Abstract
Background. Plasmodium falciparum malaria resistant to chloroquine and pyrimethamine originated in limited foci and migrated to Africa. It remains unresolved whether P. falciparum resistance to sulfadoxine, which is conferred by mutations in dihydropteroate synthase (DHPS), evolved following a similar pattern.
Methods. The dhps locus of 893 P. falciparum isolates from 12 countries in Asia, the Pacific Islands, Africa, and South America was sequenced. Haplotypes of 6 microsatellite loci flanking the dhps locus were determined to define the genetic relationships among sulfadoxine-resistant lineages.
Results. Six distinct sulfadoxine-resistant lineages were identified. Highly resistant lineages appear to have originated only in Southeast Asia and South America. Two resistant lineages found throughout Southeast Asia have been introduced to East Africa, where they appear to have spread.
Conclusions. The infrequent selection of parasites highly resistant to sulfadoxine and the subsequent migration of resistant lineages from Asia to Africa are similar to the patterns observed in chloroquine and pyrimethamine resistance. These findings strongly suggest that the global migration of resistant parasites has played a decisive role in the establishment of drug-resistant P. falciparum parasites, and that similar patterns may be anticipated for the spread of artemisinin resistance.
Despite a dramatic decrease in incidence of malaria during the last decade, there are still 250 million cases annually, of which nearly 1 million are fatal. The vast majority of these deaths are caused by Plasmodium falciparum, the most virulent malaria parasite species. The antifolate drug combination sulfadoxine/pyrimethamine (SP), although infrequently used for treating acute malaria due to resistance, is still commonly used for intermittent preventive treatment during pregnancy throughout sub-Saharan Africa [1]. SP is also being piloted for intermittent preventive treatment in infants, which has been shown to prevent malaria in pilot studies [2]. However, rising resistance to SP due to the spread of resistant P. falciparum in many endemic regions is a growing concern [3].
Sulfadoxine, a structural analogue of p-aminobenzoic acid, and pyrimethamine, the other component of SP, inhibit dihydropteroate synthase (DHPS) and dihydrofolate reductase (DHFR), respectively, in the P. falciparum folate synthesis pathway. Mutations at the dhps and dhfr loci confer resistance to sulfadoxine and pyrimethamine, respectively [4, 5], and SP treatment failure is strongly associated with the presence of at least 3 mutations in dhfr and 2 in dhps [6, 7]. In the case of pyrimethamine resistance, highly resistant parasites that harbor 3 or more mutations in the dhfr gene emerged in limited geographical foci, probably 1 in Asia and 2 in South America, which then spread worldwide [3, 8–11]. In Africa, almost all parasites harboring pyrimethamine-resistant dhfr alleles with 3 mutations are descendants of resistant parasites introduced from Southeast Asia [12–15].
Mutations associated with sulfadoxine resistance have been observed to occur at 5 amino acid (aa) positions within the dhps gene: positions 436, 437, 540, 581, and 613, with the wild-type represented by the aa sequence SAKAA at these loci [5, 16, 17]. A single mutation at aa position 437, which results in the SGKAA mutant (mutations are underlined), increases the median inhibitory concentration (IC50) of sulfadoxine by 4.8-fold in this parasite, whereas the IC50 of sulfadoxine in the AGEAA triple mutant is 9.8-fold greater than that of the wild-type parasite [17].
In contrast to the knowledge of pyrimethamine-resistant dhfr, studies are limited on the geographical origins and spread of sulfadoxine-resistant dhps. A recent study analyzing P. falciparum parasites from 27 African countries has shown the occurrence of as many as 5 distinct sulfadoxine-resistant dhps lineages in Africa. The flanking microsatellite (MS) haplotype associated with one of these lineages matched that of the K1 P. falciparum laboratory isolate that originated in Southeast Asia. This observation raises the possibility that sulfadoxine resistance was introduced from Asia to Africa in a similar way to pyrimethamine. In contrast, a study from Cambodia, which described 2 major lineages with high sulfadoxine resistance, has shown independent origins of sulfadoxine resistance in Asia and Africa [19]. However, sampling in this study was limited to Cambodia and just 2 African sites (Cameroon and Kenya).
Geographically comprehensive characterization of sulfadoxine-resistant dhps is necessary for an accurate understanding of the way in which resistance against the commonly used antimalarial drugs chloroquine, pyrimethamine, and sulfadoxine emerged and spread among populations of P. falciparum. In this study, we sequenced the dhps gene of 893 isolates of P. falciparum from 12 countries from Asia, the Pacific Islands, Africa, and South America and determined the haplotypes of MS markers flanking the dhps locus in parasites bearing mutant dhps alleles. We identified 6 independent sulfadoxine-resistant lineages worldwide, among which highly resistant lineages were found only in Southeast Asia and South America. More important, we obtained evidence for the migration of 2 Southeast Asian lineages to Africa, where it has since spread.
