Abstract
Percutaneous coronary intervention (PCI) has become an effective therapy to treat obstructive coronary artery diseases (CAD). However, one of the major drawbacks of PCI is the occurrence of restenosis in 5–25% of all initially treated patients. Restenosis is defined as the re-narrowing of the lumen of the blood vessel, resulting in renewed symptoms and the need for repeated intervention. To identify genetic variants that are associated with restenosis, a genome-wide association study (GWAS) was conducted in 295 patients who developed restenosis (cases) and 571 who did not (controls) from the GENetic Determinants of Restenosis (GENDER) study. Analysis of ∼550 000 single nucleotide polymorphisms (SNPs) in GENDER was followed by a replication phase in three independent case–control populations (533 cases and 3067 controls). A potential susceptibility locus for restenosis at chromosome 12, including rs10861032 (Pcombined = 1.11 × 10−7) and rs9804922 (Pcombined = 1.45 × 10−6), was identified in the GWAS and replication phase. In addition, both SNPs were also associated with coronary events (rs10861032, Padditive = 0.005; rs9804922, Padditive = 0.023) in a trial based cohort set of elderly patients with (enhanced risk of) CAD (PROSPER) and all-cause mortality in PROSPER (rs10861032, Padditive = 0.007; rs9804922, Padditive = 0.013) and GENDER (rs10861032, Padditive = 0.005; rs9804922, Padditive = 0.023). Further analysis suggests that this locus could be involved in regulatory functions.
INTRODUCTION
Percutaneous coronary intervention (PCI) for unblocking a narrowed coronary artery is a widely used technique for treating patients with angina or an acute coronary event. Initially, PCI was performed only with balloon catheters, but technical advances made it possible to improve patient outcome by the placement of bare metal stents (BMS), or later, drug eluting stents (DES) at the site of blockage (1–3). Patients undergoing PCI may suffer from a re-narrowing of the treated lesion, which is called restenosis, with renewed symptoms and the need for repeated intervention, typically within 3 to 6 months (4). Restenosis occurs in ∼5–25% of all treated patients depending on individual characteristics and the techniques used (1–3,5,6), thereby still causing a significant clinical and economic burden for patients and society.
So far, the etiological basis of restenosis is only partly understood. The injury induced by PCI within the vascular wall causes segmental thrombus formation and subsequent invasion of macrophages and polymorphonuclear leukocytes in the blood vessel. This process is followed by release of numerous growth factors from blood cells and stretched smooth muscle cells that lead to the proliferation of smooth muscle cells in the treated lesion (4,7,8).
In order to prevent restenosis, numerous systemic drugs have been studied for their inhibitory effect on smooth muscle cell proliferation but the results have been inconclusive (9). A new solution has been the development of DES (10,11). DES are made by applying a drug (such as sirolimus or paclitaxel) on a coronary stent. The drug is released directly into the (by PCI) injured area and is thereby preventing the inflammatory response and smooth muscle cell proliferation at the site of the coronary intervention. Although DES have decreased the incidence of restenosis, restenosis still occurs in all instances (12,13). Moreover, although several clinical factors, lesion-related, procedural and biological markers have been shown to be associated with an elevated risk of restenosis (4,14–16), these associations have not been consistently replicated in all studies.
There is evidence that systemic factors can explain part of the risk of restenosis independently of conventional clinical factors or procedures. For instance, in patients with multilesion interventions, the risk that a lesion develops restenosis is 2.5 times higher when a companion lesion developed restenosis, independently of clinical factors (17). The influence of genetic polymorphisms in the development of restenosis has been investigated by means of candidate gene approaches (18–21), with interesting, although sometimes controversial or inconsistent results. For instance, an insertion/deletion polymorphism in the angiotensin-converting enzyme was associated with restenosis within a French population (22), but not in a German or a Dutch population (23,24).
We conducted the first genome-wide association study (GWAS) to identify new genetic risk factors for restenosis in a subpopulation of the GENetic DEterminants of Restenosis (GENDER) study. This was followed by a replication phase in three independent restenosis case–control populations (replication I, replication II and replication III). In a secondary step, we tested if the most associated single nucleotide polymorphisms (SNPs) were also associated with other relevant clinical outcomes such as cardiovascular events and all-cause mortality in GENDER and in the PROspective Study of Pravastatin in the Elderly at Risk (PROSPER) population. This additional validation effort may add worthwhile information with regard to the genetic factors involved in the pathophysiology of restenosis.
RESULTS
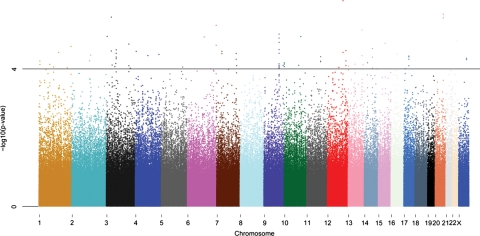
The overall GWAS results are summarized in a Manhattan plot (Fig. 1). We found that 91 SNPs were associated with restenosis at P-value <10−4 assuming the additive model (Supplementary Material, Table S1). In the replication stage, these 91 SNPs were genotyped in three restenosis populations, two BMS cohort (replication I and replication II) and a DES cohort (replication III) (Table 1). Associations found in the discovery set were replicated for an intergenic region on chromosome 12 (Table 2). This was specially found for replication I (Table 2). In replication I, rs10861032 showed an association with restenosis [P = 3.99 × 10−4; odds ratio (OR) = 3.06, 95% confidence interval (CI) (1.64–5.70)]; nominally significant at P-value <0.05 even after Bonferroni correction (0.05/91 SNPs = 5.49 × 10−4). Using the Fisher's trend combined P-value method, we observed that rs10861032 C allele was potentially associated with higher risk of restenosis (Padditive = 1.11 × 10−7) in all four populations combined. We computed I2 as a measurement of heterogeneity and we obtained a value of 77.04% for rs10861032 which can be interpreted as the percentage of total variation across studies due to heterogeneity (25). A similar association for all four combined populations was observed for rs9804922 (Padditive = 1.45 × 10−6) for the risk allele (T).
Figure 1.
