Abstract
Previously, we have shown that a heteroplasmic mutation in mitochondrial DNA-encoded complex I ND5 subunit gene resulted in an enhanced tumorigenesis through increased resistance to apoptosis. Here we report that the tumorigenic phenotype associated with complex I dysfunction could be reversed by introducing a yeast NADH quinone oxidoreductase (NDI1) gene. The NDI1 mediated electron transfer from NADH to Co-Q, bypassed the defective complex I and restored oxidative phosphorylation in the host cells. Alternatively, suppression of complex I activity by a specific inhibitor, rotenone or induction of oxidative stress by paraquat led to an increase in the phosphorylation of v-AKT murine thymoma viral oncogene (AKT) and enhanced the tumorigenesis. On the other hand, antioxidant treatment can ameliorate the reactive oxygen species-mediated AKT activation and reverse the tumorigenicity of complex I-deficient cells. Our results suggest that complex I defects could promote tumorigenesis through induction of oxidative stress and activation of AKT pathway.
INTRODUCTION
Mitochondria have been shown to play an important role in regulating both programmed cell death and cell proliferation (1). Alterations in oxidative phosphorylation (OXPHOS) resulting from mitochondrial dysfunction have long been hypothesized to be involved in tumorigenesis. Specifically, it has been postulated that the switch in adenosine triphosphate (ATP) production from mitochondrial oxidative phosphorylation to glycolysis is one of the characteristics of cancer cells (2).
As the entry point for most electrons into the respiratory chain, NADH dehydrogenase or complex I has been suggested as the rate-limiting step in overall electron transfer. It plays a central role in oxidative phosphorylation (3). Recently, mtDNA mutations in genes encoding complex I subunits have been found in various cancer cells (4). Yet, the role of these mtDNA mutations in tumorigenesis remains largely unclear, primarily because most of the mtDNA mutations identified in tumors are not adapted to grow in the lab. To overcome this limitation, we have developed a novel approach to isolate mtDNA encoding complex I subunit mutations in cultured cells (5). Cells are grown under conditions that induce the bioenergetic switch from mitochondrial oxidative phosphorylation to glycolysis, by gradually inhibiting complex I respiration and mimicking tumor cell's ATP production. Using this strategy, cells carrying mtDNA mutations in complex I subunit genes, ND4, ND5 and ND6, were generated (6–8). Interestingly, one of the ND5 mutations isolated by this method was similar to a natural ND5 mutation found in colorectal cancer (9). Cell lines carrying this colorectal cancer-related ND5 mutation have been tested for tumorigenicity using an in vivo nude mice assay and an in vitro anchorage dependence assay. Surprisingly, cells with the heteroplasmic mtDNA mutation for ND5 showed significantly enhanced tumor growth (10).
In order to confirm the role of complex I in tumorigenesis, here we investigated the effects of rescued complex I function in cells carrying heteroplasmic ND5 mutations on tumorigenesis. In contrast to mammalian cells, Saccharomyces cerevisiae lack complex I, instead they have a nucleus-encoding NADH protein quinone oxidoreductase (NDI1) gene (11). The NDI1 protein has been shown to be active in bacteria, Chinese hamster and human cells, and coupled to the downstream portion of host respiratory chain (12).
By introducing the NDI1 gene into cells carrying heteroplasmic ND5 mutation, we found that with the restoration of respiration and mitochondrial functions, the tumorigenic potential could be reversed. We also demonstrated that such regulation might be mediated by the alterations in metabolic reaction, redox status and reactive oxygen species (ROS) production, through v-AKT murine thymoma viral oncogene (AKT) pathway.
