SUMMARY
Translation of Hepatitis C viral proteins requires an internal ribosome entry site (IRES) located in the 5′ untranslated region of the viral mRNA. The core domain of the Hepatitis C virus (HCV) IRES contains a four-way helical junction that is integrated within a predicted pseudoknot. This domain is required for positioning the mRNA start codon correctly on the 40S ribosomal subunit during translation initiation. Here we present the crystal structure of this RNA, revealing a complex double-pseudoknot fold that establishes the alignment of two helical elements on either side of the four-helix junction. The conformation of this core domain constrains the open reading frame’s orientation for positioning on the 40S ribosomal subunit. This structure, representing the last major domain of HCV-like IRESs to be determined at near-atomic resolution, provides the basis for a comprehensive cryo-electron microscopy-guided model of the intact HCV IRES and its interaction with 40S ribosomal subunits.
INTRODUCTION
Hepatitis C virus (HCV) infects over 170 million people worldwide and if untreated, can lead to liver cirrhosis and hepatocellular carcinoma(Webster et al., 2009). Translation of viral proteins requires the 5′ untranslated region (UTR) of genomic RNA, a 341-nucleotide (nt) region that includes an internal ribosome entry site (IRES; Figure 1A) (Tsukiyama-Kohara et al., 1992; Wang et al., 1993). This structured RNA element directly and specifically interacts with human 40S ribosomal subunits and eukaryotic initiation factor 3 (eIF3) to drive cap-independent translation initiation (Kieft et al., 2001; Pestova et al., 1998; Sizova et al., 1998). The 5′ UTR of HCV RNA contains four domains of significant secondary structure, three of which constitute the IRES (Figure 1A). While the apical portion of domain (dom) III provides high-affinity binding sites for 40S ribosomal subunits and eIF3 (Kieft et al., 2001; Sizova et al., 1998), the pseudoknot domain at the base of domain III (IIIe–f) (Wang et al., 1995) binds at the solvent side of the 40S-subunit platform (Boehringer et al., 2005; Spahn et al., 2001). From here, this domain orients domain IV and the open reading frame (ORF) of the RNA toward the mRNA binding cleft, placing the AUG start codon in the P-site where it base pairs with the initiator tRNA anticodon (Berry et al., 2010).
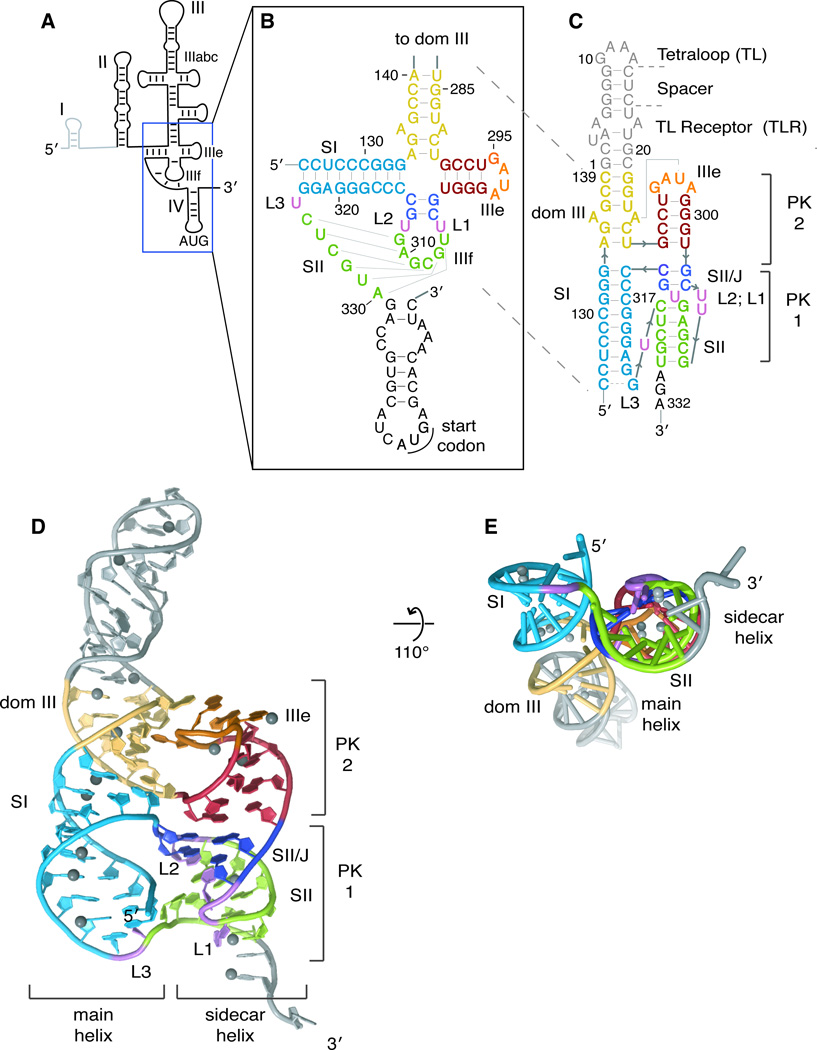
Figure 1. Structure of the HCV IRES pseudoknot domain.
(A) Secondary structure cartoon of the HCV IRES with names of major domains indicated. Dom I is shown in light grey as it is not considered part of the IRES and is not present in the reporter constructs used here. (B) Pseudoknot-domain secondary structure, as conventionally drawn. Loops and stems are indicated by L and S, respectively. (C) Crystallization construct of pseudoknot domain, with secondary structure redrawn according to the crystal structure. Nucleotides from the IRES are numbered as in the IRES and the crystallization-module nucleotides are numbered 1–21. Crystal structure shown (D) head-on and (E) at a 110° angle, looking down the sidecar helix. Ni2+ ions are shown as grey spheres. Secondary structural elements are colored consistently in b–e (dom III, yellow; SI, light blue; IIIe, red; SII/J, deep blue; and SII, green) and the crystallization module (tetraloop, spacer, tetraloop receptor) is shown in grey. The ‘main’ and ‘sidecar’ helices consisting of dom III + SI and IIIe+SII/J+SII, respectively, are labeled. Pseudoknot (PK) 1 is composed of loop IIIf and downstream sequence and PK2 is formed between the IIIe tetraloop and the main helix of dom III. See also Figure S1.
The pseudoknot domain is located at the center of the HCV IRES (Boehringer et al., 2005; Spahn et al., 2001), connecting domains II and III with the AUG-containing domain IV. The pseudoknot consists of three base-paired stems, SI, SII and SII/J, linked by three predicted single-uridine loops, L1-L3, and by a four-way junction between SI, SII/J, IIIe and dom III (Figures 1B and 1C). SII is proposed to comprise six base pairs between nucleotides in loop IIIf and downstream of the 3′ end of SI to generate a pseudoknot; base pairing throughout SII of the pseudoknot contributes to AUG-positioning and translation initiation activity (Berry et al., 2010). While SII of the HCV IRES pseudoknot domain is not necessary for IRES-40S subunit binding, it is absolutely required for efficient translation activity by mediating a downstream step to correctly orient domain IV (Berry et al., 2010; Kieft et al., 2001). This domain is the most highly structured region of the IRES (Kieft et al., 1999), and is therefore at both the structural and functional core of the IRES. Despite its importance, the molecular structure of this critical domain is unknown.