Manhattan plot of the GWAS performed in the GENDER study. P-values were obtained by the Cochrane–Armitage trend test for 556 099 SNPs and 866 individuals (295 cases and 571 controls) are plotted in –log10 scale according to their chromosomal position. A horizontal black line indicates an additive P-value of 10−4.
Table 1.
Baseline clinical characteristics of GENDER (discovery population), replication I, replication II and replication III
| Variables | GENDERa, n = 866 | P-valueb | Replication Ia, n = 265 | P-valueb | Replication IIc, n = 1445 | P-valueb | Replication IIIc, n = 1890 | P-valueb |
|---|---|---|---|---|---|---|---|---|
| Age (years) | 62 ± 11 | 64.5 ± 11.35 | 64 ± 11 | 66 ± 11 | ||||
| Sex (male) | 636 (73.44%) | 0.50 | 182 (68.68%) | 0.001 | 1110 (76.81%) | 0.78 | 1478 (79.04%) | 0.67 |
| Stentingd | 584 (67.43%) | 0.95 | 265 (100%) | 1 | 1445 (100%) | 1 | 1890 (100%) | 1 |
| Diabetes | 117 (20.43%) | 0.75 | 97 (36.60%) | <0.001 | 301 (20.83%) | 0.97 | 508 (26.88%) | 0.58 |
| Current smokere | 216 (24.94%) | 0.40 | 147 (55.47%) | 0.05 | 453 (31.34%) | 0.41 | 276 (14.60%) | 0.98 |
| Hypercholesterolemia | 520 (60.04%) | 0.84 | 198 (74.72%) | 0.93 | 624 (43.18%) | 0.21 | 1341(70.95%) | 0.51 |
| Total occlusion | 154 (17.78%) | 0.45 | 14 (5.28%) | 0.31 | 232 (16.05%) | 0.18 | 144 (7.62%) | 0.045 |
| Residual stenosis (>20%) | 112 (12.93%) | 0.07 | 11 (4.15%) | 0.53 | 54 (3.37%) | 0.92 | 48 (2.54%) | 0.08 |
n, number of individuals in each cohort.
aEndpoint: clinical restenosis.
bP-values computed between cases and controls for each variable.
cEndpoint: TLR within 1 year after PCI.
dGENDER: replication I and replication II are BMS cohorts. Replication III is a DES cohort.
eIn replication I, the data represented in this table are ever smoke and not current smoker.
Table 2.
Association results for rs10861032 and rs9804922 in GENDER (discovery set), replication I, replication II and replication III
| Population | N (case/control) | Chr | SNP | Position | Allelesa | MAF (case/control) | Padditiveb | OR (CI 95%)c |
|---|---|---|---|---|---|---|---|---|
| GWAS | 295/571 | 12 | rs10861032 | 102436636 | C/T | 0.212/0.133 | 3.29E−05 | 1.75 (1.35–2.27) |
| Replication I | 78/187 | 0.192/0.099 | 3.98E−04 | 3.06 (1.64–5.70) | ||||
| Replication II | 275/1170 | 0.159/0.147 | 4.94E−01 | 1.09 (0.85–1.41) | ||||
| Replication III | 180/1710 | 0.219/0.177 | 4.89E−02 | 1.31 (1.00–1.7) | ||||
| Combined | 1.11E−07 | |||||||
| GWAS | 295/571 | 12 | rs9804922 | 102437572 | T/C | 0.114/0.049 | 1.03E−06 | 2.48 (1.72–3.60) |
| Replication I | 78/187 | 0.077/0.043 | 0.063 | 2.26 (0.97–2.14) | ||||
| Replication II | 275/1170 | 0.077/0.074 | 8.83E−01 | 1.03 (0.72–1.46) | ||||
| Replication III | 180/1710 | 0.119/0.092 | 1.03E−01 | 1.33 (0.95–1.87) | ||||
| Combined | 1.45E−06 |
Combined P-values are computed by means of Fisher's trend method. Positions are based on hg18 build.
aThe first allele is the minor allele. MAF, minor allele frequency.
bResults of the Cochran–Armitage test.
cOR of the minor allele from the two by two allele frequency table.
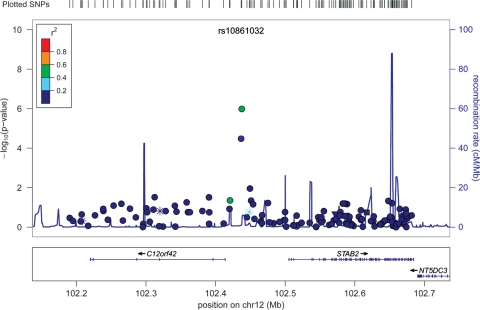
In addition, both SNPs were also associated with all-cause mortality in GENDER (rs10861032, Padditive = 0.005; rs9804922, Padditive = 0.031) and in the PROSPER study (rs10861032, Padditive = 0.007; rs9804922, Padditive = 0.013) (Tables 3 and 4), and also with coronary events (rs10861032, Padditive = 0.005; rs9804922, Padditive = 0.023) in PROSPER. Both rs10861032 and rs9804922 are located on an intergenic region. An open reading frame (C12orf42), a hypothetical protein of unknown function, is located 22.7 kb upstream of both SNPs, the gene STAB2 (Stabiline2) is located 68.5 kb downstream and the gene NT5DC3 253.5 kb downstream (Fig. 2). Non-genotyped SNPs in the 610-quad array were imputed from HapMap CEU reference panel (release 22) within a 500 kb window centered on rs10861032. Analysis on 566 imputed and 140 genotyped SNPs within this region revealed three more imputed SNPs that were associated with restenosis, rs4147305 [P = 1.72 × 10−4, OR = 2.1 95% CI (3.14–1.41)], rs17034045 [P = 1.72 × 10−4 OR = 2.1 95% CI (3.14–1.41)] and rs10861033 (P = 8.91 × 10−4 OR = 1.55 95% CI [2.02–1.20)]. These SNPs are in LD with rs10861032 (r2 = 0.37, r2 = 0.37 and r2 = 0.85, respectively).
Table 3.
