type VSD-mKate, a second component of charge movement emerged in the positive membrane potential range, indicating that a more depolarized membrane potential was required to move the S4 charges in the position 208 mutant protein.
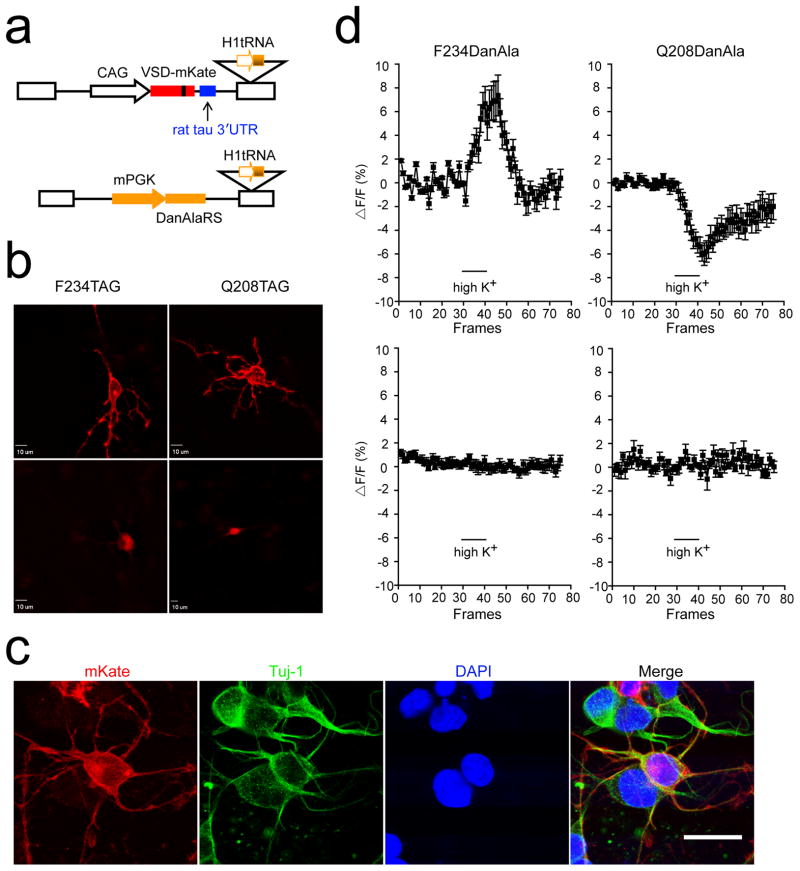
With these two positions for DanAla incorporation in hand, we next used the lentiviral vectors described in strategy 2 to incorporate DanAla into VSD-mKate(F234TAG) and VSD-mKate(Q208TAG) expressed in HCN-A94 cells. After replacing the GFP(TAG) and TyrRS genes with the VSD-mKate(TAG) and DanAlaRS genes, respectively, only low level expression of VSD-mKate with DanAla incorporated was detected in HCN-A94 cells. To enhance the expression levels of VSD-mKate, we substituted the CMV promoter with the CMV early enhancer/chicken β actin (CAG) promoter and added a rat tau 3′UTR element after the VSD-mKate gene in the lentiviral vector (Fig. 5a). The CAG promoter functions in neural stem/progenitor cells during differentiation and the rat tau 3′UTR enhances transgene expression in neuronal cells.[25] This genetic optimization significantly increased VSD-mKate expression. After differentiation into neurons, VSD-mKate with DanAla incorporated at position 234 or 208 clearly localized to the cell membrane (Fig. 5b). When no DanAla was added to the growth medium, red fluorescence appeared throughout the entire cell body, consistent with what was observed in HEK93 cells. To confirm the neural identity of the differentiated cells, Tuj-1 specific antibody was applied for immunostaining. Differentiated cells with mKate expression at the cell membrane all showed Tuj-1 positive signals (Fig. 5c), indicating that DanAla incorporation into VSD-mKate did not affect the differentiation of HCN-A94 into neurons.
Figure 5.