RESULTS
Rescue of complex I defects by yeast NDI1
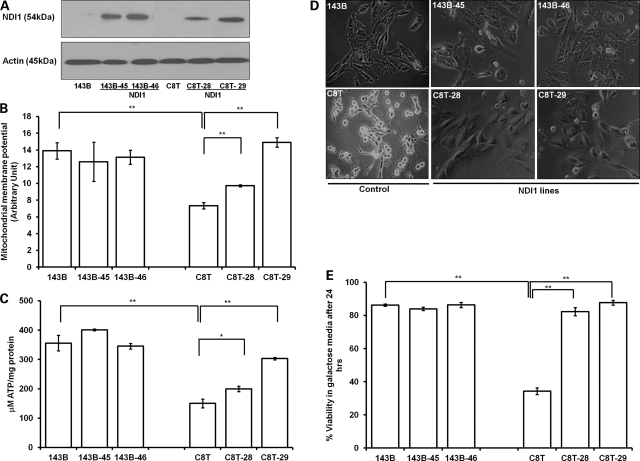
To introduce yeast NDI1 gene in C8T cells with heteroplasmic ND5 mutation and control 143B cells, several gene transfer methods including chemical and electroporation were utilized. Relatively high infection efficiency and expression of NDI1 protein were achieved only with a lentiviral-mediated gene delivery system. Two clones for each transformed lines (indicated as 143B-45, -46, C8T-28, -29) were selected, and the NDI1 expression was confirmed by western analysis (Fig. 1A). The heteroplasmic status of the ND5 gene in C8T, C8T-28 and -29 was verified by sequencing and compared with wild-type 143B using Mutation Surveyor software program (13) (Supplementary Material, Fig. S1A). The presence of the wild-type ND5 gene in C8T, C8T-28 and C8T-29 when compared with 143B were 29, 29 and 26%, respectively, as calculated by Mutation Surveyor program (Supplementary Material, Fig. S1B).
Figure 1.
Rescue of complex I defects by yeast NDI1. (A) Western blot analysis of yeast NDI1 expression from the cell lysates of independent clones of 143B (NDI1 lines: 143B-45 and 143B- 46) and C8T (NDI1 lines: C8T-28 and C8T-29) cells using anti-NDI1 antibody (1:1000 dilution). (B) MMP was determined by TMRM and presented after normalized with nuclear stain (Hoechst) fluorescence reading. (C) The total cellular ATP contents was measured in different cells with a luciferase-based ATP detection kit and normalized with total protein used/assay. (D) Morphology of cells analyzed under Nikon eclipse inverted microscope after 24 h of growth in galactose media. (E) Cellular viability after 24 h of growth in galactose media was quantitated using Vi-Cell cell viability analyzer based on the trypan blue exclusion method. Data represent mean ± SE of three independent experiments. *P< 0.05, **P< 0.01.
It has been shown that the NDI1 protein localizes into the mitochondria and incorporates into the host oxidative phosphorylation system (12,14,15). However, one concern over the replacement of NDI1 over complex I is the lack of proton pumping ability of NDI1 which contributes to establishment of mitochondrial membrane potential (MMP). An appropriate MMP is not only essential for ATP production, but also important for regulation of calcium uptake and ROS production (16). To determine whether MMP was recovered in the C8T cell with NDI1, cells were labeled with an MMP indicator, tetra-methyl-rhodamine methyl ester (TMRM) and fluorescence was measured. As shown, the MMP was reduced by 47% in C8T compared with 143B (Fig. 1B). The NDI1 expression significantly improved MMP (32 and 103% increase in C8T-28 and -29, respectively) in these mutant cells, but no significant changes were recorded in wild-type 143B cells. Further, a total ATP level was measured by luciferase-based enzyme assay. Similarly, no significant difference was detected with 143B cells; however, we found that the NDI1 expression increased the ATP contents in NDI1 transformants (C8T-28 and -29) by 32 and 102%, respectively, compared with C8T cells (Fig. 1C).
We then analyzed the growth capacity in galactose media where cells are forced to rely predominantly on oxidative phosphorylation as a source of ATP (14). Consistent with the above observation, with NDI1 expression, C8T-28 and -29 cells totally recovered their growth capacity in galactose media (Fig. 1D and E).
Taken together, our results indicated that mitochondrial defects associated with ND5 mutation were rescued with yeast NDI1 expression, which incorporated into the oxidative phosphorylation system in C8T cells.