Association results with coronary events and all-cause mortality for rs10861032 and rs9804922 in PROSPER
| Padditive | HR (95% CI) | |
|---|---|---|
| rs10861032 (MAF = 0.14) | ||
| Coronary events | 0.005 | 1.25 (107–1.46) |
| All-cause mortality | 0.007 | 1.25 (1.06–1.47) |
| rs9804922 (MAF = 0.06) | ||
| Coronary events | 0.023 | 1.30 (1.04–1.62) |
| All-cause mortality | 0.013 | 1.33 (1.06–1.67) |
Adjusted for sex, age, country and pravastatin used.
MAF, minor allele frequency; HR, hazard ratio.
Table 4.
Association results for all-cause mortality for rs10861032 and rs9804922 in GENDER
| Padditive | HR (95% CI) | |
|---|---|---|
| rs10861032 (MAF = 0.16) | ||
| All-cause mortality | 0.005 | 1.39 (1.10–1.74) |
| rs9804922 (MAF = 0.07) | ||
| All-cause mortality | 0.031 | 1.42 (1.03–1.94) |
Adjusted for sex and age.
MAF, minor allele frequency; HR, hazard ratio.
Figure 2.
Description of the loci on chromosome 12 associated with restenosis in the GWAS. On the y-axis, the –log10 (P-value) is depicted. The most significant SNP in the meta-analysis (rs10861032) is plotted in purple. LD is based on the HapMap CEU sample and is color-code as red (r2 from 0.8 to 1.0), orange (0.6–0.4), green (0.6–0.4), white blue (0.4 to 0.2) and dark blue (0.2 to 0). Recombination rate is depicted in light blue. Asterisks display TF site. The plot was generated using LocusZoom (53).
Inspection of the UCSC genome browser (http://genome.ucsc.edu) database revealed that rs10861032 maps 110 bp downstream and rs9804922 457 bp upstream to a DNase I hypersensitivity site (chr12:102436746–102437115). In addition, the promoter 2.0 Prediction Server predicts with high scores that the surrounding region of the restenosis associated SNP could be a promoter region. Moreover, the is-rSNP algorithm (26) for in silico detection of genetic variation which affect the ability of a transcription factor (TF) to bind to DNA predicts that both SNPs alter the binding affinity of TFs to the DNA (Supplementary Material, Table S2). Alleles of rs10861032 alter the binding affinity of Htlf (Helicase-like TF) (P = 4.4 × 10−3) and Pax4 (P = 5.7 × 10−3). Members of Htlf family have helicase and ATPase activities and are thought to regulate transcription of certain genes by altering the chromatin structure around those genes (27). Pax4 is a member of the paired box (PAX) family of TFs. These genes play critical roles in cancer growth (28).
Alleles of rs9804922 alter the binding affinity of TP53 (4.8 × 10−4) and HoxA (3.3 × 10−4). TP53 is a tumor suppressor protein that regulates the cell cycle known to be very important in the process of restenosis (sirolimus and paclitaxel work by inhibiting the cell cycle) (29,30). HoxA belongs to the cluster A of the called homeobox genes which may regulate gene expression, morphogenesis and differentiation (31).
We have also checked if both SNPs are expression quantitative loci (eQTLs) and regulate the expression level of genes at the locus (cis-regulation). This experiment was performed using cis expression-genotype data derived from 1469 human whole blood samples reflecting primary leukocyte gene expression (32). However, in a window of 1 Mb surrounding rs10861032 and rs9804922, no significant eQTL effect was detected.
DISCUSSION
To our knowledge, this is the first GWAS that investigates the association between genetic markers and restenosis. Restenosis is a complex phenotype in which the individual contribution of genes to the development of the disease might probably be relatively small and therefore difficult to detect (16). Using the GENDER population as a discovery set, we found several genomic regions that were associated with restenosis. However, this study presents some potential caveats. First, we combined patients that underwent PCI either by balloon angioplasty alone, BMS or DES as restenosis was and still is the major drawback of PCI in all these three cohort populations. The fact that we found similar results in the different cohorts independently of whether they used BMS or DES indicates that it is unlikely that the involvement of chromosome 12q23.3 region in the pathophysiology of restenosis is based only on the inhibitory effect on smooth muscle cell proliferation of the drugs coating the DES surface, which is only expected in subjects with DES. Secondly, it is also important to point out that in this study we have combined cohorts with two clinical endpoints [clinical restenosis (discovery set and replication I) and target lesion revascularization (TLR, replication II and replication III)]. Although both endpoints are highly comparable, it should be noted that clinical restenosis, although nowadays considered the most clinically relevant endpoint, is somewhat a broader endpoint for restenosis than TLR. The fact that the replication is mainly in replication I could indicate that the top SNPs are mainly associated with clinical restenosis and not with TLR. Finally, it must be noticed that none of the detected SNPs shows traditional GWAS threshold significance. Nevertheless, the fact that similar association trends were observed in some of these regions when using other data sets suggests that these regions could be indeed potentially involved in the restenosis phenotype. From these SNPs, a region on chromosome 12q23.3 comprising rs10861032 and rs9804922 was replicated in three other restenosis populations, two BMS cohorts and a DES cohort (Pcombined = 1.11 × 10−7 and Pcombined = 1.45 × 10−6, respectively, when considering all four populations). Interestingly, both SNPs (rs10861032 and rs9804922) are also associated with all-cause mortality in GENDER and in PROSPER and also with coronary events in PROSPER (no data available for this endpoint in GENDER), which shows that probably this region on chromosome 12 may play a more general role in the development of coronary artery diseases. These findings require further research to disentangle whether the SNPs associated not only with restenosis but also with cardiovascular events and all-cause mortality are for instance the result of a pleiotropic effect of single alleles affecting multiple phenotypes.
The associated locus on 12q23.3 is an intergenic region flanked upstream by C12orf42 and downstream by the gene STAB2 located 68.5 kb and the gene NT5DC3 located 253.5 kb away from rs10861032 (Fig. 2). The STAB2 gene encodes for a transmembrane receptor protein which may function in angiogenesis, lymphocyte homing, cell adhesion and receptor scavenging (33–35). All these processes are described to be of importance in the development of restenosis and coronary events (16), thereby making it a very plausible candidate. NT5DC3 encodes for a protein involved in the progression of pancreatic cancer (36), therefore it is a gene involved in cell proliferation, an important biological process for the development of restenosis.