DanAla genetically incorporated into the VSD of CiVSP optically reports membrane depolarization of neurons differentiated from HCN-A94 cells. (a) Modified lentiviral vector for enhancing the expression of VSD-mKate(TAG) in HCN-A94 cells. The CMV promoter was replaced with the CAG promoter to drive expression of the VSD-mKate(TAG) gene, and the 3′UTR sequence from the rat tubulin was added. The orthogonal pair was delivered using strategy 2 described in Fig. 1a. (b) Fluorescence images of VSD-mKate(F234TAG) and VSD-mKate(Q208TAG) expressing neurons differentiated from HCN-A94 cells grown in the presence (top) and absence (bottom) of DanAla. Scale bar, 10 μm. (c) Immunostaining of HCN-A94 cells incorporating DanAla into VSD-mKate(F234TAG) on day 6 after differentiation with Tuj-1 specific antibody. Similar immunostaining results were obtained for HCN-A94 cells incorporating DanAla into VSD-mKate(Q208TAG). The presence of mKate fluorescence on cell membrane and Tuj-1 signal indicate that DanAla incorporation into VSD-mKate did not alter HCN-A94 differentiation into neuronal cells. Scale bar, 10 μm. (d) DanAla fluorescence change upon membrane depolarization induced by high K+ shock. The duration of K+ shock is indicated by a black bar. Each frame encompassed 500 ms. Top panels show DanAla channel fluorescence changes in cells with DanAla incorporated into VSD-mKate at position 234 (left) and position 208 (right); bottom panels depict the DanAla channel fluorescence change in cells in the same imaging field without DanAla incorporation as the negative control. Error bars represent s.e.m.; n = 6 for site 234; n = 5 for site 208.
Next, high-K+ depolarization was applied to depolarize the membrane potential of differentiated neurons expressing VSD-mKate with DanAla incorporated at Phe234 or Gln208, and the DanAla fluorescence intensity was monitored. As shown in Fig. 5d, for the F234DanAla mutant, membrane depolarization induced a significant fluorescence increase of DanAla. In control cells in the same imaging field that had no VSD-mKate(F234DanAla) expressed, the residual free DanAla inside cells showed no fluorescence change in response to membrane depolarization. These results indicate that the DanAla incorporated at position 234 of CiVSP’s VSD experienced a polar to less polar microenvironment change upon membrane depolarization. In contrast to site 234, DanAla incorporated at site 208 showed a fluorescence decrease when the membranes of the differentiated cells were depolarized, indicating that the microenvironment of this site became more polar. Taken together these results demonstrate that genetically incorporated DanAla is able to optically report the conformational change of VSDs in response to membrane depolarization of HCN-A94 differentiated cells with spatial and temporal accuracy.
DISCUSSION
We previously developed a general method to express orthogonal prokaryotic tRNAs in eukaryotic cells by using a type-3 Pol III promoter in mammalian cells[7] and a posttranscriptionally cleavable Pol III promoter in yeast.[4] Here we demonstrate that this strategy can be extended to neural stem cells. The type-3 H1 promoter that drives the expression of human H1RNA was successfully used to express both and of E. coli origin in HCN-A94 cells throughout their neural differentiation program. Other members of the type-3 class of polymerase III promoter, such as U6, 7SK and MRP/7-2, should also work in a similar manner. Among the three strategies used for constructing lentiviral vectors to drive expression of the orthogonal tRNA/synthetase pair and the gene of interest, the second strategy employing the H1-tRNA cassette inserted in the 3′-LTR yielded the most efficient amino acid incorporation at the UAG amber codon site. The key feature of this strategy is that while only a single H1-tRNA cassette is inserted in the lentiviral vector, it generates two genomically incorporated copies of the transcription unit after lentiviral reverse transcription and integration into the host cells. The third strategy employing four tandem repeats of the H1-tRNA cassette resulted in lower amino acid incorporation, possibly because repeated promoter and tRNA sequences within the same lentiviral construct resulted in recombination during the production of transducible lentivirus, leading to deletion of the H1-tRNA cassette. Similar lentiviral genetic instability has been reported when a promoter was repeatedly used to express shRNAs encoded in the same lentiviral vector.[26] Insertion of a single H1-tRNA cassette in the 3′-LTR circumvents this problem while maintaining increased gene dosage upon integration into the host cell. Notably, amino acid incorporation efficiency by the orthogonal tRNA/synthetase increases with progression of HCN-A94 cell differentiation. It has been reported that CMV, PGK and CAG promoters all drive transgene expression more efficiently at the late stages of mouse embryonic stem cell differentiation.[27]