Cryo-electron microscopy (cryo-EM) reconstructions have revealed that the IRES binds to ribosomes in an elongated conformation in which domain III binds on the solvent side of the 40S subunit and domain II reaches toward the interface surface and into the E-site (Boehringer et al., 2005; Spahn et al., 2001). Significant progress has also been made towards determining the structures of individual domains of the HCV IRES RNA at high resolution, revealing the molecular basis for certain aspects of IRES function (Collier et al., 2002; Kieft et al., 2002; Lukavsky et al., 2003; Lukavsky et al., 2000; Rijnbrand et al., 2004; Zhao et al., 2008). However, the lack of any HCV IRES pseudoknot-domain structure has prevented high-resolution modeling of the complete IRES. Moreover, due to its high conservation and a critical role in viral translation, the pseudoknot domain is a desirable drug target. Detailed structural information about this domain would therefore greatly aid the design of new HCV therapeutics.
Here we report the crystal structure of the HCV IRES pseudoknot domain at 3.6 Å resolution. The structure consists of a complex four-way junction of non-parallel, coaxially stacked helices that, together with a non-canonical tertiary interaction between a tetraloop and neighboring helix, control the orientation of the start codon-containing mRNA strand via the SII helix. This structure reveals the molecular basis for pseudoknot-domain-mediated start-codon positioning by the HCV IRES.
RESULTS
Structural Overview
After screening a large panel of designed crystallization constructs, we chose a construct containing the core of the pseudoknot domain and a tetraloop/tetraloop receptor (TL/TLR) as a crystallization module (Figure 1C) (Ferre-D'Amare et al., 1998). We crystallized this RNA and determined its structure at 3.6 Å resolution (Table 1). Phase information was obtained from a multi-wavelength anomalous dispersion (MAD) experiment exploiting the anomalous scattering of cocrystallized nickel (II) ions, which bound predominantly at guanine N7 positions in the major groove of the RNA (Figure S1). Unambiguous repeating ridges were present in the experimentally determined electron density map, corresponding to the electron-rich phosphate groups of the RNA backbone (Figure S1). These ridges allowed A-form helices to be placed into the electron density with confidence, even though density for individual base pairs was not resolved (see Experimental Procedures). The engineered tetraloop and tetraloop receptor interact within the crystal as intended, with neighboring molecules contacting one another in a head-to-tail manner throughout the crystal, giving rise to the four-fold screw axis in the P41212 space group (Figure S1). The crystal structure clearly shows a pseudoknot topology, with base pairing between the IIIf loop and downstream sequence forming the predicted SII (Figure 1C and 1D) (Berry et al., 2010; Fletcher and Jackson, 2002; Rijnbrand et al., 1997; Wang et al., 1995). Globally, the pseudoknot domain folds to form two sets of coaxial helices: the “main helix” of dom III stacks on top of SI, while the IIIe helix stacks on top of the two base-pair SII/J helix and SII to form a “sidecar helix” that runs alongside the main helix (Figure 1D). Note that SII/J was previously referred to as SI/J (Berry et al., 2010), but has been renamed due to its stacking with SII in the structure. While the formation of SII is similar to a classic H-type pseudoknot, the intervening four-way junction and third stem (SII/J) generate a complex and unusual RNA fold. The IIIe tetraloop within the sidecar helix forms a base pair with an upstream sequence in the main helix, constituting a pseudoknot itself, making this RNA domain a double-pseudoknot (Figures 1C and 1D). While the main and sidecar helices are each roughly coaxially stacked, the axes do not run parallel to one another. The helical stacks are tilted by ~40 degrees with respect to one another (Figure 1E), contrary to the crystal structure of the other four-way junction (JIIIabc) in the HCV IRES (Kieft et al., 2002), although JIIIabc adopts multiple conformations in solution (Melcher et al., 2003).
Table 1.
Data collection and refinement statistics
| Native | Ni2+ peak | Ni2+ remote | |
|---|---|---|---|
| Data collection | |||
| X-ray Source | ALS 8.3.1 | ALS 8.3.1 | ALS 8.3.1 |
| Wavelength | 1.1159 | 1.4855 | 1.4370 |
| Space group | P 41212 | P 41212 | P 41212 |
| Cell dimensions | |||
| a, b, c (Å) | 113.39, 113.39, 88.22 | 112.75, 112.75, 86.85 | |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | |
| Resolution (Å) | 50-3.55(3.64-3.55)* | 50-4.70 (4.82-4.70) | 50-4.70 (4.82-4.70) |
| Rsym | 6.8 (75.7) | 5.9 (91.3) | 5.4 (77.9) |
| I / σ | 17.46 (3.02) | 23.81 (3.44) | 26.38 (4.01) |
| Completeness (%) | 99.5 (99.8) | 99.8 (100.0) | 99.8 (100.0) |
| Redundancy | 11.2 (8.7) | 15.2 (15.5) | 15.1 (15.4) |
| Refinement | |||
| Resolution (Å) | 3.55 | ||
| No. reflections | 82099 | ||
| Rwork / Rfree | 0.221/0.254 | ||
| No. atoms | |||
| RNA | 1796 | ||
| Ligand/ion | 17 | ||
| B-factors | |||
| RNA | 148.5 | ||
| Ligand/ion | 165.8 | ||
| r.m.s. deviations | |||
| Bond lengths (Å) | 0.008 | ||
| Bond angles (°) | 0.769 |
Values in parentheses are for highest-resolution shell.
Structure of the four-way junction
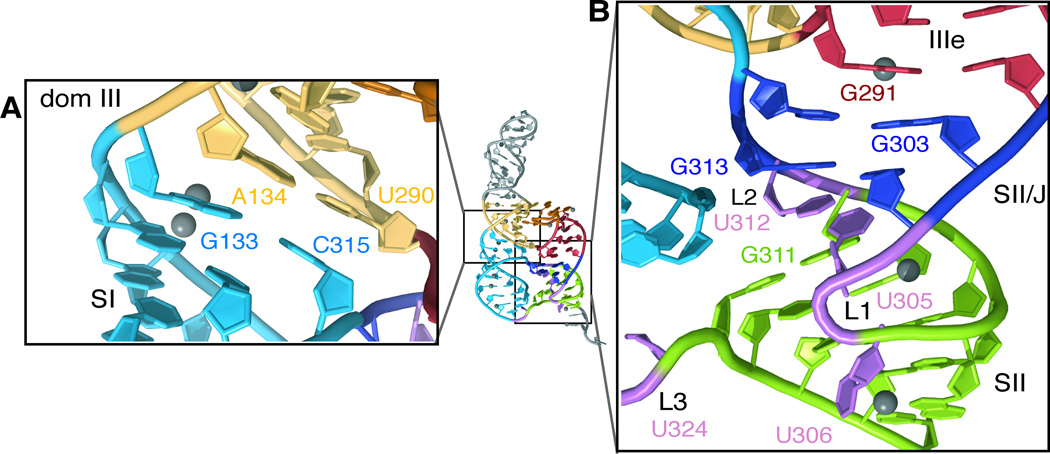
The four-way junction connects two sets of globally stacked helices with multiple contacts in between. The SI and dom III helices adopt a perfect coaxial stack, with the top base pair of SI directly underneath the bottom base pair of dom III (Figure 2A). By contrast, the coaxial stacking between IIIe, SII/J and SII in the sidecar helix is more complex. The base pairs of SII/J are underwound and shifted slightly ajar from those in the IIIe stem, facilitated by cross-strand purine-purine stacking between G291 and G303 (Figure 2B). A single U (U312 in L2) inserts between G311 and G313 at the respective termini of SII and SII/J, allowing continuous base stacking between these two stems (Figure 2B) and preserving the overall coaxial arrangement of SII with IIIe (Figure 1E). The base of U305 also stacks underneath the SII/J base pairs, such that U305 and U312 may participate in a U-U wobble base pair. Given that the IRES retains 85% of wild type (WT) translation activity when both of these uridines are mutated simultaneously to adenosines (Berry et al., 2010), it appears that this potential U-U base pair is not functionally required.
Figure 2. Structure of the IIIef four-way junction.