The top associated SNP in this study, rs10861032, is in low linkage disequilibrium (LD) (r2< 0.2) with SNPs in STAB2, NT5DC3 and C12orf42 (Fig. 2). Moreover, rs10861032 flanks upstream to a DNaseI hypersensitivity site (chr12:102436746–102437115), a universal feature of active cis-regulatory sequences, including promoters, insulators, enhancers, boundary elements and locus control regions (37,38). Furthermore, the region flanked by rs10861032 and rs9804922 is likely to be a promoter region as predicted by the promoter 2.0 Prediction Server (39). In addition, the is-rSNP algorithm (26) predicts that alleles of both SNPs alter the binding affinity of several TFs to the DNA, making them likely to be regulatory SNPs. Furthermore, it has recently been shown that ‘gene desert’ regions found by GWAS can be involved in cis-regulatory functions (40–42). This could also be the case of rs10861032 and rs9804922 in the regulation of the expression of STAB2 or NT5DC3, indicating that the region, associated with restenosis on chromosome 12, might be involved in the regulation of the expression of these two genes. However, all these bioinformatic predictions would require wet lab confirmation; in fact, analysis of gene expression and genetic variation on a 1 Mb region surrounding both rs10861032 and rs9804922 in 1469 whole blood samples (32) did not find any significant eQTL effect. Nevertheless, this result does not preclude cis-regulating effect in the case of restenosis, as this analysis was performed in whole blood and not yet in a restenosis population and eQTL effects are often cell-type specific (43). It might be possible that the associated SNPs affect gene expression in a more specific tissue such as coronary or even carotid tissue. These tissues should be further investigated in order to disentangle the possible regulatory functions of rs10861032 and rs9804922 in the development of restenosis.
In conclusion, we have performed the first GWAS to look for genetic variants associated with the development of restenosis after PCI in the GENDER study followed by three independent replication steps and we have identified association for rs10861032 at the 12q23.3 region. The SNPs, rs10861032 and rs9804922, are also associated with all-cause mortality in GENDER and in PROSPER and also with coronary events in PROSPER which indicates that this region might play an important role in the broader range of coronary events. Further research will be needed to disentangle the biological implication of this region in restenosis.
MATERIALS AND METHODS
We investigated the association between genetic variation and clinical restenosis in patients from four different cohort studies, a Dutch population (GENDER), an American population (replication I) and two populations from Germany, a BMS population (replication II) and a DES population (replication III). The GWAS was performed in the GENDER study (discovery set), whereas the replication was performed in replication I, replication II and replication III. In addition, the PROSPER and the GENDER studies were used to check whether the top associated SNPs found in the discovery set and validated in the replication cohorts are involved in other relevant clinical outcomes such as all-cause mortality and cardiovascular events.
All studies were approved by the medical ethical committees of the participating hospitals, had independent clinical event committees who adjudicated the endpoints in a blinded way. Blood samples were collected at the index procedure for DNA isolation after having obtained written informed consent from the patient and the trials were conducted in concordance with the Declaration of Helsinki.
Genome-wide association study
The main characteristics of the GENDER population have been described previously (1). Briefly, 3104 consecutive symptomatic patients treated successfully by PCI for angina were included in four referral centers for interventional cardiology in the Netherlands. The follow-up protocol included a phone contact or a medical visit at the outpatient clinic at 30 days and around 9 months after stent placement. Clinical restenosis was defined as renewed symptoms requiring target vessel revascularization either by repeated PCI or coronary artery bypass graft surgery, by death from cardiac causes or myocardial infarction not attributable to another coronary event than the target vessel (1). Within the 9-month follow-up period, 346 patients developed clinical restenosis. Clinical restenosis was not angiographically confirmed.
The GWAS was performed in 325 cases of restenosis (all cases with enough quality DNA to perform the experiment) and 630 matched controls. Cases and controls were matched by gender, age and some possible confounding clinical variables for restenosis in the GENDER study, such as total occlusion, diabetes, current smoking and residual stenosis (Table 1).
All-cause mortality long-term follow-up data were collected in March 2011 and are defined as death from any cause and the information was collected from death certificates. Mean time follow-up was 9.5 years (SD = 3 years) and 239 (27.9%) patients died during the study. From the patients analysed in the GWAS, only nine subjects (1.0%) were not possible to long-term follow-up.
Genotyping and quality control
In the discovery stage, we conducted the genotyping using Illumina Human 610-Quad Beadchips and the infiniumII assay following manufacturer's instructions. These beadchips contain 620 901 SNPs and copy number variants covering 89% of the common genetic variation in the European population at r2> 0.8. Initially, 955 samples (325 cases and 630 controls) from the GENDER study were genotyped. After excluding bad-performing samples (call rate < 0.98 and manually checked sample quality by means of B-allele frequency and LogR ratio), genotype cluster definitions for each SNP were determined using the ‘Cluster all SNPs’ option in Illumina BeadStudio Genotype module version 3.2. Genotype calls were made when a genotype yielded a quality metric (Gencall score) of 0.15 or higher. The final raw data set released from Beadstudio (eliminating intensity probes only) contained reliable called genotypes for 592 186 SNPs and 941 samples (321 cases and 620 controls), with 14 samples not released due to inadequate quality of genotypes. The remaining samples have a call rate ≥0.99. Additional quality-control measurements were then performed using PLINK (44). Seven samples (four cases and three controls) were excluded due to sex discrepancies between the recorded sex and the inferred sex by the X-chromosome genotypes. We checked for the presence of population substructure in the GENDER study by means of multidimensional scaling (MDS). An identical by state distance matrix was calculated for each pair of individuals along with the 940 individuals from the human genome diversity project-centre d'Etude du polymorphisme humain (HGDP-CEPH) panel (45). We observed that the vast majority of the individuals fell in the same cluster along with the European population, but 67 individuals were outside this cluster and were considered genetic outliers (see Supplementary Material, Fig. S1). One sample was eliminated because it showed a close genetic relationship with another sample from the GENDER study. Furthermore, we excluded SNPs for further analyses with a call rate lower than 95% (n = 1731), with a minor allele frequency lower than 1% (n = 34250) or with a significant deviation from Hardy–Weinberg equilibrium (HWE) in controls (P< 0.00001). The final data set consisted of 866 (295 cases, 571 controls) individuals and 556 099 SNPs that passed all quality-control criteria.