Fluorophores have been chemically attached to ion channels using engineered Cys residues and sulfhydryl reactive fluorophore conjugates. Together with voltage-clamp fluorometry,[28] which detects fluorescence changes upon application of different voltages to a cell membrane, they have provided valuable information on conformational changes of voltage- or ligand-gated channels.[29–34] This pioneering technique has been largely used in Xenopus oocytes, and only extracellular amino acid residues that are accessible for Cys labeling can be readily modified and then studied using fluorescence spectroscopy. Moreover, inconsistent behaviors of VSDs in Xenopus oocytes compared to more natural mammalian cells have been noted. For instance, the gating charge movement of CiVSP in mammalian PC12 cells is 10-times faster than in Xenopus oocytes, possibly due to differences in lipid environments.[24] Mammalian and neuronal cells are native hosts for mammalian ion channels, receptors and transporters, and the genetic encoding method reported here should enable such proteins to be more quantitatively studied in these cells. In addition, instead of being limited to certain extracellular residues, genetically encoding Uaas with fluorogenic properties allows spatial precision for fluorophore incorporation without the limitation inherent to traditional chemical modification strategies. For instance, the S4 Phe234 position chosen in this study resides on the cytoplasmic face of the lipid bilayer and is not readily accessible for Cys-mediated labeling. Moreover, in comparison to Cys labeling and fluorescent protein tagging, genetically encoded fluorescent Uaas permit introduction of the fluorophore closer to the protein backbone, so that the fluorophore follows domain movements closer.[35] Therefore, the method reported here should significantly expand our ability to investigate ion channel and other membrane protein behavior on the molecular level with spatial and temporal precision and in their native cellular environments.
A physical-chemical model for how the VSD transfers gating charges on S4 across the low dielectric membrane bilayer is fundamental for understanding the switch-like responses of ion channels to membrane voltage changes. One model suggests that the conserved basic residues on S4 move up to 15–20 Å sweeping across the membrane.[36] A second model posits that there is very little physical movement of S4 and instead a reorganization of the electric field around the S4 charges through changes in the local water structure and/or membrane bilayer deformation.[30, 37–39]
By genetically incorporating DanAla into the VSD of CiVSP in HCN-A94 cells, we showed that DanAla fluorescence changes optically reported VSD conformational changes in response to membrane depolarization in differentiated neurons. Fluorescence increases and decreases were respectively observed at opposite ends of S4, suggesting that the microenvironments of the two ends of S4 change in opposing manners upon membrane depolarization. The experimentally measured increase in fluorescence observed for DanAla incorporated a position 234 of the VSD of CiVSP represents the first measurement of a fluorescence change of a residue on the intracellular end of S4. While the observed fluorescence increase is consistent with either of the above models, it provides an experimental platform for more spatially and temporally defined fluorescent experiments aimed at resolving how charge moves in response to membrane depolarization.
The fluorescence decrease observed for DanAla incorporated at position 208 is more intriguing. Residue 208 sits on the S3–S4 loop near the extracellular surface of the lipid bilayer. The observed decrease in DanAla fluorescence suggests that this position along S3–S4 is sequestered from solvent possibly buried within the hydrophobic membrane in the closed VSD state and then moves to a more hydrophilic environment upon membrane depolarization and VSD opening. The appearance of a second component on the Q-V curve for the VSD(Gln208DanAla) mutant lends support for this model. Mutagenic replacement of a hydrophilic Gln with a substantially more hydrophobic DanAla residue should make it more difficult to “pull” the DanAla side chain from a buried hydrophobic layer. This “difficulty” would likely be evidenced by the requirement for a more positive membrane voltage to exert force on the VSD transiting to the open conformation. While preliminary in nature, these latter results are consistent with a gating model involving a large S4 sweep across the membrane. In contrast, small changes in S4 position or localized membrane reconstructuring would be unlikely to effectuate such a pronounced fluorescence change in DanAla that already resides on the extracellular side of the transmembrane region. Nonetheless, a systematic fluorescence mapping of VSD residues at more loop sites and throughout the transmembrane helices is needed to provide a sufficient amount of data to better infer the global conformational changes of VSD in response to voltage changes of the membrane potential.[40]
In summary, we developed lentiviral vectors for the long-term genetic incorporation of Uaas into translated proteins in neural stem cells. Uaa incorporation does not change the overall differentiation process of the neural progenitor HCN-A94 cells, and Uaa incorporation efficiency increases as the differentiation program ensues. Moreover, a genetically encoded fluorescent Uaa optically reports the conformational change of a VSD in response to membrane polarization in neurons differentiated from HCN-A94 cells, setting the stage for the systematic functional mapping of global conformation changes of VSDs in response to membrane voltage changes.