(A) Close-up of coaxial stacking between the main stem of dom III (yellow) and SI (blue). (B) Close-up of complex coaxial stacking within the sidecar helix, between IIIe (red), SII/J (deep blue) and SII (green). Note that the base of U324 (L3) is omitted from the crystallographic model, as no density was observed for this nucleotide apart from the sugar-phosphate backbone. Ni2+ ions are shown as grey spheres.
A role for sequence non-specific base stacking by U312 (L2) is consistent with a previous mutational study that found that the sequence of L2 is not critical for robust translation activity by the HCV IRES; either deletion of L2 or lengthening of the loop by two nucleotides severely inhibited translation by the IRES (29% and 11% of WT, respectively) (Berry et al., 2010). Functionally, L2 (U312) stands in contrast to U305 of L1, which can be deleted without detrimental effect, and especially L3 (U324), which can be deleted or lengthened by three nts while retaining >70% of WT IRES-mediated translation activity (Berry et al., 2010). These findings are consistent with the observed structural roles for L1 and L3, as both are substantially more solvent-exposed than L2. Indeed, L3 is so flexible even in the context of the crystal that, while the path of the sugar-phosphate backbone between SI and SII is clearly defined, no electron density is observed for the uridine of L3 (U324).
Tertiary interactions of the IIIe tetraloop
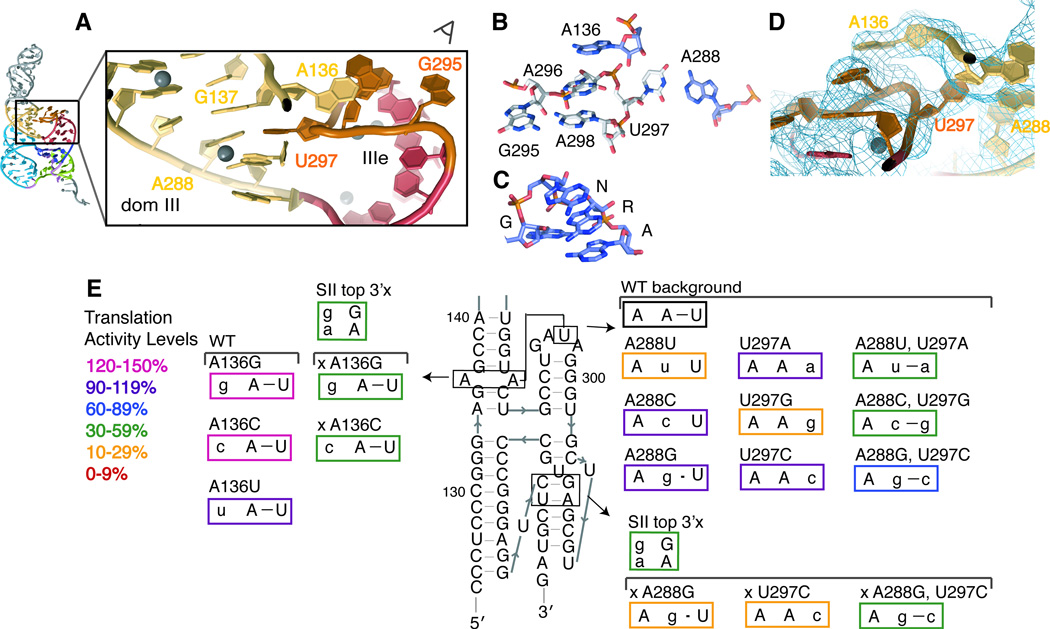
The primary sequence of the IIIe tetraloop is conserved as GA[U/C]A between HCV genotypes and HCV-like IRESs (Easton et al., 2009). The crystal structure (Figure 3A) reveals that the G and A form a base pair at the bottom of the tetraloop, as they would in a canonical GNRA tetraloop. Instead of the sheared G-A pair observed for GNRA-type tetraloops, however, the IIIe tetraloop contains a Saenger type X G-A base pair (Leontis and Westhof, 2001) between the Watson-Crick face of A298 and the sugar edge (N3/N2) of G295 (Figures 3B and 3C). Previous sequence alignments revealed that the sequence of the bulged purine at position 288 covaries as a Watson-Crick base-pair partner with the pyrimidine at nt 297, the third position in this tetraloop. This observation led to the proposal of an interhelical base pair between nts 288 and 297(Easton et al., 2009). Consistent with this prediction, the atypical pyrimidine in the third position flips out of the tetraloop and base pairs with A288, one of two bulged purines in the main stem of dom III.
Figure 3. Structure of the IIIe tetraloop tertiary interaction.
(A) Close-up view of the interaction between the IIIe tetraloop and the main stem of dom III. Ni2+ ions are shown as grey spheres. (B) Stick representation of the four tetraloop nucleotides and the two dom III adenosines which directly participate in tertiary interactions with the tetraloop, viewed along the sight line in (A), contrasted against (C) a canonical GNRA tetraloop, taken from PDB 1HMH. (D) 2Fo-Fc electron density map contoured at 1.0 σ around the tetraloop structure, viewed in a similar orientation as in (B). Density for each nucleotide in this tertiary interaction returns when it is omitted from the model during refinement. (E) Mutational analysis of tetraloop tertiary interactions. Mutated nucleotides are shown in lowercase and mutants are boxed in colors according to their translation activity, relative to WT, as defined in the legend. Those mutations which were made in the disrupted pseudoknot background of the SII top 3′x mutant are named beginning with an ‘x.’
The flipping out of U297 from the tetraloop leaves only three nts participating in the tetraloop structure, in contrast to the canonical four. The stacking interactions generally fulfilled by the ‘N’ nt of the GNRA tetraloop appears to be furnished by the sequence-distant A136, the other bulged purine in the dom III stem, which flips over from the neighboring strand to lie on top of A296 and A298 (Figures 3A, 3B and 3D). To test this model and the resulting sequence-independence at position 136, site-specific mutations were made in an HCV IRES-firefly luciferase (FF luc) reporter construct and tested for translation activity in a rabbit reticulocyte translation extract system that faithfully recapitulates IRES-driven translation(Berry et al., 2011). When mutated to any of the other three nucleotides, translation activity remained at least 99% of WT, confirming a lack of sequence requirement at this position (Table 2 and Figure 3E).
Table 2.
Translation activities of IRES mutants
| Mutant | Translation Activity a | Mutant | Translation Activity a |
|---|---|---|---|
| WT | 100 ± 7 %b | WT | 100 ± 7 %b |
| SII penult 5′x | 19 ± 4 % | A136G | 134 ± 16 % |
| SII penult 3′x | 74 ± 11 % | A136C | 134 ± 6 % |
| SII penult comp | 61 ± 5 % | A136U | 99 ± 10 % |
| SII ult 5′x | 103 ± 4 % | A288U | 20 ± 3 % |
| SII ult 3′x | 75 ± 7 % | U297A | 104 ± 8 % |
| SII ult comp | 70 ± 8 % | A288U,U297A | 32 ± 9 % |
| SII end comp | 50 ± 1 %c | A288C | 98 ± 10 % |
| SI end GU | 98 ± 12 % | U297G | 24 ± 3 % |
| SI end AU | 93 ± 7 % | A288C,U297G | 50 ± 4 % |
| SI penult 5′x | 38 ± 4 % | A288G | 107 ± 5 % |
| SI penult 3′x | 87 ± 3 % | U297C | 118 ± 12 % |
| SI penult comp | 109 ± 4 % | A288G,U297C | 93 ± 8 % |
| SI ult 5′x | 106 ± 4 % | SII top 3′x | 40 ± 9 %c |
| SI ult 3′x | 84 ± 4 % | x A288G | 22 ± 4 % |
| SI ult comp | 100 ± 4 % | x U297C | 10 ± 3 % |
| x A288G, U297C | 45 ± 12 % | ||
| x A136G | 50 ± 3 % | ||
| x A136C | 32 ± 1 % |
reported as the mean and standard deviation between four translation reactions from two independent transcriptions.