MATERIALS AND METHODS
Study Sites
Blood samples were obtained from P. falciparum–infected patients in all age groups, unless otherwise stated, living in 12 malaria-endemic countries (Table 1) as follows:
Bangladesh: Samples were collected from malaria patients in a Bandarban district hospital in 2007 and in 6 malaria-endemic villages in Bandarban in 2008. This study was approved by the Bangladesh Medical Research Council and the local health regulatory body in Bandarban, Bangladesh.
Cambodia: Samples were collected from infected individuals during a cross-sectional survey of rural villages in Chumkiri, Kampot province, in 2004. The study was approved by the National Center for Parasitology, Entomology, and Malaria Control of Cambodia.
Thailand: Samples were collected from malaria patients at town clinics located in the western border of Tak, Kanchanaburi, and Ratchburi provinces from 2001 to 2002 [10]. The study was approved by the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University.
Papua New Guinea: Samples were collected from infected individuals at Town and Wirui Clinic in Wewak in 2002 and 2003 [20]. The study was approved by the National Department of Health Medical Research Advisory Committee of Papua New Guinea.
Solomon Islands: Samples were collected from infected individuals during cross-sectional surveys in northeastern Guadalcanal Island from 1995 to 1996. The study was approved by the Ethics Committee of the Solomon Islands for Medical Research.
Vanuatu: Samples were collected from infected individuals during cross-sectional surveys in rural villages located in 4 islands; Gaua, Santo, Pentecost, and Malakula, in 1996 and 1998. The study was approved by the Vanuatu Department of Health.
Kenya: Samples were collected from infected individuals during cross-sectional surveys at 4 villages in Kisii District in 1998 and 2002 [15]. The study was approved by the Kenyan Ministry of Health and Education.
Tanzania: Samples were collected from infected individuals during cross-sectional surveys in the Rufiji River Delta in eastern coastal Tanzania in 1998 and 2003. The study was approved by the Ethics Committee of the National Institute for Medical Research of Tanzania.
Malawi: Samples were collected from infected individuals during cross-sectional surveys at 2 primary schools of Salima district in 2000 [21]. The study was approved by the local ethics committee of the Malaria Control Programme and Malawi Ministry of Health.
Republic of the Congo: Samples were collected from patients with malaria in Pointe-Noire, Brazzaville, and Gamboma in 2006. The study was approved by the Ministry of Research and Ministry of Health of the Republic of the Congo.
Ghana: Samples were collected from infected children during cross-sectional surveys in 3 villages near Winneba, a western coastal region, in 2004 [15]. This study was approved by the Ministry of Health/Ghana Health Service.
Brazil: Samples were collected from infected individuals in the eastern part of Acre state in 1985–1986, 1999, and 2004–2005. The study protocol was approved by the ethics review board of the Institute of Biomedical Sciences, University of São Paulo.
Table 1.
Plasmodium falciparum Isolates From 12 Countries
| Country | Area | No. of isolates | Year of sampling |
| Southeast Asia | |||
| Bangladesh | Bandarban | 135 | 2007–2008 |
| Cambodia | Chumkiri, Kampot | 36 | 2004 |
| Thailand | Tak, Kanchanaburi, Ratchburi | 78 | 2001–2002 |
| Pacific Islands | |||
| Papua New Guinea | East Sepik | 113 | 2002–2003 |
| Solomon Islands | Guadalcanal Island | 48 | 1995–1996 |
| Vanuatu | Gaua, Santo, Pentecost, Malakula | 109 | 1996, 1998 |
| Africa | |||
| Kenya | Kisii | 77 | 1998, 2002 |
| Tanzania | Rufiji River Delta | 55 | 1998, 2003 |
| Malawi | Salima | 71 | 2000 |
| Republic of Congo | Pointe-Noire, Brazzaville, Gamboma | 42 | 2006 |
| Ghana | Winneba | 91 | 2004 |
| South America | |||
| Brazil | Acre | 38 | 1985–1986, 1999, 2004–2005 |
In all study areas, informed consent was obtained from individual patients or their guardians and antimalarial treatment was provided.
dhps Genotyping and Flanking MS Haplotyping
In Bangladesh, Cambodia, Papua New Guinea, Vanuatu, Kenya, Malawi, Republic of Congo, and Ghana, finger-prick blood samples (75 μL) were spotted onto chromatographic filter paper (ET31CHR; Whatman). In Thailand, the Solomon Islands, Tanzania, and Brazil, venous blood was collected in ethylenediaminetetraacetic acid–containing tubes. Parasite genomic DNA was extracted using QIAamp DNA Mini Kits (QIAGEN). An EZ1 BioRobot (QIAGEN) was used to extract DNA from samples from the Republic of Congo. A region of dhps (aa 406–642), encompassing all known polymorphic sites (aa 436, 437, 540, 581, and 613), was amplified by nested polymerase chain reaction (PCR) as previously reported [22]. The amplified products were subjected to direct sequencing using a BigDye Terminator version 1.1 Cycle Sequencing Kit in an ABI 377 DNA sequencer (Applied Biosystems). Sequences showing 2 or more peaks at the same position on the electropherogram were considered mixed infections and were excluded from further analysis. We also included data of dhps alleles previously obtained from Papua New Guinea [23] and Malawi [21].