Reversal of tumorigenesis after complex I functional recovery
In order to further investigate the role of complex I deficiency in tumorigenesis, we sought to analyze the tumorigenic potential of complex I defective C8T cells and its NDI1 transformants with restored mitochondrial functions. It is known that normal cells are obligatorily anchorage dependent, and grow in vitro by attaching to a solid surface, while transformed cells can grow unattached demonstrating their anchorage independence capacity (17). Also, a parallel relationship between the anchorage dependence and in vivo tumorigenesis has been well established (18). Therefore, growth of cells suspended in soft agar is a convenient way to test the tumorigenesis. As we observed previously, heteroplasmic C8T cells formed more colonies in soft agar than the control 143B cells (10). Interestingly, we found a significant reduction in the number of colony-forming units on soft agar assay with the expression of the NDI1 gene in C8T cells (C8T-28 and -29), which recovered from mitochondrial defects caused by ND5 mutation (Fig. 2A and B). We then continued to explore if the recovery of the complex I-dependent function was also accompanied with the change in motility and invasive potential of cells. Hence, we analyzed the cell migration capacity using the chemo attractant-based cell migration method (migration from serum-free media to complete media) (19). As shown in Figure 2C and D, we found that, with the rescued complex I functions, number of cells migrated through membrane pores from serum-free to complete media were significantly less in NDI1 (C8T-28 and -29) lines compared with C8T cells.
Figure 2.
Reversal of tumorigenesis after complex I function recovery. (A) Equal number of cells (four replicates for each line) were suspended in 0.27% agar in growth media and seeded on 60 mm dish contained 0.4% solid agar. The colonies were analyzed after 3 weeks and representative pictures of one of the three experiments were shown. (B) The colonies >0.1 mm were scored with alpha imager colony-counting software. (C) Cell migration was analyzed using trans-well assay (four wells/line) and cells that were migrated to the lower surface were stained and imaged. (D) Total four independent fields per well were selected for counting migrated cells using NIS-element software. Representative images (n= 4) are shown and quantitation represents mean ± SE of three independent experiments.*P< 0.05, **P< 0.01. (E) Different percentage of mutation in COX1 subunit (G6930A) of complex IV was used for colony-forming units on soft agar assay and imaged after 3 weeks. Representative pictures of one of the three experiments are shown.
To test whether the increase in tumorigenicity we observed with C8T cells is complex I specific rather than the defects in mitochondrial function in general, we performed the colony formation assay with cells carrying a complex IV subunit COX1 mutation at G6930A with different heteroplasmy (ranging from 0, 35, 50, 70 and 100%). Although G6930A mutation was shown to cause mitochondrial dysfunction measured by respiration and ATP production (20), we did not see any increase in the colony formation with different COX1 heteroplasmy (Fig. 2E). It suggests that complex I defects may initiate some signals which could play a role in tumorigenesis.
Alterations in metabolism, redox status and ROS production
To outline the signaling from complex I defect to tumorigenesis, several biochemical/metabolic changes in complex I defective and restored cells were analyzed. One of the immediate effects of complex I dysfunction on metabolism is the accumulation of its substrate NADH. The NAD+/NADH ratio is an indicator for redox status which in turn is a major modulator for many important signaling pathways (21). To investigate the effect of complex I defect on redox status, we measured both NAD+ and NADH levels. As expected, we found a 44% increase in the NADH level (Fig. 3A) and 32% decrease in the NAD+/NADH ratio (Fig. 3C) in C8T cells compared with control 143B. Expression of NDI1 decreased NADH levels in C8T-28 and -29 (27 and 32%, respectively) than C8T and in 143B-45 and -46 (22 and 16%, respectively) than 143B (Fig. 3A). Interesting, no significant changes were recorded in NAD levels in both C8T and 143B cells with the introduction of NDI (Fig. 3B). Overall NAD+/NADH ratio increased by 28 and 21% in 143B-45 and -46 when compared with 143B, while 33 and 49% in C8T-28 and -29 when compared with C8T cells (Fig. 3C). Metabolic switch from oxidative phosphorylation to ‘energy inefficient’ glycolytic pathway is a hallmark for cancer development (22). To maintain the ATP level without normal mitochondrial respiration, the glycolytic pathway is accelerated and lactate would thus accumulate (23). To determine whether the glycolysis process was altered accompanied with the changes in complex I-associated oxidative phosphorylation activity, we measured the extracellular lactate levels in these cells. We found an increase by 43% of the lactate level in complex I defective C8T cells compared with 143B cells, and the lactate level was significantly reduced in NDI1 lines C8T-28 and -29 by 39 and 48%, respectively (Fig. 3D). However, NDI1 expression did not change the lactate secretion in 143B cells.
Figure 3.