We applied the genomic-control method on the GWAS data and found that there was only a slight inflation of the genomic-control parameter (λ = 1.01581), which implies a high genetic homogeneity between the cases (n = 295) and the controls (n = 571). Given that the inflation factor was found to be minimal, all the statistics results are reported without genomic-control correction. At the tail end of the quantile–quantile plot (Q–Q plot), the P-values from the Cochrane–Armitage trend test deviate from the null distribution expected under the hypothesis of no association (Supplementary Material, Fig. S2), which indicates that several modest associations were present.
Replication stage
Replication I
The CardioGene Study was an IRB-approved, prospective cohort study of 358 patients enrolled at the time of BMS implantation to treat de novo, previously untreated native coronary artery lesions at William Beaumont Hospital (Royal Oak, Michigan, USA) and the Mayo Clinic (Rochester, Minnesota, USA). Patients were followed for 1 year to determine in-stent restenosis (ISR) outcomes. Enrolment began in February 2002 and was closed in September 2003, prior to the approval and clinical use of DES in the USA. Additionally, 104 individuals were enrolled with historical in-stent restenosis in bare metallic stents, with two or more episodes of restenosis in native coronary arteries. The protocol was approved by the NHLBI IRB as well as the IRB at each of the clinical enrolment sites. Informed consent was provided by each patient. Standardized case report forms were used to collect baseline clinical data and outcome information in follow-up (46).
For the clinical phenotype, consecutive patients presenting to the cardiac catheterization laboratories of the clinical enrolment sites were approached for participation in the study. Follow-up clinical evaluation was performed via patient interview and review of all available medical records at 6 months and 12 months post-stent. ISR was defined as clinical restenosis (46), which was defined by ischemic symptoms after stent implantation and evidence of flow limitation in the treated vessel by either invasive or non-invasive testing. Follow-up angiography was not specifically performed for the CardioGene Study. Any available angiographic data performed as part of each patient's clinical care was recorded.
Genotyping and quality control
Genotypes were assayed using the Affymetrix Genome-Wide Human SNP Array 6.0 platform, and genotypes were called using the Birdseed algorithm. For this analysis, the final sample with genotype data consisted of 265 samples, with 78 in-stent restenosis cases and 187 stented no-restenosis controls, from European ancestry participants among which call rates and deviations from HWE for all SNPs were calculated. Genotype call rates were 95% or greater for all samples. Of these, 35 samples were removed in data cleaning steps for sex mismatch, first degree relative of an included individual and genetic outlier based upon allele sharing and principal components analysis. Genetic analyses were conducted using an additive model, using logistic regression to evaluate the association between the allele dosage and the trait of interest. We adjusted the analysis for age and gender. Genomic control was not applied. Data management and statistical analysis used R, ProbABEL (47) and PLINK software (44).
Replication II and replication III
Patients of replication II and replication III cohorts presented ischemic symptoms or evidence of myocardial ischemia in the presence of ≥50% de novo stenosis located in native coronary vessels. They were treated with PCI and stent implantation at Deutsches Herzzentrum München or 1. Medizinische Klinik rechts der Isar der Technischen Universität München. The main characteristics of the cohorts and the protocols of stent placement and post-stenting therapy have been described previously (23,48). Briefly, replication II included 1445 patients treated with implantation of BMS and replication III consisted of 1890 patients treated with implantation of DES. TLR within 1 year after PCI was considered the primary endpoint for both cohorts. Re-hospitalization for repeat angiography was scheduled between 6 and 8 months or earlier if non-invasive evaluation or clinical presentation suggested the presence of ischemia. The secondary endpoint was defined as a diameter stenosis 50% at follow-up angiography at 6 months.
Genotyping and quality control
Initially, 3657 samples were genotyped by means of iPLEX assays. All SNPs showing a P-value <10−4 in the Cochran–Armitage trend test (additive model) in the discovery set (n = 91) were selected for replication. Four assays were designed using MassArray design software (Sequenom, San Diego, CA, USA). Genotyping was performed using iPLEX assays with the use of the MassARRAY methodology (Sequenom), with alleles discriminated by mass spectrometry, following manufacturer's instructions. Four SNP pairs were in complete LD (r2 = 1) and thus we ascertained one tagSNP from each pair and the other was discarded.
Two SNPs did not fit the assay and four more SNPs failed in the experiment. Samples with a call rate <75% per iPLEX or with a call rate <90% when considering all iPLEXes were removed for further analysis. SNPs with call rate <90% or out of HWE (P-value<0.001 in controls) were also removed. Duplicate samples (∼2.5%) showed identical genotypes. Blanks and positive controls were added in each experiment. Finally, 79 SNPs and 3335 samples (2880 controls and 455 cases) passed all quality criteria and were further analyzed.
In order to know if the most associated SNPs in the discovery set contain information to detect the presence of population substructure, we performed a MDS extracting the genotypes of these SNPs from the HGDP-CEPH panel (45), which contains samples belonging to 52 populations all over the world. The MDS did not show any cluster (data not shown) in the HGDP-CEPH panel, not even between populations from different continents, thus indicating that population substructure cannot be considered a confounding factor in the replication cohorts when considering these SNPs.