WT error is the mean deviation of WT from all duplicate translation reactions.
previously reported (Berry et al., 2010)
To test the functional importance of the observed tertiary interactions surrounding the IIIe tetraloop, the A288:U297 interhelical base pair was systematically disrupted by point mutations or restored by compensatory mutations in HCV IRES-FF luc reporter RNAs. When purine/pyrimidine identity is maintained (A288G, U297C, and A288G:U297C), all mutant IRESs maintain near WT translation activity (>90%; Table 2 and Figure 3E). This result stands in contrast to the previous observation that the disruption of this base pair leads to ~50% translation activity in a genotype 1a HCV IRES (Easton et al., 2009) (see Discussion).
We hypothesized that this interhelical A288-U297 base pair stabilizes the orientation of SI and SII within the pseudoknot-domain structure. This interaction may be required for full IRES function when the pseudoknot is intrinsically less stable, as it may be in other HCV genotypes or HCV-related IRESs like PTV-1(Easton et al., 2009), or under different translation conditions. To test this idea, we made use of a previously identified destabilized mutant (SII top 3′x) that disrupts the top two base pairs of SII, but maintains 40% translation activity of WT (Berry et al., 2010). In the background of this destabilized pseudoknot, disruption of the proposed A288:U297 base pair from either side leads to a drop of translation activity from 40% to 22% or 10% (Table 2 and Figure 3E). The compensatory mutation, A288G:U297C, restores translation activity to 45%, confirming the functional importance of the interhelical IIIe base pair in a compromised pseudoknot domain. Given that the compromised pseudoknot was important to reveal interactions that stabilize the IIIe tertiary interaction, we reexamined the sequence-independence of the flipped A136 in this context. As predicted, both A136G and A136C mutations are still well tolerated in the background of the destabilized mutant (Table 2 and Figure 3E).
In a WT IRES background, alteration of purine/pyrimidine identity to disrupt the interhelical A288-U297 base pair leads to substantially impaired translation activity in certain cases. Both A288U and U297G mutants yield ~20% translation activity of WT, while A288C and U297A maintain WT-like translation activity (Table 2 and Figure 3E). We have shown, however, that Watson-Crick base pairing is not required for high translation activity in a fully stable IRES, suggesting that the severely compromised mutants may arise from RNA misfolding. Furthermore, both of the compensatory mutants in which purine/pyrimidine identity has been switched (A288U:U297A and A288C:U297G) have only modest translation activity (32% and 50%, respectively). This suggests that the purine-pyrimidine orientation of the interhelical base pair is important for the formation of the correct tertiary structure, most likely due to cross-strand stacking between A288 and G137 (Figure 3A).
Probing solution structure and dynamics
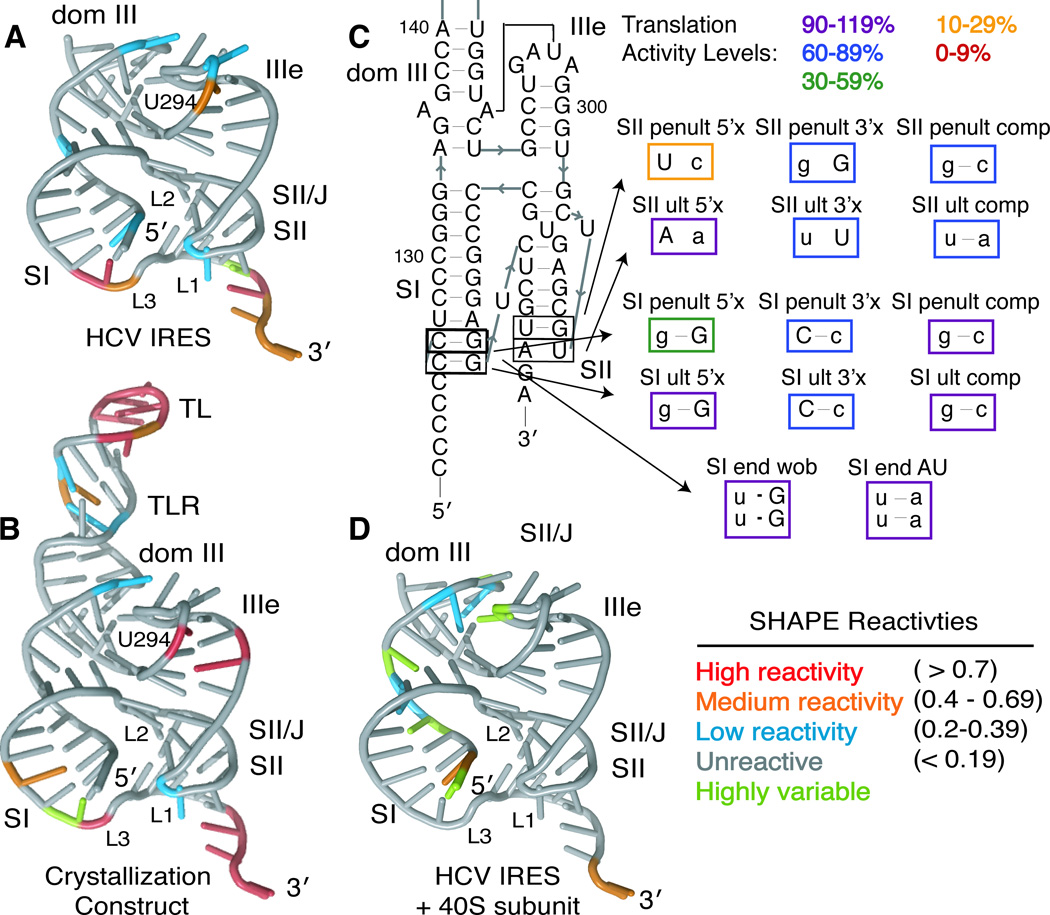
To examine the structure of the pseudoknot domain in the context of the full-length HCV IRES in solution, we utilized SHAPE (Selective 2′-hydroxyl acylation analyzed by primer extension) chemistry to analyze the flexibility of each nucleotide in the folded RNAs (McGinnis et al., 2009). The reactivities of the majority of nucleotides within the crystallization construct and full-length IRES were reproducible between experiments (Figure S2). SHAPE reactivities of the pseudoknot domain both alone and within the full-length HCV IRES are in good agreement with the crystal structure and with each other (Figures 4A, 4B and S2). The GAAA tetraloop and the tetraloop receptor of the crystallization construct have high and moderate SHAPE reactivities, respectively; this is expected since the SHAPE experiments are conducted at concentrations well below the high micromolar Kd of the TL/TLR interaction (Qin et al., 2001). The IIIe tetraloop has low to moderate reactivity in both the full-length IRES and crystallization construct, consistent with involvement in a constraining tertiary interaction (Mortimer and Weeks, 2007). Indeed, U294, part of a wobble base pair just beneath the tetraloop is more flexible than any of the nucleotides in the tetraloop itself. The L3 loop is much more reactive than L1 or L2, indicating flexibility in this loop and consistent with the poorly resolved electron density for the L3 uridine base.
Figure 4. Solution structural probing and pseudoknot-domain dynamics.
SHAPE modification reactivities of individual nucleotides from the (A) full-length HCV IRES or (B) the crystallization construct are mapped onto the crystal structure, colored according to reactivity (see legend). (C) Mutational analysis of the ultimate and penultimate predicted base pairs of SI and SII. Mutated nucleotides are shown in lowercase and mutants are boxed in colors according to their translation activity, relative to WT, as defined in the legend. (D) SHAPE modification reactivities of individual nucleotides from the full-length HCV IRES in the presence of 40S ribosomal subunits are shown on the crystal structure, colored according to reactivity (see legend). See also Figure S2.