Alteration in metabolism, redox status and ROS production. For the redox molecules measurement, freshly seeded cells were grown in triplicate for 24 h; later (A) NADH, (B) NAD and (C) ratio of NAD to NADH were measured in the cell lysate according to the manufacturer's instruction. Values were calculated from standard curve and normalized with mg protein used/well. (D) For lactate measurement, cells were cultured for 48 h in complete media and extracellular lactate level was measured in the media and calculated for equal number of cells. (E) For mitochondrial ROS levels, cells were counted in Vi-Cell cell viability analyzer and equal number of cells was stained with mitochondrial superoxide indicator (MitoSOX™ red) and fluorescence was measured on fluorescence plate reader. (F) The H2O2 levels in isolated mitochondria were measured using the HRP-linked fluorometric assay and values were calculated from standard curve and normalized with per mg protein used per assay. Data are presented as mean ± SE of three independent experiments. *P< 0.05, **P< 0.01.
Mitochondria are the major source of ROS production, important signaling molecules which regulate cell death and proliferation (24,25). Therefore, we investigated whether there was an alteration in mitochondrial ROS with the changes in oxidative phosphorylation. Mitochondrial-specific generation of superoxide was first detected by Mitosox, which was rapidly and selectively targeted to the mitochondria and oxidized by superoxide (26). As shown in Figure 3E, significant increases in C8T were recorded compared with 143B cells, at the same time NDI1-expressing lines of both wild-type 143B and C8T cells showed reduction in the mitochondrial ROS level. To validate our measurement of mitochondrial generated ROS, we utilized another detection method Amplex Red assay in the isolated mitochondrial preparations. This assay employs horseradish peroxidase (HRP) to trap emitted H2O2 with high selectivity and affinity (27). As shown in Figure 3F, ROS levels in isolated mitochondria preparations measured by Amplex Red showed a pattern similar to that determined by Mitosox (Fig. 3E). We also measured the mitochondrial superoxide in complex IV mutants carrying different percentage of mutation in the COX1 subunit gene. With the exception of cells carrying homoplasmic mutation (100%) in COX1 subunit which exhibited reduced ROS production, no significant differences among others cells were recorded (Supplementary Material, Fig. S2).
Complex I defect and AKT signaling
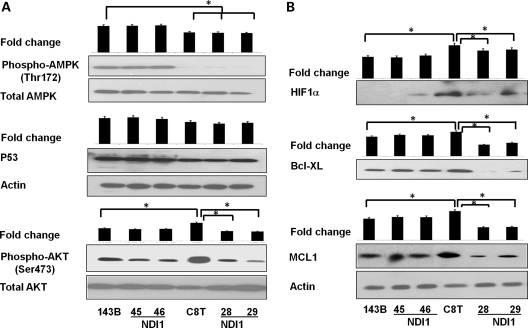
Despite the increasing knowledge of oncogenic signaling in cancer cells, very few pathways have been established directly from mitochondrial defects to tumorigenesis. We began with testing several molecules which play important roles in tumorigenesis and had been shown to be regulated by redox or ROS signaling. Signaling of adenosine monophosphate-activated protein kinase (AMPK), AKT and P53 was analyzed with the immuno-blotting method (Fig. 4A). Among them we found a significant increase in the AKT phosphorylation (Ser473) level in tumorigenic C8T cells compared with 143B cells; conversely, a drastic decrease in AKT phosphorylation was observed in NDI1-expressing C8T-28 and -29 cells where tumorigenic feature was reversed (Fig. 4A).
Figure 4.
AKT signaling activation. (A) A total of 40 μg whole cell protein was resolved by SDS–PAGE and the level of different proteins were analyzed using immunoblotting with primary antibodies against (A) phospho AMPK (Thr172), total AMPK, p53, phospho-AKT (Ser473), total AKT and (B) HIF1α, BCL-XL and MCL1 (all used as 1:1000 dilution in 5% BSA). β-Actin (1:2000) was used as a loading control in different cell lines. One of representative images of each experiment is shown (n= 3). Fold changes in the protein level were determined by densitometry of western blots using Image J software, normalized with their respective controls and shown as histograms relative to 143B. *P< 0.05.
To confirm that AKT signaling plays a role in complex I defect-dependent tumorigenesis, we verified the expression of some key factors downstream of AKT activation. As shown in Figure 4B, we found the up-regulation of HIF1α and anti-apoptotic proteins BCL-XL and MCL1 in C8T cells, whereas significant decrease in these protein levels in C8T-28 and -29 lines. The involvement of the AKT pathway was also examined with a pathway-specific real-time polymerase chain reaction array analysis. Transcription analysis of C8T and its NDI1 line (C8T-29) after normalizing with 143B revealed that many genes associated with the PI3K/AKT pathway were altered in C8T-29 compared with C8T cells (Supplementary Material, Fig. S3). In particular, transcription of several downstream targets of the AKT pathway involved in cell proliferation and anti-apoptosis was up-regulated in C8T cells but reversed in the C8T-29 line.