Nucleotide length variations of 6 MS markers (determined by the number of TA repeats) located at −2.9, −1.5, −.13, .8, 4.3, and 7.7 kilobase (kb) flanking dhps were measured as described previously [19, 24]. In brief, seminested PCR was performed using fluorescent end-labeled primers. Size variations of the amplified products were determined by electrophoresis on a DNA sequencer and analyzed with GeneScan software (Applied Biosystems). When 2 or more alleles were detected, these isolates were considered to be mixed infections and were excluded from further analysis. Only isolates with resistant dhps alleles were characterized for the MS markers. It has been well documented that wild-type parasites show extensive MS variation reflecting a lack of selective sweeps [18, 19] and are not useful in determining resistance lineages.
Determination of Lineages of Sulfadoxine-Resistant dhps Alleles
Lineages of sulfadoxine-resistant dhps alleles were determined based on the MS haplotypes composed of the 6 MS loci flanking dhps and genetic distances between the 6 dhps flanking MS markers. STRUCTURE 2.3.3 [25, 26] was used to assign individual isolates from all populations to a predetermined number of clusters based on MS haplotypes. STRUCTURE is a Bayesian model–based clustering program that makes no a priori assumptions regarding the number of clusters present or the genetic relatedness of samples from the same region or presumed population. Rather, STRUCTURE can be used to calculate the most probable number of clusters (K) in the sample set and then assign probabilities of membership to each cluster for each individual, regardless of geographic origin. For each run, a burn-in period of 30 000 steps was followed by 1 × 106 iterations under the admixture model and the assumption of uncorrelated allele frequencies among populations. For each K of 1–10 clusters, 10 runs were performed. ΔK was calculated as described in Evanno et al [27] and used to choose the best approximation of the “true” K. In addition, estimated log probabilities (Ln P(D)) were averaged across runs and compared to determine the posterior probability of each K. Once the best approximation of K was determined, a visual output for the run of highest Ln P(D) was generated using the program Distruct [28], which allowed the rearrangements of the STRUCTURE output such that the clustering of individuals assigned by STRUCTURE could be compared with their geographic origins. A haplotype network was constructed based on polymorphisms at the 6 MS loci using the median-joining network method in NETWORK version 4.6 (http://www.fluxus-engineering.com/sharenet.htm) [29]. This method assumes no recombination events and stepwise mutation in a data set. In this study, we used MS polymorphism of sulfadoxine-resistant parasites, which have emerged recently, and hence recombination events and multistep mutations seem less frequent.
RESULTS
Geographical Distribution of dhps Alleles
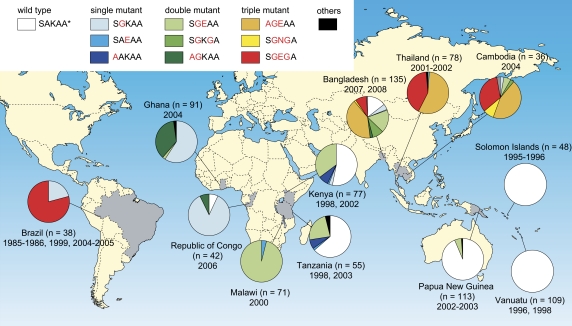
The dhps alleles were successfully determined for 893 isolates. We identified 16 dhps alleles, 10 of which constitute > 99% of all samples: the wild-type allele (SAKAA; 41%); single mutants (SGKAA, SAEAA, and AAKAA; 14%); double mutants (SGEAA, SGKGA, and AGKAA; 22%); and triple mutants (AGEAA, SGEGA, and SGNGA; 23%). Nearly all parasites (97%) harbored the wild-type allele in the Pacific Islands, whereas, in Thailand, all but 2 isolates harbored triple mutants (AGEAA and SGEGA); the exception harbored double and quadruple mutants (SGEGA and AGEGA) (Figure 1). The prevalences of these triple mutants in other Southeast Asian countries were 89% in Cambodia and 53% in Bangladesh. The distribution of dhps alleles was remarkably different between West/Central Africa and East Africa. Nearly all parasites (95%) in West/Central Africa harbored either the SGKAA or AGKAA mutants, whereas the wild-type (40%) and SGEAA (52%) alleles were predominant in East Africa, where alleles with a single mutation are rare (8%).
Figure 1.
Geographical distribution of dhps alleles in 893 Plasmodium falciparum isolates from 12 countries. Capital letters next to the shaded boxes denote amino acid residues at positions 436, 437, 540, 581, and 613, with mutations identified in red. Other mutations (underlined) include alleles of CAKAA, FAKAA, HAKAA, SGKAG, AGKGA, and AGEAE with frequencies of <0.2%.