The SHAPE reactivities also suggest that the termini of both SI and SII are flexible and that the predicted terminal base pair of each stem may not form in solution. Consistent with the SHAPE data, electron density at the terminus of SII suggests that nts U306 and A330 are splayed apart in the crystal (Figure S1); while full base pairing is observed in SI, the B-factors at the terminus of SI are very high (>250 Å2) and base pairing may be partially driven by crystal packing (Figure S1). Functionally, disruption of the ultimate base pair in each stem is less deleterious than disruption of the penultimate base pair (Table 2 and Figure 4C). This confirms that the predicted ultimate base pair of both SI and SII are not necessary for a fully functional pseudoknot domain and that L1 and L3 may each be 2 nts in length (Figure 1C). Based on the previous finding that switching purine/pyrimidine identity at the eighth and ninth predicted base pairs of SI substantially decreased translation activity, we tested the translation activity of mutants in which these GC base pairs were changed to either GU or AU base pairs. These mutants, which maintain purine/pyrimidine identity, retain >90% translation activity of WT (Table 2 and Figure 4C). Rather than a specific tertiary interaction requiring the GC sequence at these positions, it seems that purine/pyrimidine identity is important in these base pairs, perhaps due to a long stretch of purines stacking favorably in SI (Tilton et al., 1983).
40S-induced structural changes
We wondered how the pseudoknot domain structure may be altered when the IRES interacts with 40S ribosomal subunits in the translation initiation pathway. To investigate this, we repeated SHAPE probing on the full-length IRES in the presence of a saturating concentration of 40S ribosomal subunits purified from HeLa cells. Reactivities in the entire IIIe stem-loop are decreased, as are those of nucleotides in the terminus of SII and in L1 and L3 (Figure 4D vs. 4A). In addition, the AUG and surrounding nucleotides show a substantial increase in flexibility (Figure S2), consistent with the unfolding of domain IV to facilitate translation initiation (Filbin and Kieft, 2011; Honda et al., 1996). We also observe that certain nucleotides in the 5′ sides of SI and dom III in the pseudoknot domain display increased flexibility in the presence of 40S ribosomal subunits, though this may be due to experimental noise (see Discussion).
Structural model of the full-length HCV IRES
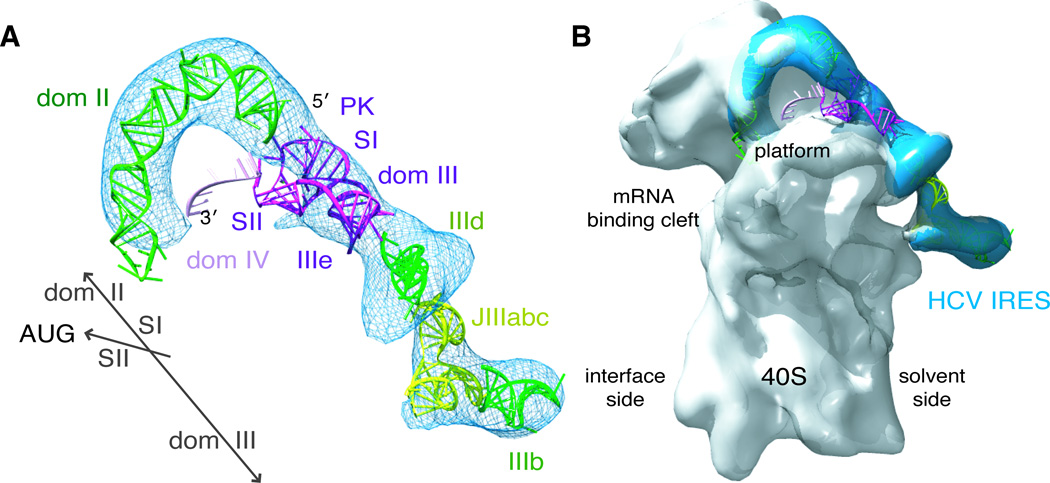
The structures of many individual domains of the HCV IRES have been solved by NMR and x-ray crystallography (Collier et al., 2002; Kieft et al., 2002; Lukavsky et al., 2003; Lukavsky et al., 2000; Rijnbrand et al., 2004; Zhao et al., 2008). These previous structures along with our structure of the pseudoknot domain can be fit into a cryo-EM difference density map of the HCV IRES (Siridechadilok et al., 2005), producing a model of the HCV IRES including all major domains bound to the 40S ribosomal subunit (Figure 5). Domains II and III bind to the 40S ribosomal subunit in an elongated fashion, with domain II extending towards the ribosomal E-site and domain III running along the solvent side of the 40S subunit (Boehringer et al., 2005; Spahn et al., 2001). The majority of the pseudoknot domain structure fits well into the cryo-EM HCV IRES density between density previously assigned to domains II and IIId (Boehringer et al., 2005; Spahn et al., 2001). In primary sequence, SI of the pseudoknot connects the rest of domain III with domain II. Thus, SI is placed along the main axis of the IRES, in line with domains II and III. As discussed above, tertiary interactions position the sidecar helix at a ~40 degree angle relative to SI. Although there is no clear density in the cryo-EM difference density map that would correspond to SII of the pseudoknot, this SI/SII orientation is well configured to place SII on top of the platform domain of the 40S subunit. The lack of clear density for SII of the pseudoknot could be due to flexibility of this region and/or an intimate interaction with the 40S ribosomal subunit. This structural model shows how the pseudoknot domain acts as a connector piece that couples the overall binding of the IRES with precise positioning of domain IV in the mRNA binding cleft.
Figure 5. Structural model of the full HCV IRES.
(A) The pseudoknot-domain structure (dark purple), along with previously determined crystal and NMR structures of dom II (1P5P), IIId (1F84), JIIIabc (1KH6) and IIIb (1KP7), was manually modeled into the into the cryo-EM difference density from an HCV IRES-eIF3 reconstruction (blue mesh) (Siridechadilok et al., 2005) and (B) overlayed with a cryo-EM reconstruction of the 40S-HCV IRES (Spahn et al., 2001). SI is placed in line between dom II and IIId, according to primary sequence. SII is placed so that it can interact with the platform domain of the 40S subunit, according to the established function of the pseudoknot domain in controlling the placement of the open reading frame in the mRNA binding cleft. Dom IV is modeled as ssRNA (light purple) downstream of the pseudoknot domain, as the hairpin unfolds upon 40S binding. This figure was generated using Chimera (Goddard et al., 2007). See also Figure S3.
DISCUSSION
The pseudoknot domain of the HCV IRES forms the structural and functional core of the RNA that engages with 40S ribosomal subunits and positions the start codon during viral translation initiation (Berry et al., 2010; Boehringer et al., 2005; Easton et al., 2009; Spahn et al., 2001; Wang et al., 1995; Wang et al., 1994). We show here that this domain fittingly adopts the most complex structure of any domain within the HCV IRES, forming a double-pseudoknot surrounding a four-way helical junction. A cryo-EM-guided model of the full-length HCV IRES and its interaction with 40S ribosomal subunits shows how the pseudoknot domain is positioned at the center of the HCV IRES and connects the start codon-containing domain IV to the two other major domains of the IRES, thus enabling correct open reading frame placement on the 40S subunit.
The structure of the pseudoknot domain is more complex than anticipated. Whereas overall configuration of stems in the crystal structure agrees with a previous computational model for the HCV IRES pseudoknot domain (Figure S1) (Lavender et al., 2010), the precise register of SI and SII differs from the prediction and the crystal structure reveals details not present in the computational model. For example, the sidecar and main helices form several specific contacts with one another at the four-way junction and the IIIe tertiary interaction. It will be of interest in the future to further investigate the conformational flexibility of these stems in silico or in solution.