PROSPER study
The protocol of PROSPER (PROspective Study of Pravastatin in the Elderly at Risk) has been described elsewhere (49). PROSPER is a prospective multicenter randomized placebo-controlled trial to assess whether treatment with pravastatin diminishes the risk of major vascular events in elderly individuals. Between December 1997 and May 1999, subjects were screened and enrolled in Scotland (Glasgow), Ireland (Cork) and the Netherlands (Leiden). Men and women aged 70–82 years were recruited if they had pre-existing vascular disease or increased risk of such disease because of smoking, hypertension or diabetes. A total number of 5804 subjects were randomly assigned to pravastatin or placebo. In this study, the predefined endpoints, coronary events, vascular events, vascular mortality and all-cause mortality were evaluated. In particular, coronary events are a combination of fatal and non-fatal myocardial infarction. Information on all-cause mortality was received by post-mortem reports, death certificates, hospital records, general practitioners’ records and/or interviews of family members or witnesses. All endpoints were adjudicated by the study endpoint committee.
Mean follow-up was 3.2 years (range 2.8–4.0) and 604 (10.4%) patients died during the study (50). The SNPs were selected from the GWAS performed in 5244 subjects of the PROSPER study from whom genotype data were available.
Statistical analysis
Statistical analysis was undertaken using R (v2.8), PLINK v1.06 (44), GenABEL and ProABEL softwares implemented in the R package (47,51). Haploview software (52) was used to infer the LD in the targeted regions. LocusZoom (53) was used to draw regional plots for associated regions.
Each SNP was tested for association using a Cochran–Armitage test (additive model). Inflation in the test statistics was assessed using the genomic-control method and a Q–Q plot was computed (54). The genotype counts of the discovery and replication stages were combined by means of the Fisher's trend combined P-value approach. All P-values are two-sided. Imputation of genotypes around the top associated SNPs were performed using MACH software (55,56).
The promoter 2.0 Prediction Server (39) was used to predict promoter regions and is-rSNP for in silico regulatory detection (26). The eQTL analysis was done following methodology described in reference (32).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by grants from the Interuniversity Cardiology Institute of the Netherlands (ICIN) and the Durrer Center for Cardiogenetic Research both Institutes of the Netherlands Royal Academy of Arts and Sciences (KNAW), the Netherlands Heart Foundation, the Center for Medical Systems Biology (CMSB), a center of excellence approved by the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research (NWO), the Netherlands Consortium for Healthy Ageing (NCHA) and the EU project HEALTH-F2-2007 223004 PHASE. J.W.J. is an established clinical investigator of the Netherlands Heart Foundation (2001D032). Part of this project was funded by a grant from the Netherlands Genomics Initiative (NGI) and Netherlands Organization for Scientific Research (NWO) within the framework of the Forensic Genomics Consortium Netherlands (FGCN) to P.d.K. CardioGene was funded in part by the National Heart, Lung and Blood Institute Division of Intramural Research. S.K.G. was supported in part by NHLBI R00HL089413. The funders had no role in study design, data collection and analysis, decision to publish or the preparation of the manuscript.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Nico Lakenberg, Yavuz Ariyurek, Dennis Kremer and Eka Suchiman for technical assistance and to Oscar Lao for valuable comments and helpful insights.
REFERENCE
- 1.Agema W.R., Monraats P.S., Zwinderman A.H., de Winter R.J., Tio R.A., Doevendans P.A., Waltenberger J., de Maat M.P., Frants R.R., Atsma D.E., et al. Current PTCA practice and clinical outcomes in The Netherlands: the real world in the pre-drug-eluting stent era. Eur. Heart. J. 2004;25:1163–1170. doi: 10.1016/j.ehj.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Roiron C., Sanchez P., Bouzamondo A., Lechat P., Montalescot G. Drug eluting stents: an updated meta-analysis of randomised controlled trials. Heart. 2006;92:641–649. doi: 10.1136/hrt.2005.061622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sigwart U., Puel J., Mirkovitch V., Joffre F., Kappenberger L. Intravascular stents to prevent occlusion and restenosis after transluminal angioplasty. N. Engl. J. Med. 1987;316:701–706. doi: 10.1056/NEJM198703193161201. [DOI] [PubMed] [Google Scholar]
- 4.Lee M.S., David E.M., Makkar R.R., Wilentz J.R. Molecular and cellular basis of restenosis after percutaneous coronary intervention: the intertwining roles of platelets, leukocytes, and the coagulation-fibrinolysis system. J. Pathol. 2004;203:861–870. doi: 10.1002/path.1598. [DOI] [PubMed] [Google Scholar]
- 5.Pache J., Kastrati A., Mehilli J., Schuhlen H., Dotzer F., Hausleiter J., Fleckenstein M., Neumann F.J., Sattelberger U., Schmitt C., et al. Intracoronary stenting and angiographic results: strut thickness effect on restenosis outcome (ISAR-STEREO-2) trial. J. Am. Coll. Cardiol. 2003;41:1283–1288. doi: 10.1016/s0735-1097(03)00119-0. [DOI] [PubMed] [Google Scholar]
- 6.Serruys P.W., de Jaegere P., Kiemeneij F., Macaya C., Rutsch W., Heyndrickx G., Emanuelsson H., Marco J., Legrand V., Materne P. A comparison of balloon-expandable-stent implantation with balloon angioplasty in patients with coronary artery disease. Benestent Study Group. N. Engl. J. Med. 1994;331:489–495. doi: 10.1056/NEJM199408253310801. [DOI] [PubMed] [Google Scholar]