Sulfadoxine-Resistant Lineages and Their Geographical Distributions
Amongst the 528 dhps mutants, 172 were excluded because of mixed MS alleles and/or unsuccessful determination of MS alleles. Thus, 356 isolates were used for further analysis. We obtained a total of 112 MS haplotypes composed of 6 MS loci flanking dhps (Supplementary Figure 1).
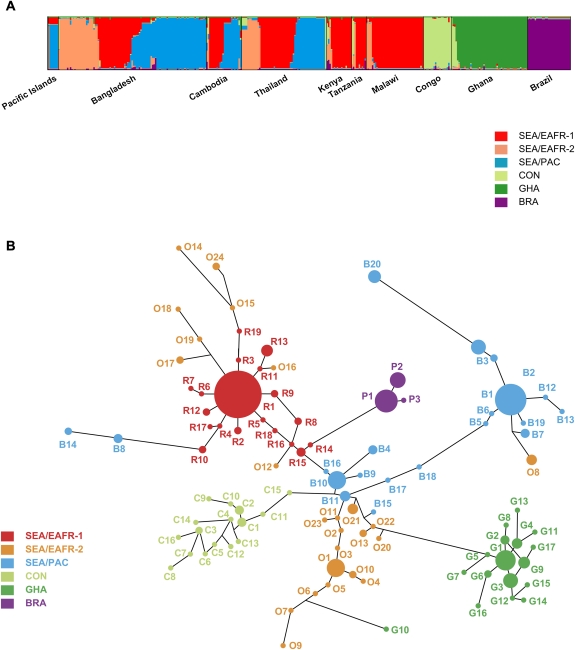
Results from the STRUCTURE Bayesian analysis provided the highest ΔK at K = 6; 2 clusters in Southeast Asia and East Africa (SEA/EAFR-1 and SEA/EAFR-2), Southeast Asia and Pacific Islands (SEA/PAC), the Republic of Congo (CON), Ghana (GHA), and Brazil (BRA) (Figure 2A). Five samples had <.5 proportion of membership to any 1 cluster and were categorized as “unassigned” (UA) ancestry. There are 8 samples that were initially assigned as SEA/EAFR-2 (U1, 10, and 12) or CON (U2, U6–U9) in the STRUCTURE analysis but had only 1 allele in common with putative ancestral alleles in each lineage (O1 and C1). We categorized these 8 samples as UA as well.
Figure 2.
Bayesian population structure analysis (A) and median-joining network diagram (B) indicating the presence of 6 lineages of sulfadoxine-resistant Plasmodium falciparum. A, Plot from highest log likelihood STRUCTURE run at K = 6. Individual isolates (n = 356) are grouped by collection site. Each individual is represented by a vertical bar displaying proportion of membership to each of 6 clusters: red (Southeast Asia and East Africa [SEA/EAFR-1], orange (SEA/EAFR-2), blue (Southeast Asia and Pacific Islands [SEA/PAC]), light green (Republic of the Congo [CON]), green (Ghana [GHA]), and purple (Brazil [BRA]). B, Haplotype network of 343 individuals comprising 99 haplotypes (Supplementary Figure 1), based on allelic variations in the 6 microsatellite loci flanking the dhps locus. Each individual was counted as belonging to its major cluster. Individuals not assigned to any clusters by Bayesian population structure analysis were excluded from this analysis (n = 13). The size of each circle corresponds to the number of individuals sharing that haplotype, and the length of an edge is proportional to the number of genetic changes between the 2 haplotypes it connects.
Analysis of the relationships among these MS haplotypes, excluding 13 UA isolates (n = 343), by the construction of a haplotype network (Figure 2B) corroborated the clustering patterns produced in the STRUCTURE analysis. However, 7 haplotypes (O14–O19 and O24) from the SEA/EAFR-2 lineage clustered with the SEA/EAFR-1 lineage in the spanning network. These haplotypes shared an equal number of MS alleles with both the SEA/EAFR-1 and SEA/EAFR-2 lineages but were assigned to SEA/EAFR-2 in the STRUCTURE analysis. Likewise, small numbers of haplotypes from the SEA/PAC (B8, B14) and GHA (G10) lineages cluster with other lineages in the haplotype network, reflecting shared alleles with multiple lineages. The network algorithm assumes no recombination events and stepwise mutation in a dataset. These assumptions are not always likely to hold for P. falciparum, which may contribute to a slight difference in clustering between network and STRUCTURE analysis.