The most striking structural feature of the HCV IRES pseudoknot domain is the IIIe tetraloop tertiary interaction. The sequence of each of the four IIIe tetraloop nucleotides is required for efficient IRES translation (Psaridi et al., 1999). In the crystal structure, base-swapping between the IIIe tetraloop and the dom III helix involves full U297 insertion into the dom III helix without disruption of the helical axis or base pairing. While the resolution of the diffraction data requires that the position of individual base pairs within A-form helices be inferred based on the position of phosphate ridges, the electron density surrounding this important IIIe tetraloop structural feature is quite clear (Figure 3d) and density for each nucleotide in this tertiary interaction returns when it is omitted from the model during refinement. The observed IIIe tetraloop conformation is distinct from that of the IIIe hairpin alone (Lukavsky et al., 2000), indicating that tertiary interactions influence tetraloop folding. This interaction explains the previous observation that magnesium ions induce protection of the IIIe tetraloop from RNase cleavage, which is indicative of tertiary structure formation (Kieft et al., 1999). In contrast to previous results that suggested a critical role for the IIIe tetraloop interaction in the HCV genotype 1a and PTV-1 IRESs (Easton et al., 2009), we find that this interaction is only required for efficient IRES-mediated translation in the context of a destabilized HCV IRES. These seemingly contradictory results may be due to variances between genotype 1a and 1b sequences, although the pseudoknot domain itself is completely conserved between these genotypes, or due to differences in the translation conditions between the studies.
SHAPE structural probing verifies that the structure observed in the crystals also forms in the full-length IRES in solution. Furthermore, the crystal structure and SHAPE data reveals that the pseudoknot-stem termini are more dynamic than previously thought. While Watson-Crick base pairing in the terminal two base pairs of SII is required for efficient IRES-mediated translation (Berry et al., 2010), electron density, solution flexibility and functional studies demonstrate here that only the penultimate base pair forms and that L2 is actually 2 nts long. Whereas enzymatic probing showed that SII is sensitive to both single-stranded and double-stranded RNases (Fletcher et al., 2002; Kolupaeva et al., 2000; Wang et al., 1995), SHAPE analysis reveals that the top five base pairs of SII form a well-folded helix under the conditions used here. In addition to the SHAPE data, we compared our crystal structure to previous chemical probing data identifying regions of protection from hydroxyl radicals in the HCV IRES, representing areas of tertiary structure (Kieft et al., 1999). Consistent with the observed structure, strong protections were reported at the four way junction itself, at the top of SII, across SII/J, and the bottom of SI where these stems approach one another, and in dom III where the IIIe tetraloop interacts.
The HCV IRES engages directly with 40S ribosomal subunits during translation initiation (Kieft et al., 2001; Pestova et al., 1998). Decreases in SHAPE reactivity upon 40S subunit binding in the IIIe stem-loop and the terminus of SII could be due to 40S-stabilized base pairing in these regions or to direct protections of the IRES RNA by the 40S subunit. The latter interpretation is consistent with previous data showing that the presence of 40S subunits protects G295 in IIIe and the predicted U306-A330 base pair in SII from DMS-modification and phosphorothioate-iodine cleavage, respectively (Kieft et al., 2001; Lukavsky et al., 2000). Increased SHAPE reactivities of nucleotides on the 5′ side of SI and dom III observed in the presence of 40S subunits may be due to increased flexibility of these helical regions upon binding to 40S subunits, or to experimental noise as this portion of the SHAPE chromatograms displayed markedly more noise in the presence than in the absence of 40S subunits (Figure S2; cf. ref. 25). In the absence of 40S subunits, the reactivities of domain IV observed in this work are inconsistent with other studies (Filbin and Kieft, 2011; Honda et al., 1996), while the SHAPE reactivities for the full-length IRES are generally in good agreement with the IRES secondary structure (Figure S2). This may be due to Mg2+ concentration or the presence of 30 nts of luciferase coding region or the 3′ SHAPE handle. Upon 40S subunit binding, we observe that nucleotides at and beyond the start codon become more reactive, whereas the nucleotides between the pseudoknot and AUG are protected. This stabilization of the upstream portion of domain IV in the mRNA binding cleft has recently been reported to depend on the apical sequence of domain IIb (Filbin and Kieft, 2011). It is possible that additional structural changes in the pseudoknot domain are not detectable due to protection by the 40S subunit. Nevertheless, aside from the unfolding of domain IV upon 40S subunit binding and potential loosening of the SI helix, our data do not support any large-scale conformational rearrangements of the pseudoknot domain upon binding to the 40S subunit.
Positioning the pseudoknot domain and other IRES domains into a cryo-EM model for the IRES-40S subunit complex (Spahn et al., 2001) provides insights into pseudoknot-domain-mediated positioning of the open reading frame in the ribosomal mRNA binding cleft. The pseudoknot domain, located between domain II of the IRES and the rest of domain III, arranges the main and sidecar helices to orient SII with respect to the overall axis of the IRES. Based on functional studies (Berry et al., 2010) and the path of mRNA through the ribosome (Yusupova et al., 2001), we infer that SII binds to the back of the platform domain. This location, similar to that occupied by the Shine-Dalgarno sequence on 30S ribosomal subunits, enables the pseudoknot domain to present domain IV to the mRNA binding cleft. Previous work using site-specific crosslinking between the IIIe tetraloop and 40S ribosomal subunits identified S3a, S5 and S16 as ribosomal proteins in the vicinity of IIIe; these results largely agree with our cryo-EM guided 40S-IRES model when compared to the 40S Tetrahymena thermophila crystal structure (Figure S3) (Laletina et al., 2006; Rabl et al., 2011). Although insertion of the photoactivatable cross-linker into the IIIe tetraloop may have disrupted the pseudoknot-domain tertiary structure, crosslinking of the IIIe tetraloop to S3a is consistent with our model and proteins S5 and S16 are reasonably nearby as well.
The majority of the pseudoknot-domain structure fits well into the HCV IRES density observed in IRES-eIF3 and IRES-40S subunit complexes (Siridechadilok et al., 2005; Spahn et al., 2001). It is less clear how the pseudoknot-domain crystal structure fits into the IRES difference density from the cryo-EM reconstruction of the IRES in complex with the 80S ribosome (Boehringer et al., 2005). The semi-parallel nature of the pseudoknot stems seen in the crystal structure makes it difficult to imagine how the structure would fit into the large, perpendicular L-shaped density proposed for the pseudoknot domain within the difference density from this complex. This bent density could reflect other conformational changes between elongating 80S and IRES-bound 80S ribosomes. Alternatively, there could be significant conformational rearrangements of the pseudoknot domain upon start-codon positioning and subunit joining. A recent computational model of the pseudoknot domain suggested that domain IV may be perpendicular to SI and SII of the pseudoknot (Lavender et al., 2010). While our structure provides no direct evidence about the relative orientations of domain IV and SII, it does seem sterically possible for domain IV to coaxially stack with SII. However, the domain IV hairpin likely unfolds upon initial 40S subunit binding (Filbin and Kieft, 2011). Our cryo-EM based model for the HCV IRES does not resolve the previous observation that the stems around the IIIabc junction may be oriented differently in solution than in the crystal structure (Boehringer et al., 2005; Kieft et al., 2002; Melcher et al., 2003), although the parallel arrangement of stems fits better into the IRES density when bound to the 40S subunit or to eIF3 than when bound to the 80S ribosome (Boehringer et al., 2005; Siridechadilok et al., 2005; Spahn et al., 2001). The fact that the IIId domain NMR structure does not completely fill the cryo-EM density assigned to it may also be a matter of incorrect segmentation of density from the IRES-eIF3 complex (Siridechadilok et al., 2005).