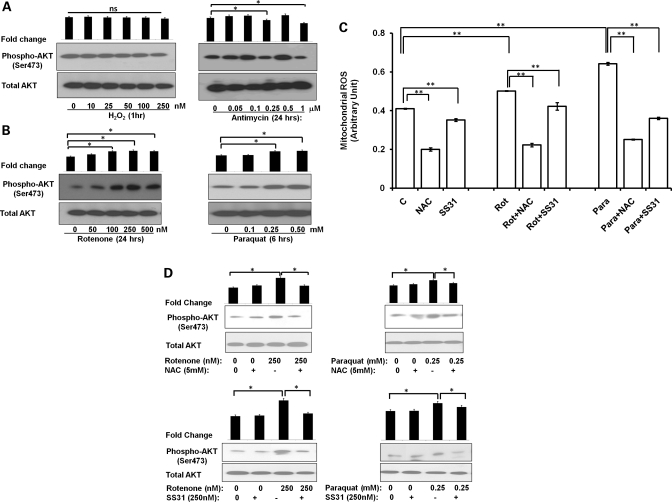
Complex I stress activates AKT signaling through ROS signaling
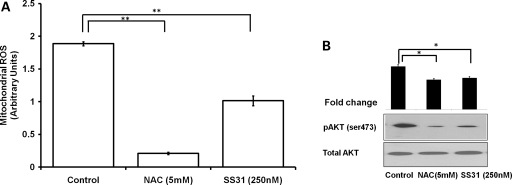
To confirm the relationship between complex I defect and tumorigenesis, we utilized pharmacological agents, known to produce both complex I-dependent or -independent mitochondrial stress. Rotenone shuts off the supply of electrons from complex I to the respiratory system and enhances the generation of free radical from complex I (28). Similarly, paraquat is reduced by complex I to form the paraquat radical cation that reacts with oxygen to form superoxide (29). Although, we did not find a significant activation in AKT phosphorylation after the general oxidizing agent H2O2 and complex III inhibitor antimycin treatment (Fig. 5A) in wild-type 143B cells, but complex I-specific stressors rotenone and paraquat induced a significant activation in AKT phosphorylation in a dose-dependent manner (Fig. 5B). To further determine whether this AKT activation is mediated by ROS, we first measured mitochondrial ROS with Mitosox in 143B cells treated with rotenone or paraquat. We found both rotenone and paraquat increased the ROS level significantly (22 and 56%, respectively) (Fig. 5C) at a concentration (250 nm and 0.25 mm, respectively) which induced AKT phosphorylation. This increase in ROS production could be abolished with the treatment of either general antioxidant such as N-acetyl cystein (NAC) or mitochondria-specific antioxidant Szeto-Schiller peptides-31: SS31 (30) (Fig. 5C). Interestingly, neutralized ROS production also reversed the AKT activation (Fig. 5D). Similarly, in C8T which is genetically defective in complex I and exhibited increased tumorigenesis, we detected an elevated mitochondrial ROS level and AKT activation compared with control 143B cells. Treatment of NAC and SS31 reduced both ROS production and AKT phosphorylation significantly in C8T cells (Fig. 6A and B). These results indicated that alteration in mitochondrial ROS is responsible for AKT activation during complex I stress.
Figure 5.
Complex I stress activates AKT signaling through ROS signaling. (A) Wild-type 143B cells were treated with different concentrations of H2O2 for 1 h or complex III inhibitor antimycin for 24 h. Cells were collected and the level of phospho-AKT (Ser473) was determined by immunoblotting. (B) Different concentrations of complex I stressors, rotenone (24 h) or paraquat (6 h) were used to determine the phospho-AKT level. (C) Mitochondrial ROS levels were measured using Mitosox assay in cells either non-treated or pretreated with antioxidants (NAC-5 mm or SS31-250 nm) for 2 h followed by treatment with 250 nm rotenone (Rot) for 24 h/0.25 mm paraquat (Para) for 6 h. Mitosox values were normalized with Hoechst fluorescence. Data are presented as mean ± SE of three independent experiments. **P< 0.01. (D) Similarly, effects of NAC/SS31 antioxidants alone or with rotenone/paraquat treatment on phospho-AKT levels were measured by immunoblotting. Representative blots of one of the three independent experiments are shown. Fold changes in the protein level were determined by densitometry and normalized by the total AKT level and shown as histograms relative to non-treated control. ns, not significant, *P< 0.05.