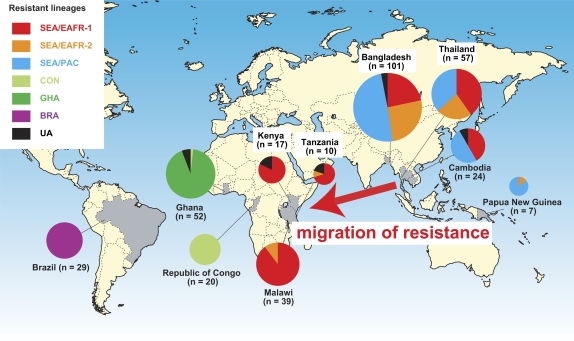
The geographical distribution of sulfadoxine-resistant dhps haplotypes of P. falciparum provides strong evidence of lineages shared by Southeast Asia and Africa (Figure 3). The lineage SEA/EAFR-1 occurred at the highest frequency in our samples and includes 3 major sulfadoxine-resistant dhps alleles (SGEAA, SGKGA, and SGEGA). Lineages SEA/EAFR-1 and SEA/EAFR-2 were both found in Asian and East African countries. In East Africa, among the 61 isolates harboring SGEAA mutants, 92% (56/61) belong to the SEA/EAFR-1 lineage and 7% (4/61) belong to the SEA/EAFR-2 lineage. These results strongly suggest that the SGEAA type of sulfadoxine-resistant alleles in East Africa migrated from Southeast Asia. A third Southeast Asian lineage, SEA/PAC, also has 1 major (AGEAA) and 2 minor (SGEAA and SGEGA) mutant dhps alleles and was shared with the Pacific Islands.
Figure 3.
Geographical distribution of 6 major sulfadoxine-resistant dhps lineages of Plasmodium falciparum. Haplotypes not assigned to any 1 cluster (n = 13) are designated “unassigned” lineage (UA). Abbreviations: SEA/EAFR, Southeast Asia and East Africa; SEA/PAC, Southeast Asia and Pacific Islands; CON, Republic of the Congo; GHA, Ghana; BRA, Brazil.
Two major lineages were found in West/Central Africa, CON (with dhps allele SGKAA), and GHA (with dhps allele AGKAA). All but 1 isolate in CON lineage were observed in the Republic of Congo. All isolates in GHA lineage were distributed only in Ghana. None of the Southeast Asian lineages were found in either the Republic of Congo or in Ghana. Likewise, the BRA lineage was exclusively found in Brazil.
DISCUSSION
This study presents strong evidence that the way in which sulfadoxine resistance arose and spread is very similar to the patterns for resistance to chloroquine [30, 31] and high-level resistance to pyrimethamine [3, 12–15], and elevates the concern that a similar pattern might be expected for other drugs such as the artemisinins. Four highly sulfadoxine-resistant (3 mutations in the dhps gene) lineages originated in only 2 geographic regions: Southeast Asia and South America. More importantly, the resistance-conferring dhps double mutant allele (SGEAA) arose in Southeast Asia and spread to East Africa, where it is now highly prevalent. In our samples, the vast majority of dhps mutants in East Africa shared lineages with Southeast Asian parasites. These findings suggest that indigenously evolved forms of sulfadoxine resistance have a minor role in East Africa, perhaps due to fitness differences among different resistant forms.
We observed 3 highly sulfadoxine-resistant lineages in Asia. The SEA/EAFR-1 and SEA/PAC lineages were the same as those in the previous reports in Asia [19, 32], sharing identical alleles at all 3 MS markers (−2.9, −1.5, and −.13 kb); 191/172/134 base pairs (bp) in SEA/EAFR-1 and 183/172/134 bp in SEA/PAC. The other lineage (SEA/EAFR-2) has not been described previously. The most prevalent MS haplotype in the SEA/EAFR-2 lineage (O1) is distinct from the most frequent haplotypes in SEA/EAFR-1 (R1) and SEA/PAC (B1), suggesting a discrete evolutionary path of the SEA/EAFR-2 lineage. In East Africa, all isolates placed in the SEA/EAFR-2 lineage (n = 5) appeared to arise from recombination between the SEA/EAFR-1 and SEA/EAFR-2 lineages.
Consistent with a previous study in Africa [18], the sulfadoxine-resistant dhps allele of Southeast Asian origin was prevalent in East Africa but not in West Africa. In contrast, the pyrimethamine-resistant dhfr mutant allele of Southeast Asian origin is widely distributed in both East and West Africa [14, 15]. The reason for this difference remains unclear. It is possible that mutant dhps alleles may have arrived in East Africa later than mutant dhfr alleles and have had less time to migrate west. The earliest record of the appearance of a dhfr triple mutant in Africa came in 1988 from Kenya [33], whereas the dhps double mutant (SGEAA) is thought to have first surfaced in Kenya in 1993–1995 [34]. Furthermore, SP use was implemented earlier in East Africa than in West Africa, and it was first-line therapy in more East African countries, so drug pressure in West Africa was likely lower than in East Africa. The K540E mutation (mainly found as part of the double dhps mutant allele SGEAA) is common in East Africa. It was recently found in West Africa as well, but at much lower levels [34].