- 7.Ross R., Masuda J., Raines E.W. Cellular interactions, growth factors, and smooth muscle proliferation in atherogenesis. Ann. N. Y. Acad. Sci. 1990;598:102–112. doi: 10.1111/j.1749-6632.1990.tb42282.x. [DOI] [PubMed] [Google Scholar]
- 8.Ross R. Atherosclerosis—an inflammatory disease. N. Engl. J. Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 9.Rajagopal V., Rockson S.G. Coronary restenosis: a review of mechanisms and management. Am. J. Med. 2003;115:547–553. doi: 10.1016/s0002-9343(03)00477-7. [DOI] [PubMed] [Google Scholar]
- 10.Arjomand H., Turi Z.G., McCormick D., Goldberg S. Percutaneous coronary intervention: historical perspectives, current status, and future directions. Am. Heart. J. 2003;146:787–796. doi: 10.1016/S0002-8703(03)00153-4. [DOI] [PubMed] [Google Scholar]
- 11.Jenkins N.P., Prendergast B.D., Thomas M. Drug eluting coronary stents. BMJ. 2002;325:1315–1316. doi: 10.1136/bmj.325.7376.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morice M.C., Serruys P.W., Sousa J.E., Fajadet J., Ban H.E., Perin M., Colombo A., Schuler G., Barragan P., Guagliumi G., et al. A randomized comparison of a sirolimus-eluting stent with a standard stent for coronary revascularization. N. Engl. J. Med. 2002;346:1773–1780. doi: 10.1056/NEJMoa012843. [DOI] [PubMed] [Google Scholar]
- 13.Stone G.W., Ellis S.G., O'Shaughnessy C.D., Martin S.L., Satler L., McGarry T., Turco M.A., Kereiakes D.J., Kelley L., Popma J.J., Russell M.E. Paclitaxel-eluting stents vs vascular brachytherapy for in-stent restenosis within bare-metal stents: the TAXUS V ISR randomized trial. JAMA. 2006;295:1253–1263. doi: 10.1001/jama.295.11.1253. [DOI] [PubMed] [Google Scholar]
- 14.Bourassa M.G., Lesperance J., Eastwood C., Schwartz L., Cote G., Kazim F., Hudon G. Clinical, physiologic, anatomic and procedural factors predictive of restenosis after percutaneous transluminal coronary angioplasty. J. Am. Coll. Cardiol. 1991;18:368–376. doi: 10.1016/0735-1097(91)90588-z. [DOI] [PubMed] [Google Scholar]
- 15.Stein B., Weintraub W.S., Gebhart S.P., Cohen-Bernstein C.L., Grosswald R., Liberman H.A., Douglas J.S., Jr, Morris D.C., King S.B., III Influence of diabetes mellitus on early and late outcome after percutaneous transluminal coronary angioplasty. Circulation. 1995;91:979–989. doi: 10.1161/01.cir.91.4.979. [DOI] [PubMed] [Google Scholar]
- 16.Agema W.R., Jukema J.W., Pimstone S.N., Kastelein J.J. Genetic aspects of restenosis after percutaneous coronary interventions: towards more tailored therapy. Eur. Heart. J. 2001;22:2058–2074. doi: 10.1053/euhj.2000.2589. [DOI] [PubMed] [Google Scholar]
- 17.Kastrati A., Schomig A., Elezi S., Schuhlen H., Wilhelm M., Dirschinger J. Interlesion dependence of the risk for restenosis in patients with coronary stent placement in in multiple lesions. Circulation. 1998;97:2396–2401. doi: 10.1161/01.cir.97.24.2396. [DOI] [PubMed] [Google Scholar]
- 18.de Maat M.P., Jukema J.W., Ye S., Zwinderman A.H., Moghaddam P.H., Beekman M., Kastelein J.J., van Boven A.J., Bruschke A.V., Humphries S.E., et al. Effect of the stromelysin-1 promoter on efficacy of pravastatin in coronary atherosclerosis and restenosis. Am. J. Cardiol. 1999;83:852–856. doi: 10.1016/s0002-9149(98)01073-x. [DOI] [PubMed] [Google Scholar]
- 19.Kastrati A., Schomig A., Seyfarth M., Koch W., Elezi S., Bottiger C., Mehilli J., Schomig K., von Beckerath N. PlA polymorphism of platelet glycoprotein IIIa and risk of restenosis after coronary stent placement. Circulation. 1999;99:1005–1010. doi: 10.1161/01.cir.99.8.1005. [DOI] [PubMed] [Google Scholar]
- 20.Monraats P.S., Pires N.M., Agema W.R., Zwinderman A.H., Schepers A., de Maat M.P., Doevendans P.A., de Winter R.J., Tio R.A., Waltenberger J., et al. Genetic inflammatory factors predict restenosis after percutaneous coronary interventions. Circulation. 2005;112:2417–2425. doi: 10.1161/CIRCULATIONAHA.105.536268. [DOI] [PubMed] [Google Scholar]
- 21.Monraats P.S., Pires N.M., Schepers A., Agema W.R., Boesten L.S., De Vries M.R., Zwinderman A.H., de Maat M.P., Doevendans P.A., de Winter R.J., et al. Tumor necrosis factor-alpha plays an important role in restenosis development. FASEB J. 2005;19:1998–2004. doi: 10.1096/fj.05-4634com. [DOI] [PubMed] [Google Scholar]
- 22.Amant C., Bauters C., Bodart J.C., Lablanche J.M., Grollier G., Danchin N., Hamon M., Richard F., Helbecque N., McFadden E.P., et al. D allele of the angiotensin I-converting enzyme is a major risk factor for restenosis after coronary stenting. Circulation. 1997;96:56–60. doi: 10.1161/01.cir.96.1.56. [DOI] [PubMed] [Google Scholar]
- 23.Koch W., Kastrati A., Mehilli J., Bottiger C., von Beckerath N., Schomig A. Insertion/deletion polymorphism of the angiotensin I-converting enzyme gene is not associated with restenosis after coronary stent placement. Circulation. 2000;102:197–202. doi: 10.1161/01.cir.102.2.197. [DOI] [PubMed] [Google Scholar]
- 24.Agema W.R., Jukema J.W., Zwinderman A.H., van der Wall E.E. A meta-analysis of the angiotensin-converting enzyme gene polymorphism and restenosis after percutaneous transluminal coronary revascularization: evidence for publication bias. Am. Heart J. 2002;144:760–768. doi: 10.1067/mhj.2002.125509. [DOI] [PubMed] [Google Scholar]
- 25.Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacIntyre G., Bailey J., Haviv I., Kowalczyk A. is-rSNP: a novel technique for in silico regulatory SNP detection. Bioinformatics. 2010;26:i524–i530. doi: 10.1093/bioinformatics/btq378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blastyak A., Hajdu I., Unk I., Haracska L. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol. Cell Biol. 2010;30:684–693. doi: 10.1128/MCB.00863-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hata S., Hamada J., Maeda K., Murai T., Tada M., Furukawa H., Tsutsumida A., Saito A., Yamamoto Y., Moriuchi T. PAX4 has the potential to function as a tumor suppressor in human melanoma. Int. J. Oncol. 2008;33:1065–1071. [PubMed] [Google Scholar]