Figure 6.
Antioxidant treatment decreases ROS and oncogenic AKT activation in C8T. (A) Complex I defective C8T cells were treated with NAC or SS31 for 2 h and mitochondrial ROS was quantitated using Mitosox assay. **P< 0.01. (B) Similarly, the level of phospho-AKT after this treatment was determined by immunoblotting and fold changes were quantitated by densitometry. Fold changes (n= 3) in the phospho-AKT level is shown as histograms relative to non-treated control. *P< 0.05.
Complex I stress, ROS and tumorigenesis
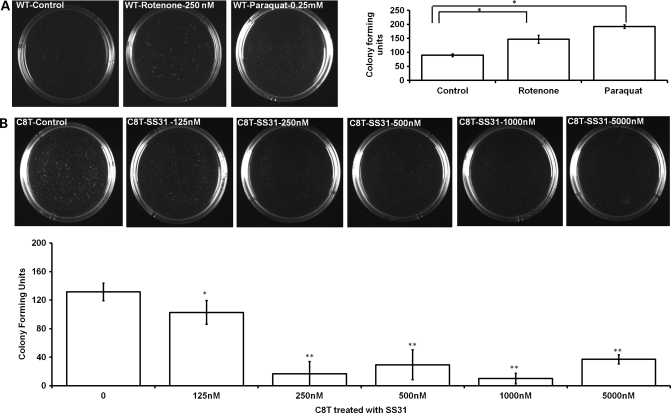
To further confirm that the effect of complex I stress on tumorigenesis was mediated by AKT activation through ROS upregulation, we performed colony-forming soft agar assay after complex I stressors rotenone or paraquat treatment in wild-type 143B cells. As expected, we found an increase in the number of colonies on soft agar plates (Fig. 7A). Similarly, to see whether the reduced ROS level in the complex I defective C8T cells would reverse its tumorigenic feature, we carried out the soft agar assay after treating C8T cells with different concentration of mitochondrial-specific antioxidant SS31. We found that antioxidant treatment can reduce the number of colonies as observed by soft agar assay (Fig. 7B). In particular, dramatic reduction in the number of colonies was observed with SS31 at 250 nm or above, capable of inhibiting of ROS accumulation and AKT activation as shown previously (Fig. 6A and B). In addition, NAC and a known AKT inhibitor (LY294002) were also found to reduce the tumorigenic potential of C8T cells (Supplementary Material, Fig. S4), further implicating the ROS-dependent AKT pathway in regulating tumorigenesis in complex I defective C8T cells.
Figure 7.
Complex I inhibition increases and antioxidant treatment decreases tumorigenesis. (A) Wild-type 143B cells were treated with complex I stressors (250 nm rotenone for 24 h/0.25 mm paraquat for 6 h) and equal numbers of viable cells were used for soft agar assay. (B) For C8T, cells were pre-treated with different concentrations of mitochondrial antioxidant (SS31) for 2 h and equal number of viable cells was analyzed for colony formation by soft agar assay and compared with non-treated C8T cells. Images were acquired and colonies were counted after 3 weeks. Values are mean ± SE of three independent experiments. *P< 0.05, **P< 0.01.