Supplementary Material
Acknowledgments
We thank Drs. Gerald Pao and Inder M. Verma for providing lentiviral vectors and helpful suggestions on lentivirus preparation, Drs. Dejan Vucinic and Terrence J. Sejnowski for the cDNA of CiVSP, Dr. Paul A. Slesinger for comments on CiVSP mutants, and Dr. Kimberly McIntyre and Eunice Mejia for technical assistance on microscopy and immunocytochemistry. L.W. thanks the support from the March of Dimes Foundation (5-FY08-110), CIRM (RN1-00577-1) and NIH (1DP2OD004744).
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors indicate no potential conflicts of interest.
Author contribution summary
Bin Shen: Experimental design, collection of data, data analysis and interpretation, manuscript writing
Zheng Xiang: Collection of data, data analysis and interpretation
Barbara Miller, Gordon Louie, Wenyuan Wang: Collection of data
Joseph P. Noel: Provision of study material, manuscript writing
Fred H. Gage: Provision of study material, manuscript writing
Lei Wang: Conception and design, data analysis and interpretation, financial support, manuscript writing
Details for chemical reagents, plasmid construction, Western blot, immunocytochemistry, and VSD model building. Fig. S1.
References
- 1.Wang L, Schultz PG. Expanding the genetic code. Angew Chem Int Ed Engl. 2005;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- 2.Wang Q, Parrish AR, Wang L. Expanding the genetic code for biological studies. Chem Biol. 2009;16:323–336. doi: 10.1016/j.chembiol.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, Brock A, Herberich B, et al. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 4.Wang Q, Wang L. New methods enabling efficient incorporation of unnatural amino acids in yeast. J Am Chem Soc. 2008;130:6066–6067. doi: 10.1021/ja800894n. [DOI] [PubMed] [Google Scholar]
- 5.Sakamoto K, Hayashi A, Sakamoto A, et al. Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells. Nucleic Acids Res. 2002;30:4692–4699. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu W, Brock A, Chen S, et al. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 7.Wang W, Takimoto JK, Louie GV, et al. Genetically encoding unnatural amino acids for cellular and neuronal studies. Nat Neurosci. 2007;10:1063–1072. doi: 10.1038/nn1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Rossi DJ, Jamieson CH, Weissman IL. Stems cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 10.Ma DK, Marchetto MC, Guo JU, et al. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat Neurosci. 2010;13:1338–1344. doi: 10.1038/nn.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murata Y, Iwasaki H, Sasaki M, et al. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 12.Gage FH, Coates PW, Palmer TD, et al. Survival and differentiation of adult neuronal progenitor cells transplanted to the adult brain. Proc Natl Acad Sci U S A. 1995;92:11879–11883. doi: 10.1073/pnas.92.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamill OP, Marty A, Neher E, et al. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 14.Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 15.Holst J, Vignali KM, Burton AR, et al. Rapid analysis of T-cell selection in vivo using T cell-receptor retrogenic mice. Nat Methods. 2006;3:191–197. doi: 10.1038/nmeth858. [DOI] [PubMed] [Google Scholar]
- 16.Takimoto JK, Adams KL, Xiang Z, et al. Improving orthogonal tRNA-synthetase recognition for efficient unnatural amino acid incorporation and application in mammalian cells. Mol Biosyst. 2009;5:931–934. doi: 10.1039/b904228h. [DOI] [PubMed] [Google Scholar]
- 17.Summerer D, Chen S, Wu N, et al. A genetically encoded fluorescent amino acid. Proc Natl Acad Sci U S A. 2006;103:9785–9789. doi: 10.1073/pnas.0603965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohout SC, Ulbrich MH, Bell SC, et al. Subunit organization and functional transitions in Ci-VSP. Nat Struct Mol Biol. 2008;15:106–108. doi: 10.1038/nsmb1320. [DOI] [PubMed] [Google Scholar]