In addition to the 2 migrated Southeast Asian lineages, we detected 2 other dhps lineages in Africa. Pearce et al [18] reported as many as 5 resistant lineages in Africa, among which 3 lineages in East, West, and South/West Africa had identical MS alleles to the migrated SEA/EAFR-1 (131/104/107 bp at .8, 4.3, and 7.7 kb), CON lineages (117/106/diverse alleles at 7.7 kb in previous study), and GHA (121/108/113 bp) in the present study.
A limited number of point mutations in dhps are considered as sufficient for conferring high resistance to sulfadoxine [5, 16, 17], implying that P. falciparum may relatively easily acquire resistance to this drug. However, the limited foci of sulfadoxine-resistant lineages observed in this study suggest that sulfadoxine resistance was not selected de novo as often as was previously believed. Moreover, highly sulfadoxine-resistant lineages, with 3 mutations in the dhps gene, appeared only in Southeast Asia and South America. Lineages with high resistance against pyrimethamine [8, 9, 11, 12, 15], as well as a lineage with resistance against chloroquine [30, 31], have also emerged from the same 2 regions—the world’s first case of chloroquine resistance was simultaneously identified in both regions around 1960 [35, 36]. Subsequently, SP was introduced as the first-line treatment of uncomplicated malaria in 1970s in both regions.
Why does the emergence of drug-resistant malaria in Asia and South America and spread throughout Africa seem to recur? The relatively early deployment of chloroquine and SP in Southeast Asia and South America may partly account for this recurrent pattern. In addition, most infections are symptomatic in these regions, and infected individuals repeatedly take antimalarial drugs, creating a situation conducive to the selection of drug resistance [37, 38]. Moreover, the parasite population genetic structure is relatively similar in Southeast Asia and South America, that is, there are lower levels of genetic diversity and a relatively small parasite population size [39] compared with Africa, which may facilitate the fixation of drug resistance [40]. Alternatively, there may be something special about Southeast Asian parasites that promotes evolution of drug resistance [41], such as differences in DNA mismatch repair [42]. The pattern of spread from Asia to Africa is likely to reflect patterns of human movement, and identifying patterns of human movement between malaria-endemic areas might inform efforts to contain resistance.
Because efficacy of SP intermittent preventive treatment in infants appears to be reduced in regions of high resistance [43], it is possible that such rapid expansion of sulfadoxine-resistant P. falciparum may erode the benefits of SP intermittent preventive treatment programs. Our findings suggest that this increase of highly resistant dhps alleles may represent the selection and migration of a limited number of parasite lineages. If this is the case, careful monitoring of the prevalence and spread of highly resistant dhfr and dhps alleles, both within East Africa and as they move into West Africa, is required to assess continued efficacy of SP-based treatments.
Moreover, our conclusions are limited to samples collected up to the mid-2000s, with a wide time span. As human movement continues to become more frequent, it is possible that increased contact between Asia and Africa may result in new patterns or increased rates of parasite migration that could have important consequences in the spread of highly SP-resistant malaria.
In conclusion, highly resistant lineages appear to have arisen only in Southeast Asia and South America. Two of these highly resistant lineages were introduced to East Africa from Southeast Asia. These findings are analogous to the patterns observed for the resistance to the other commonly used antimalarial drugs, chloroquine and pyrimethamine. It is reasonable to anticipate that a similar pattern of spread may occur with the newly emerging forms of artemisinin-resistant malaria reported in western Cambodia, and aggressive measures should be taken to prevent this from occurring [44, 45]. Rapid sharing of reliable information on patterns of emerging and spreading resistance, such as via the WorldWide Antimalarial Resistance Network’s interactive maps (http://www.wwarn.org), will aid in this effort [46, 47].
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://www.oxfordjournals.org/our_journals/jid/).
Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments.
The authors thank all participants in this study; Drs Anders Björkman, Anna Färnert, Ingegerd Ruth, Hikota Osawa, Miki Sakurai, Hiroshi Ohmae, Ilomo Hwaihwanje, Aung Swi Prue Marma, Mawuli Dzodzomenyo, Willis Akhwale, and Akira Kaneko for their kind cooperation in the field; and Toshiyuki Hayakawa for his analytical advice.
Financial support.
This work was supported by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (23659211, 23590498, 22406012, 18073013, 18GS03140013) and from the Ministry of Health, Labor, and Welfare of Japan (H20-Shinkou-ippan-013). M. V. and C. V. P. are supported by the Howard Hughes Medical Institute and by the Bill and Melinda Gates Foundation through their support of the Worldwide Antimalarial Resistance Network (WWARN).