DISCUSSION
Metabolic switch from oxidative phosphorylation to glycolysis has long been observed as a bioenergetic signature of cancer cells (2). However, whether mitochondrial dysfunction and/or mtDNA mutations play a role in tumorigenesis is still not very clear (31). On the one hand, there have been expanding reports on mitochondrial dysfunction and mutations in both nuclear and mitochondrial genomes in various cancer cells (3,4). In particular, mutations in the nuclear DNA-encoded subunits of respiratory complex II or succinate dehydrogenase (32) and other components of tricarboxylic acid (TCA) cycle, such as fumarate hydratase (33) and isocitrate dehydrogenase (34), have been identified in different cancers. In such cases, the resulting accumulation of metabolites was suggested as oncogenic signals. On the other hand, some well-established tumor suppressors and oncogenes were shown to regulate energy substrate utilization and metabolic pathways (35). Loss of tumor suppressor p53 directly down-regulated OXPHOS capacity at respiratory complex IV (36) and at the same time increased glycolysis (37). Activation of oncogenes, i.e. C-MYC, was shown to enhance aerobic glycolysis by upregulating the expression of glucose transporters and several other genes encoding enzymes involved in glycolysis (38). However, contrary to Warburg's original hypothesis, some of the recent studies demonstrated that even during oncogenic (C-MYC and RAS) activation, such bioenergetic switch can happen without damaging OXPHOS machinery (25). Therefore, it becomes an important issue in cancer research whether mtDNA mutations and/or mitochondrial dysfunction sufficient and/or necessary for tumorigenesis.
With the establishment of the approach to generate cybrids by transferring mtDNA from one nuclear background to another, it became feasible to study the role of mtDNA mutation in disease progression. The effect of a pathogenic ATPase6 (T8993G) mtDNA mutation was first tested with standard tumorigenesis assays and reported for enhancing tumor growth (39). Later, mtDNA mutations identified in cancer cells were also analyzed with both cybrids (10,40) and introduction of nuclear-transcribed and mitochondrial-targeted gene methods (41). These cancer-specific mutations again exhibited increased anchorage-independent growth and/or metastasis. In all cases, upregulation of ROS production which may evoke oncogenic signaling and apoptotic resistance were implicated in mediating the signal from mtDNA mutations to tumorigenesis.
Although mutations all over the mitochondrial genome were found in various cancer cells, more mutations have been identified in the genes encoding complex I subunits (4). Mutations in the ND6 gene identified in high metastatic potential Lewis lung carcinoma caused the reduction in complex I activity, ROS overproduction and increased metastatic potential when the mutant mtDNA transferred to a cancer cell nuclear background with low metastatic potential (40). The expression of a mutant mtDNA-encoded ND2 gene detected in the head and neck squamous cell carcinomas was found simultaneously to stimulate aerobic glycolysis, ROS production and malignant tumor growth (41). Previously, we also reported that complex I defective cells carrying heteroplasmic mutation in ND5 subunit were defective in mitochondrial functions and showed increased tumorigenic potential, probably mediated by accumulation of mitochondrial ROS and/or resistance to apoptosis (10).
In the present study, we took a genetic rescue approach to assess the role of complex I dysfunction in tumorigenesis. Advantage was taken with the availability of a yeast NADH dehydrogenase (NDI1) gene which previously was shown functionally complemented the complex I defects in mammalian cells (12,14).
One important finding of our study is the role of AKT activation in mediating the signal from complex I to tumorigenesis, probably through alterations in the ROS level and redox status. The AKT/PI3K pathway plays a central role in integrating various cellular stimuli to a broad range of cellular functions (42). Upon activation, AKT enhances several metabolic activities and promotes cell survival and growth. It increases the expression of nutrient transporters, enabling increased uptake of glucose, amino acids and other nutrients (43). In addition, through effects on gene expression and enzyme activity, AKT enhances glycolysis and HIF1α stabilization which coordinates glucose consumption and lactate production (44). AKT further regulates an array of downstream targets involving cell survival, cell growth and cell proliferation, including members of the BCL-2 family and FOXO family, mTOR complex 1 and GSK-3β (45,46). We found that correlated with the activation of AKT, the protein levels of HIF1α, BCL-XL and MCL1 were higher in complex I defective C8T cells. Conversely, in C8T cells with NDI1 where complex I function was partially restored, both AKT activation and upregulation of these downstream proteins were reversed. Pathway focused gene expression analysis further suggested that compared with the parent cells with normal complex I function, AKT activating pathways and anti-apoptotic pathways were up-regulated in complex I defective C8T cells, and those pathways which negatively regulates AKT such as pTEN were downregulated, and these alterations were again reversed when complex I function was partially restored (Supplementary Material, Fig. S3).