- 19.Wu S, Zhang Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007;35:3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Wang Q, Ni F, et al. Structure of the full-length Shaker potassium channel Kv1.2 by normal-mode-based X-ray crystallographic refinement. Proc Natl Acad Sci U S A. 2010;107:11352–11357. doi: 10.1073/pnas.1000142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long SB, Tao X, Campbell EB, et al. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 22.Dimitrov D, He Y, Mutoh H, et al. Engineering and characterization of an enhanced fluorescent protein voltage sensor. PLoS ONE. 2007;2:e440. doi: 10.1371/journal.pone.0000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villalba-Galea CA, Sandtner W, Starace DM, et al. S4-based voltage sensors have three major conformations. Proc Natl Acad Sci U S A. 2008;105:17600–17607. doi: 10.1073/pnas.0807387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lundby A, Mutoh H, Dimitrov D, et al. Engineering of a genetically encodable fluorescent voltage sensor exploiting fast Ci-VSP voltage-sensing movements. PLoS ONE. 2008;3:e2514. doi: 10.1371/journal.pone.0002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brun S, Faucon-Biguet N, Mallet J. Optimization of transgene expression at the posttranscriptional level in neural cells: implications for gene therapy. Mol Ther. 2003;7:782–789. doi: 10.1016/s1525-0016(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 26.ter Brake O, t Hooft K, Liu YP, et al. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol Ther. 2008;16:557–564. doi: 10.1038/sj.mt.6300382. [DOI] [PubMed] [Google Scholar]
- 27.Hong S, Hwang DY, Yoon S, et al. Functional analysis of various promoters in lentiviral vectors at different stages of in vitro differentiation of mouse embryonic stem cells. Mol Ther. 2007;15:1630–1639. doi: 10.1038/sj.mt.6300251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mannuzzu LM, Moronne MM, Isacoff EY. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 1996;271:213–216. doi: 10.1126/science.271.5246.213. [DOI] [PubMed] [Google Scholar]
- 29.Cha A, Bezanilla F. Characterizing voltage-dependent conformational changes in the Shaker K+ channel with fluorescence. Neuron. 1997;19:1127–1140. doi: 10.1016/s0896-6273(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 30.Glauner KS, Mannuzzu LM, Gandhi CS, et al. Spectroscopic mapping of voltage sensor movement in the Shaker potassium channel. Nature. 1999;402:813–817. doi: 10.1038/45561. [DOI] [PubMed] [Google Scholar]
- 31.Smith PL, Yellen G. Fast and slow voltage sensor movements in HERG potassium channels. J Gen Physiol. 2002;119:275–293. doi: 10.1085/jgp.20028534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng J, Zagotta WN. Gating rearrangements in cyclic nucleotide-gated channels revealed by patch-clamp fluorometry. Neuron. 2000;28:369–374. doi: 10.1016/s0896-6273(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 33.Dahan DS, Dibas MI, Petersson EJ, et al. A fluorophore attached to nicotinic acetylcholine receptor beta M2 detects productive binding of agonist to the alpha delta site. Proc Natl Acad Sci U S A. 2004;101:10195–10200. doi: 10.1073/pnas.0301885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savalli N, Kondratiev A, Toro L, et al. Voltage-dependent conformational changes in human Ca(2+)- and voltage-activated K(+) channel, revealed by voltage-clamp fluorometry. Proc Natl Acad Sci U S A. 2006;103:12619–12624. doi: 10.1073/pnas.0601176103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bezanilla F. Ion channels: from conductance to structure. Neuron. 2008;60:456–468. doi: 10.1016/j.neuron.2008.10.035. [DOI] [PubMed] [Google Scholar]
- 36.Jiang Y, Ruta V, Chen J, et al. The principle of gating charge movement in a voltage-dependent K+ channel. Nature. 2003;423:42–48. doi: 10.1038/nature01581. [DOI] [PubMed] [Google Scholar]
- 37.Ahern CA, Horn R. Focused electric field across the voltage sensor of potassium channels. Neuron. 2005;48:25–29. doi: 10.1016/j.neuron.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 38.Posson DJ, Ge P, Miller C, et al. Small vertical movement of a K+ channel voltage sensor measured with luminescence energy transfer. Nature. 2005;436:848–851. doi: 10.1038/nature03819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chanda B, Asamoah OK, Blunck R, et al. Gating charge displacement in voltage-gated ion channels involves limited transmembrane movement. Nature. 2005;436:852–856. doi: 10.1038/nature03888. [DOI] [PubMed] [Google Scholar]
- 40.Swartz KJ. Sensing voltage across lipid membranes. Nature. 2008;456:891–897. doi: 10.1038/nature07620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.