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.World Health Organization. World Malaria Report 2008. Geneva, Switzerland: World Health Organization, 2008. [Google Scholar]
- 2.Grobusch MP, Egan A, Gosling RD, Newman RD. Intermittent preventive therapy for malaria: progress and future directions. Curr Opin Infect Dis. 2007;20:613–20. doi: 10.1097/QCO.0b013e3282f1ae3b. [DOI] [PubMed] [Google Scholar]
- 3.Mita T, Tanabe K, Kita K. Spread and evolution of Plasmodium falciparum drug resistance. Parasitol Int. 2009;58:201–9. doi: 10.1016/j.parint.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci U S A. 1988;85:9114–8. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Triglia T, Cowman AF. Primary structure and expression of the dihydropteroate synthetase gene of Plasmodium falciparum. Proc Natl Acad Sci U S A. 1994;91:7149–53. doi: 10.1073/pnas.91.15.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kublin JG, Dzinjalamala FK, Kamwendo DD, et al. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002;185:380–8. doi: 10.1086/338566. [DOI] [PubMed] [Google Scholar]
- 7.Plowe CV. The evolution of drug-resistant malaria. Trans R Soc Trop Med Hyg. 2009;103(Suppl 1):S11–4. doi: 10.1016/j.trstmh.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nair S, Williams JT, Brockman A, et al. A selective sweep driven by pyrimethamine treatment in Southeast Asian malaria parasites. Mol Biol Evol. 2003;20:1526–36. doi: 10.1093/molbev/msg162. [DOI] [PubMed] [Google Scholar]
- 9.McCollum AM, Mueller K, Villegas L, Udhayakumar V, Escalante AA. Common origin and fixation of Plasmodium falciparum dhfr and dhps mutations associated with sulfadoxine-pyrimethamine resistance in a low-transmission area in South America. Antimicrob Agents Chemother. 2007;51:2085–91. doi: 10.1128/AAC.01228-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mita T, Tanabe K, Takahashi N, et al. Independent evolution of pyrimethamine resistance in Plasmodium falciparum isolates in Melanesia. Antimicrob Agents Chemother. 2007;51:1071–7. doi: 10.1128/AAC.01186-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Z, Griffing SM, de Oliveira AM, et al. Decline in sulfadoxine-pyrimethamine-resistant alleles after change in drug policy in the Amazon region of Peru. Antimicrob Agents Chemother. 2008;52:739–41. doi: 10.1128/AAC.00975-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roper C, Pearce R, Nair S, Sharp B, Nosten F, Anderson T. Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004;305:1124. doi: 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- 13.McCollum AM, Poe AC, Hamel M, et al. Antifolate resistance in Plasmodium falciparum: multiple origins and identification of novel dhfr alleles. J Infect Dis. 2006;194:189–97. doi: 10.1086/504687. [DOI] [PubMed] [Google Scholar]
- 14.Maiga O, Djimde AA, Hubert V, et al. A shared Asian origin of the triple-mutant dhfr allele in Plasmodium falciparum from sites across Africa. J Infect Dis. 2007;196:165–72. doi: 10.1086/518512. [DOI] [PubMed] [Google Scholar]
- 15.Mita T, Tanabe K, Takahashi N, et al. Indigenous evolution of Plasmodium falciparum pyrimethamine resistance multiple times in Africa. J Antimicrob Chemother. 2009;63:252–5. doi: 10.1093/jac/dkn482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P, Read M, Sims PF, Hyde JE. Sulfadoxine resistance in the human malaria parasite Plasmodium falciparum is determined by mutations in dihydropteroate synthetase and an additional factor associated with folate utilization. Mol Microbiol. 1997;23:979–86. doi: 10.1046/j.1365-2958.1997.2821646.x. [DOI] [PubMed] [Google Scholar]
- 17.Triglia T, Wang P, Sims PF, Hyde JE, Cowman AF. Allelic exchange at the endogenous genomic locus in Plasmodium falciparum proves the role of dihydropteroate synthase in sulfadoxine-resistant malaria. Embo J. 1998;17:3807–15. doi: 10.1093/emboj/17.14.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pearce RJ, Pota H, Evehe MS, et al. Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria. PLoS Med. 2009;6:e1000055. doi: 10.1371/journal.pmed.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vinayak S, Alam MT, Mixson-Hayden T, et al. Origin and evolution of sulfadoxine resistant Plasmodium falciparum. PLoS Pathog. 2010;6:e1000830. doi: 10.1371/journal.ppat.1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mita T, Kaneko A, Hombhanje F, et al. Role of pfmdr1 mutations on chloroquine resistance in Plasmodium falciparum isolates with pfcrt K76T from Papua New Guinea. Acta Trop. 2006;98:137–44. doi: 10.1016/j.actatropica.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Bwijo B, Kaneko A, Takechi M, et al. High prevalence of quintuple mutant dhps/dhfr genes in Plasmodium falciparum infections seven years after introduction of sulfadoxine and pyrimethamine as first line treatment in Malawi. Acta Trop. 2003;85:363–73. doi: 10.1016/s0001-706x(02)00264-4. [DOI] [PubMed] [Google Scholar]