Due to its high sequence conservation across genotypes and its essential function for viral propagation, the HCV IRES is an attractive, although challenging, drug target (Berry et al., 2011; Gallego and Varani, 2002; McHutchinson et al., 2006). We have previously proposed that the pseudoknot domain in particular would be a promising drug target, as small disruptions in its conformation block translation activity without blocking association of 40S subunits, and we have validated the pseudoknot domain as a drug target using complementary 2′OMe-oligonucleotides to disrupt the structure (Berry et al., 2010). The crystal structure solved here sets the stage for future work to search for small molecules that specifically interact with and disrupt the pseudoknot domain, either through computational docking of small molecule libraries or through binding-based assays (Hermann and Westhof, 2000; Seth et al., 2005). The surface of the figure eight-shaped double-pseudoknot presents two large cavities (Figure S1) that could serve as initial binding pockets for a small molecule to disrupt the structure. This crystal structure represents a significant step forward in our understanding of the molecular basis of this critical process for viral translation by HCV.
EXPERIMENTAL PROCEDURES
RNA Crystallization
For cloning, transcription and purification of RNA for crystallography, see Supplemental Experimental Procedures. Crystals were grown using the hanging drop vapor diffusion method; RNA was mixed in a 1:1 ratio with well solution (0.5–0.6 M Li2SO4, 30–37 mM MgCl2, 1–2 mM spermine, 50 mM MES, pH 6.5) with an additional 10 mM NiCl2 added to the drop before equilibration over well solution at 18°C. Crystals appeared after 2–5 d and were either harvested directly or dehydrated by placing the drop above well solution supplemented with 5% PEG3350 for 16 h before harvesting. Crystals were cryo-protected in well solution supplemented with 10 mM NiCl2 and 35% ethylene glycol and were flash cooled in liquid nitrogen. The crystals belonged to space group P41212 and contained 1 molecule per asymmetric unit with 84% solvent content.
Data Collection and Phasing
X-ray diffraction was collected at beamline 8.3.1 at the Advanced Light Source (Lawrence Berkeley National Laboratory) under cryo-conditions. Native data were collected using 1° oscillations and 1 s and 10 s exposures at 1.116 Å. A two-wavelength MAD experiment was conducted (peak: 1.486 Å, between the peak and inflection; remote: 1.437 Å) using the inverse beam method, and alternations between peak and remote wavelengths every 1°. Data were processed in XDS(Kabsch, 2010). Twelve initial nickel sites were located using AutoSHARP(Vonrhein et al., 2007) and initial phase estimates (FOM = 0.57) were improved with density modification within AutoSHARP and using DM(Cowtan, 1994) (FOM = .80). Data measurement and refinement statistics are summarized in Table 1.
Model Building and Refinement
Density for the tetraloop and tetraloop receptor could be unambiguously assigned in the initial solvent flattened experimental maps, and additional regions of A-form helical density were assigned to other helical elements of the pseudoknot domain to produce an initial model, built in COOT (Emsley and Cowtan, 2004). While 3.6Å is a modest resolution, the repeating electron-rich phosphates and regular structure of A-form helices made it much easier to model RNA with confidence at this resolution than to model protein. Unambiguous ridges corresponding to phosphates were evident in the experimentally determined electron density maps, allowing the register of A-form helices to be determined. The initial model, with hydrogens explicitly modeled, was refined against the 3.55 Å native dataset using Phenix (Adams et al., 2010) through iterative rounds of energy minimization, individual coordinate refinement, torsion angle simulated annealing, group atomic displacement factor (ADP) refinement (1 ADP group per residue), and manual rebuilding in COOT with the help of B-factor sharpening. Although individual base pairs were not initially identifiable within the electron density, the use of B-factor sharpening of 2Fo-Fc maps revealed holes in electron density between base pairs, further confirming their positions. In later rounds of refinement, base pairs were restrained once they were unambiguously identified in the model, nickel occupancies were refined, the relative stereochemistry/xray weights were optimized and stringent acceptable r.m.s. deviations were enforced (bonds = 0.01, angles = 1.5); the interhelical A288:U297 base pair was never restrained. One round of translation, libration, screw (TLS) refinement was used with 6 TLS groups consisting of SI, dom III, crystallization module, IIIe, SII/J and SII. Electron density maps were generated in Phenix with missing F(obs) reflections replaced with F(calc) values.
All 84 nucleotides in the crystallization construct could be built into the electron density, with the exception of a single nucleotide (U324) for which electron density for only the sugar-phosphate backbone could be seen. Thus, the base of U324 was omitted from the model. In addition, the density around U306 in L2 was somewhat ambiguous, as two symmetry-related copies of this position are placed directly next to each other in three-dimensional space and there is additional density that likely belongs to unaccounted for ligands (Figure S1). The position of U306 has been modeled to the best of our ability, and while other conformations are possible, in no case would a base pair between U306 and A330 be consistent with the observed electron density. Treatment of Nickel ions during refinement is described in Supplemental Experimental Procedures. The final rms deviations of the model from ideal bond lengths and angles are ~0.008 Å and ~0.769°, respectively, and the final overall B-factor is 149 Å2 (Table 1), which is not uncommonly high for RNA structures at modest resolution.
In vitro translation reactions
Site-directed mutagenesis of the WT pKB84 HCV IRES-FF luc reporter construct, luciferase RNA transcription, purification, and in vitro translation reactions in salt-adjusted RRL were performed as previously described (Berry et al., 2010).
SHAPE Modification Chemistry
The crystallization construct with 5′- and 3′-handles and full-length IRES with a 3′-handle attached were transcribed, purified and annealead as described in Supplemental Experimental Methods (Wilkinson et al., 2006). Folded RNA samples were placed at room temperature for 10 min before reaction with 1-methyl-7-nitroisatoic anhydride (1M7). For the full-length IRES, 40S ribosomal subunits (7.5 pmol) or 40S storage buffer was added to folded RNAs and 9 µL reactions were brought to final concentrations of 50 mM Hepes 7.5, 90 mM KCl, 2.3 mM MgCl2 and 1.0 mM DTT, heated at 37°C for 10 min for 40S activation and incubated at room temperature for 10 min prior to reaction with 1M7. The 40S ribosomal subunits were in 1.5-fold excess of IRES RNA at concentrations >250-fold the Kd of the interaction to ensure binding (Kieft et al., 2001).
SHAPE modification, reverse transcription, capillary electrophoresis, and data processing and normalization were performed largely as previously described (McGinnis et al., 2009), with minor modifications (see Supplemental Experimental Procedures).
HIGHLIGHTS.
-
-
Crystal structure of the most complex HCV IRES domain reveals its functional core
-
-
A tertiary base pair creates a double pseudoknot encompassing a four-helix junction
-
-
Final domain to be solved facilitates comprehensive IRES model bound to 40S subunit
-
-
Orientation of two helical elements directs start-codon positioning by the IRES
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Doudna Laboratory for helpful discussions and critical reading of the manuscript; D. Sashital (UC Berkeley) help with crystallographic data collection; B. Wiedenheft (UC Berkeley) for assistance with cryo-EM models; M. Jinek (UC Berkeley) for invaluable assistance with data processing and structure refinement; T. Chen, C. Onak (UC Berkeley), and N. Husain (California Institute of Technology) for assistance with the screening of initial crystallography constructs; James Holton and George Meigs at Beamline 8.3.1 at the Advanced Light Source (Lawrence Berkeley National Laboratory) for expert assistance with data collection and processing; E. Westhof (University of Strasbourg) for helpful discussion; N. Echols (Lawrence Berkeley National Laboratory) for assistance with refinement in Phenix; and Richard Shan at Quintara Biosciences for generously running capillary electrophoresis on our samples. HeLa cytoplasmic lysate was a kind gift from the laboratory of R. Tjian (UC Berkeley). S.A.M. is a fellow of the Leukemia and Lymphoma Society. This work was supported by a Program project grant from the National Institutes of Health and a research gift generously provided by Gilead, Inc. (to J.A.D.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Atomic coordinates and structure factors have been deposited with the Protein Data Bank under accession code 3T4B.