- 29.Gallo R., Padurean A., Jayaraman T., Marx S., Roque M., Adelman S., Chesebro J., Fallon J., Fuster V., Marks A., Badimon J.J. Inhibition of intimal thickening after balloon angioplasty in porcine coronary arteries by targeting regulators of the cell cycle. Circulation. 1999;99:2164–2170. doi: 10.1161/01.cir.99.16.2164. [DOI] [PubMed] [Google Scholar]
- 30.Giannakakou P., Robey R., Fojo T., Blagosklonny M.V. Low concentrations of paclitaxel induce cell type-dependent p53, p21 and G1/G2 arrest instead of mitotic arrest: molecular determinants of paclitaxel-induced cytotoxicity. Oncogene. 2001;20:3806–3813. doi: 10.1038/sj.onc.1204487. [DOI] [PubMed] [Google Scholar]
- 31.Argiropoulos B., Humphries R.K. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 32.Dubois P.C., Trynka G., Franke L., Hunt K.A., Romanos J., Curtotti A., Zhernakova A., Heap G.A., Adany R., Aromaa A., et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S.J., So I.S., Park S.Y., Kim I.S. Thymosin beta4 is involved in stabilin-2-mediated apoptotic cell engulfment. FEBS Lett. 2008;582:2161–2166. doi: 10.1016/j.febslet.2008.03.058. [DOI] [PubMed] [Google Scholar]
- 34.Park S.Y., Kim S.Y., Jung M.Y., Bae D.J., Kim I.S. Epidermal growth factor-like domain repeat of stabilin-2 recognizes phosphatidylserine during cell corpse clearance. Mol. Cell Biol. 2008;28:5288–5298. doi: 10.1128/MCB.01993-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris E.N., Baggenstoss B.A., Weigel P.H. Rat and human HARE/stabilin-2 are clearance receptors for high- and low-molecular-weight heparins. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;296:G1191–G1199. doi: 10.1152/ajpgi.90717.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yatsuoka T., Furukawa T., Sunamura M., Matsuno S., Horii A. TU12B1-TY, a novel gene in the region at 12q22-q23.1 frequently deleted in pancreatic cancer, shows reduced expression in pancreatic cancer cells. Oncol. Rep. 2004;12:1263–1268. [PubMed] [Google Scholar]
- 37.Sabo P.J., Hawrylycz M., Wallace J.C., Humbert R., Yu M., Shafer A., Kawamoto J., Hall R., Mack J., Dorschner M.O., et al. Discovery of functional noncoding elements by digital analysis of chromatin structure. Proc. Natl Acad. Sci. USA. 2004;101:16837–16842. doi: 10.1073/pnas.0407387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabo P.J., Humbert R., Hawrylycz M., Wallace J.C., Dorschner M.O., McArthur M., Stamatoyannopoulos J.A. Genome-wide identification of DNaseI hypersensitive sites using active chromatin sequence libraries. Proc. Natl Acad. Sci. USA. 2004;101:4537–4542. doi: 10.1073/pnas.0400678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knudsen S. Promoter2.0: for the recognition of PolII promoter sequences. Bioinformatics. 1999;15:356–361. doi: 10.1093/bioinformatics/15.5.356. [DOI] [PubMed] [Google Scholar]
- 40.Visel A., Zhu Y., May D., Afzal V., Gong E., Attanasio C., Blow M.J., Cohen J.C., Rubin E.M., Pennacchio L.A. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hardy J., Singleton A. Genomewide association studies and human disease. N. Engl. J. Med. 2009;360:1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerrits A., Li Y., Tesson B.M., Bystrykh L.V., Weersing E., Ausema A., Dontje B., Wang X., Breitling R., Jansen R.C., de H.G. Expression quantitative trait loci are highly sensitive to cellular differentiation state. PLoS Genet. 2009;5:e1000692. doi: 10.1371/journal.pgen.1000692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Myers R.M. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 46.Ganesh S.K., Skelding K.A., Mehta L., O'Neill K., Joo J., Zheng G., Goldstein J., Simari R., Billings E., Geller N.L., et al. Rationale and study design of the CardioGene Study: genomics of in-stent restenosis. Pharmacogenomics. 2004;5:952–1004. doi: 10.1517/14622416.5.7.949. [DOI] [PubMed] [Google Scholar]
- 47.Aulchenko Y.S., Struchalin M.V., van Duijn C.M. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoppmann P., Erl A., Turk S., Tiroch K., Mehilli J., Schomig A., Kastrati A., Koch W. No association of chromosome 9p21.3 variation with clinical and angiographic outcomes after placement of drug-eluting stents. JACC Cardiovasc. Interv. 2009;2:1149–1155. doi: 10.1016/j.jcin.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 49.Shepherd J., Blauw G.J., Murphy M.B., Cobbe S.M., Bollen E.L., Buckley B.M., Ford I., Jukema J.W., Hyland M., Gaw A., et al. The design of a prospective study of Pravastatin in the Elderly at Risk (PROSPER). PROSPER Study Group. PROspective Study of Pravastatin in the Elderly at Risk. Am. J. Cardiol. 1999;84:1192–1197. doi: 10.1016/s0002-9149(99)00533-0. [DOI] [PubMed] [Google Scholar]
- 50.Shepherd J., Blauw G.J., Murphy M.B., Bollen E.L., Buckley B.M., Cobbe S.M., Ford I., Gaw A., Hyland M., Jukema J.W., et al. Pravastatin in elderly individuals at risk of vascular disease (PROSPER): a randomised controlled trial. Lancet. 2002;360:1623–1630. doi: 10.1016/s0140-6736(02)11600-x. [DOI] [PubMed] [Google Scholar]
- 51.Aulchenko Y.S., Ripke S., Isaacs A., van Duijn C.M. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 52.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 53.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCarthy M.I., Abecasis G.R., Cardon L.R., Goldstein D.B., Little J., Ioannidis J.P., Hirschhorn J.N. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 55.Li Y., Willer C., Sanna S., Abecasis G. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.