- 22.Reeder JC, Rieckmann KH, Genton B, Lorry K, Wines B, Cowman AF. Point mutations in the dihydrofolate reductase and dihydropteroate synthetase genes and in vitro susceptibility to pyrimethamine and cycloguanil of Plasmodium falciparum isolates from Papua New Guinea. Am J Trop Med Hyg. 1996;55:209–13. doi: 10.4269/ajtmh.1996.55.209. [DOI] [PubMed] [Google Scholar]
- 23.Mita T, Kaneko A, Hwaihwanje I, et al. Rapid selection of dhfr mutant allele in Plasmodium falciparum isolates after the introduction of sulfadoxine/pyrimethamine in combination with 4-aminoquinolines in Papua New Guinea. Infect Genet Evol. 2006;6:447–52. doi: 10.1016/j.meegid.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Roper C, Pearce R, Bredenkamp B, et al. Antifolate antimalarial resistance in southeast Africa: a population-based analysis. Lancet. 2003;361:1174–81. doi: 10.1016/S0140-6736(03)12951-0. [DOI] [PubMed] [Google Scholar]
- 25.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubisz M, Falush D, Stephens M, Pritchard J. Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour. 2009;9:1322–32. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–20. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 28.Rosenberg N. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–8. [Google Scholar]
- 29.Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 30.Wootton JC, Feng X, Ferdig MT, et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–3. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- 31.Ariey F, Fandeur T, Durand R, et al. Invasion of Africa by a single pfcrt allele of South East Asian type. Malar J. 2006;5:34. doi: 10.1186/1475-2875-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alam MT, Vinayak S, Congpuong K, et al. Tracking origins and spread of sulfadoxine-resistant Plasmodium falciparum dhps alleles in Thailand. Antimicrob Agents Chemother. 2011;55:155–64. doi: 10.1128/AAC.00691-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Certain LK, Briceno M, Kiara SM, Nzila AM, Watkins WM, Sibley CH. Characteristics of Plasmodium falciparum dhfr haplotypes that confer pyrimethamine resistance, Kilifi, Kenya, 1987–2006. J Infect Dis. 2008;197:1743–51. doi: 10.1086/588198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naidoo I, Roper C. Following the path of most resistance: dhps K540E dispersal in African Plasmodium falciparum. Trends Parasitol. 2010;26:447–56. doi: 10.1016/j.pt.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 35.Moore DV, Lanier JE. Observations on two Plasmodium falciparum infections with an abnormal response to chloroquine. Am J Trop Med Hyg. 1961;10:5–9. doi: 10.4269/ajtmh.1961.10.5. [DOI] [PubMed] [Google Scholar]
- 36.Harinasuta T, Suntharasamai P, Viravan C. Chloroquine-resistant falciparum malaria in Thailand. Lancet. 1965;2:657–60. doi: 10.1016/s0140-6736(65)90395-8. [DOI] [PubMed] [Google Scholar]
- 37.Hastings IM. The origins of antimalarial drug resistance. Trends Parasitol. 2004;20:512–8. doi: 10.1016/j.pt.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Plowe CV, Kublin JG, Dzinjalamala FK, et al. Sustained clinical efficacy of sulfadoxine-pyrimethamine for uncomplicated falciparum malaria in Malawi after 10 years as first line treatment: five year prospective study. BMJ. 2004;328:545. doi: 10.1136/bmj.37977.653750.EE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson TJ, Haubold B, Williams JT, et al. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–82. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- 40.Ariey F, Duchemin JB, Robert V. Metapopulation concepts applied to falciparum malaria and their impacts on the emergence and spread of chloroquine resistance. Infect Genet Evol. 2003;2:185–92. doi: 10.1016/s1567-1348(02)00099-0. [DOI] [PubMed] [Google Scholar]
- 41.Rathod PK, McErlean T, Lee PC. Variations in frequencies of drug resistance in Plasmodium falciparum. Proc Natl Acad Sci U S A. 1997;94:9389–93. doi: 10.1073/pnas.94.17.9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castellini MA, Buguliskis JS, Casta LJ, et al. Malaria drug resistance is associated with defective DNA mismatch repair. Mol Biochem Parasitol. 2011;177:143–7. doi: 10.1016/j.molbiopara.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gosling RD, Gesase S, Mosha JF, et al. Protective efficacy and safety of three antimalarial regimens for intermittent preventive treatment for malaria in infants: a randomised, double-blind, placebo-controlled trial. Lancet. 2009;374:1521–32. doi: 10.1016/S0140-6736(09)60997-1. [DOI] [PubMed] [Google Scholar]
- 44.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–20. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 45.Dondorp AM, Nosten F, Yi P, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–67. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plowe CV, Roper C, Barnwell JW, et al. World Antimalarial Resistance Network (WARN) III: molecular markers for drug resistant malaria. Malar J. 2007;6:121. doi: 10.1186/1475-2875-6-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sibley CH, Barnes KI, Plowe CV. The rationale and plan for creating a World Antimalarial Resistance Network (WARN) Malar J. 2007;6:118. doi: 10.1186/1475-2875-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.