SUPPLEMENTAL INFORMATION
Supplemental Information includes three figures and Supplemental Experimental Procedures and can be found with this article online at doi:.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry KE, Peng B, Koditek D, Beeman D, Pagratis N, Perry JK, Parrish J, Zhong W, Doudna JA, Shih IH. Optimized high-throughput screen for hepatitis C virus translation inhibitors. J. Biomol. Screen. 2011;16:211–220. doi: 10.1177/1087057110391665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry KE, Waghray S, Doudna JA. The HCV IRES pseudoknot positions the initiation codon on the 40S ribosomal subunit. RNA. 2010;16:1559–1569. doi: 10.1261/rna.2197210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure. 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Collier AJ, Gallego J, Klinck R, Cole PT, Harris SJ, Harrison GP, Aboul-Ela F, Varani G, Walker S. A conserved RNA structure within the HCV IRES eIF3-binding site. Nat. Struct. Biol. 2002;9:375–380. doi: 10.1038/nsb785. [DOI] [PubMed] [Google Scholar]
- Cowtan K. An automated procedure for phase improvement by density modifcation. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- Easton LE, Locker N, Lukavsky PJ. Conserved functional domains and a novel tertiary interaction near the pseudoknot drive translational activity of hepatitis C virus and hepatitis C virus-like internal ribosome entry sites. Nucleic Acids Res. 2009;37:5537–5549. doi: 10.1093/nar/gkp588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ferre-D'Amare AR, Zhou K, Doudna JA. A general module for RNA crystallization. J. Mol. Biol. 1998;279:621–631. doi: 10.1006/jmbi.1998.1789. [DOI] [PubMed] [Google Scholar]
- Filbin ME, Kieft JS. HCV IRES domain IIb affects the configuration of coding RNA in the 40S subunit's decoding groove. RNA. 2011 doi: 10.1261/rna.2594011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher SP, Ali IK, Kaminski A, Digard P, Jackson RJ. The influence of viral coding sequences on pestivirus IRES activity reveals further parallels with translation initiation in prokaryotes. RNA. 2002;8:1558–1571. [PMC free article] [PubMed] [Google Scholar]
- Fletcher SP, Jackson RJ. Pestivirus internal ribosome entry site (IRES) structure and function: elements in the 5' untranslated region important for IRES function. J. Virol. 2002;76:5024–5033. doi: 10.1128/JVI.76.10.5024-5033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego J, Varani G. The hepatitis C virus internal ribosome-entry site: a new target for antiviral research. Biochem. Soc. Trans. 2002;30:140–145. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Hermann T, Westhof E. Rational drug design and high-throughput techniques for RNA targets. Comb. Chem. High Throughput Screen. 2000;3:219–234. doi: 10.2174/1386207003331652. [DOI] [PubMed] [Google Scholar]
- Honda M, Brown EA, Lemon SM. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Cryst D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Grech A, Jubin R, Doudna JA. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat. Struct. Biol. 2002;9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- Kolupaeva VG, Pestova TV, Hellen CU. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J. Virol. 2000;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laletina ES, Graifer DM, Malygin AA, Shatskii IN, Karpova GG. Molecular environment of the subdomain IIIe loop of the RNA IRES element of hepatitis C virus on the human 40S ribosomal subunit. Bioorg Khim. 2006;32:311–319. doi: 10.1134/s1068162006030101. [DOI] [PubMed] [Google Scholar]
- Lavender CA, Ding F, Dokholyan NV, Weeks KM. Robust and generic RNA modeling using inferred constraints: a structure for the hepatitis C virus IRES pseudoknot domain. Biochemistry. 2010;49:4931–4933. doi: 10.1021/bi100142y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- McGinnis JL, Duncan CD, Weeks KM. High-throughput SHAPE and hydroxyl radical analysis of RNA structure and ribonucleoprotein assembly. Methods Enzymol. 2009;468:67–89. doi: 10.1016/S0076-6879(09)68004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHutchinson JG, Bartenschlager R, Patel K, Pawlotsky JM. The face of future hepatitis C antiviral drug development: Recent biological and virologic advances and their translation to drug development and clinical practice. J. Hepatol. 2006;44:411–421. doi: 10.1016/j.jhep.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Melcher SE, Wilson TJ, Lilley DM. The dynamic nature of the four-way junction of the hepatitis C virus IRES. RNA. 2003;9:809–820. doi: 10.1261/rna.5130703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psaridi L, Georgopoulou U, Varaklioti A, Mavromara P. Mutational analysis of a conserved tetraloop in the 5' untranslated region of hepatitis C virus identifies a novel RNA element essential for the internal ribosome entry site function. FEBS Lett. 1999;453:49–53. doi: 10.1016/s0014-5793(99)00662-6. [DOI] [PubMed] [Google Scholar]
- Qin PZ, Butcher SE, Feigon J, Hubbell WL. Quantitative analysis of the isolated GAAA tetraloop/receptor interaction in solution: a site-directed spin labeling study. Biochemistry. 2001;40:6929–6936. doi: 10.1021/bi010294g. [DOI] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- Rijnbrand R, Thiviyanathan V, Kaluarachchi K, Lemon SM, Gorenstein DG. Mutational and structural analysis of stem-loop IIIC of the hepatitis C virus and GB virus B internal ribosome entry sites. J. Mol. Biol. 2004;343:805–817. doi: 10.1016/j.jmb.2004.08.095. [DOI] [PubMed] [Google Scholar]
- Rijnbrand R, van der Straaten T, van Rijn PA, Spaan WJ, Bredenbeek PJ. Internal entry of ribosomes is directed by the 5' noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J. Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth PP, Miyaji A, Jefferson EA, Sannes-Lowery KA, Osgood SA, Propp SS, Ranken R, Massire C, Sampath R, Ecker DJ, et al. SAR by MS: discovery of a new class of RNA-binding small molecules for the hepatitis C virus: internal ribosome entry site IIA subdomain. J. Med. Chem. 2005;48:7099–7102. doi: 10.1021/jm050815o. [DOI] [PubMed] [Google Scholar]
- Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. Specific interaction of eukaryotic translation initiation factor 3 with the 5' nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- Tilton RF, Jr, Weiner PK, Kollman PA. An analysis of the sequence dependence of the structure and energy of A- and B-DNA models using molecular mechanics. Biopolymers. 1983;22:969–1002. doi: 10.1002/bip.360220316. [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- Wang C, Le SY, Ali N, Siddiqui A. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5' noncoding region. RNA. 1995;1:526–537. [PMC free article] [PubMed] [Google Scholar]
- Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Sarnow P, Siddiqui A. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J. Virol. 1994;68:7301–7307. doi: 10.1128/jvi.68.11.7301-7307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster DP, Klenerman P, Collier J, Jeffery KJ. Development of novel treatments for hepatitis C. Lancet Infect. Dis. 2009;9:108–117. doi: 10.1016/S1473-3099(09)70020-9. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Han Q, Kissinger CR, Hermann T, Thompson PA. Structure of hepatitis C virus IRES subdomain IIa. Acta Crystallogr. D Biol. Crystallogr. 2008;64:436–443. doi: 10.1107/S0907444908002011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.