Our results further indicated that activation of AKT was probably due to the alterations in ROS production and redox status resulted from complex I dysfunction. Because complex I is the major site for mitochondrial ROS production, complex I mutants were shown to exhibit increased oxidative stress (3). Reducing equivalent NADH is generated through glycolysis and the TCA cycle. One of the immediate consequences of complex I dysfunction was the accumulation of its substrate, NADH, and this metabolic alteration led to a lower NAD to NADH ratio. The elevated ROS level and altered redox status are common characteristics of many tumors and have been shown to regulate many pathways involved in cell growth and cell death (21,45). A recent study also suggested that elevated levels of NADH in respiration defective cells could activate AKT through a NADH/NADPH redox modification mechanism (47).
We found that the complex I defective C8T cells exhibited more mitochondrial ROS, and incorporation of NDI1 in the host respiratory system in C8T cells not only significantly improved the mitochondrial function such as membrane potential and ATP generation, but also reduced oxidative stress. It is interesting to note that although introduction of NDI1 in wild-type 143B did not improve mitochondrial function in terms of membrane potential and ATP production, it decreased ROS production and increased the NAD/NADH ratio as in C8T cells. Accordingly, fewer colonies were formed in soft agar with 143B after NDI1 expression.
To exclude the possibility of a role of NDI1 on tumorigenesis independent of complex I activity, and to further confirm the role of complex I stress, a complementary pharmacological approach was also taken to test the effect of complex I defect and its effect on ROS production, AKT activation and tumorigenesis. It is interesting to note that C8T-28 and C8T-29 showed quite different responses in some of the measurements. To find out if there were any changes in the level of heteroplasmy in C8T-28 and C8T-29 cells which could contribute to the phenotypes we observed with the NDI-1 transformants, we analyzed the level of heteroplasmy in these cells. We found no significant shift in the heteroplasmy in the NDI1-expressing C8T-28 and C8T-29 lines compared with C8T cells (Supplementary Material, Fig. S1A and B). Therefore, we believe that NDI1 expression is the primary reason for the improvement of mitochondrial functions and reduction in tumorigenesis in these heteroplasmic C8T-28 and -29 cells. The differences between C8T-28 and C8T-29 lines may be due to the expression level of NDI1 protein in these cells (Fig. 1A). Also, we cannot completely rule out the possibility of any alterations in the nuclear genome as they may also contribute to the changes we observed. Taken together, a signaling pathway from complex I dysfunction to ROS overproduction/redox alteration to AKT activation to metabolic switch/cell survival to tumorigenesis has been established.
In summary, among several approaches, the Warburg effect or metabolic switch in cancer cells can also be achieved by activation of signaling pathways initiated by oncogenes/loss of tumor suppressors or mitochondrial dysfunction through oxidative stress and redox alteration (Supplementary Material, Fig. S5A). In particular, the ROS-dependent activation of AKT pathway resulted from complex I defect could lead to enhancement of glycolysis and cell survival which contributed to the increased tumorigenesis (Supplementary Material, Fig. S5B).
MATERIALS AND METHODS
Cell culture
The 143B (ATCC CRL 8303) is a human osteosarcoma-derived cell line. The C8T cell line used in this study was generated and characterized as described previously (8,10). All the cell lines used in the present work were grown as monolayer in Dulbecco's modified Eagle's medium (Cellgro-Meditech,Inc., Herdon, VA, USA) supplemented with 10% fetal bovine serum (FBS).
Generation of stable NDI1 transgenic lines
The construction of the NDI1-expressing plasmid pHook (NDI1) has been described previously (15). From this construct, full-length yeast NDI1 gene (∼1.5 kb) was amplified, cloned in lentiviral vector (pLenti6.3 vector, Invitrogen, Carlsbad, CA, USA) and confirmed by sequencing. To generate lentiviral supernatant, the ViraPower lentivirus system was used according to the manufacturer's instructions (Invitrogen). The 143B and C8T cells were infected with concentrated lentiviral supernatant (108–109 IU/ml) and selected with 0.5 μg/ml blasticidin for 3 weeks. Several independent single clones were selected by immunoblotting and two clones for each line (143B: -45, -46; C8T:-28, -29) were used for further study. To confirm the presence of heteroplasmic mutation in the ND5 gene of 143B, C8T-28 and C8T-29, cells were sequenced periodically using ND5-specific primers as described previously (8).
Western blotting
Proteins were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) under reducing conditions. The resolved proteins were transferred electrophoretically to polyvinylidene fluoride membrane. After incubating with 5% dry milk in tris-buffered saline with tween (TBS-T) [150 mm NaCl, 50 mm Tris–HCl (pH 7.4), 0.05% Tween 20] for at least 1 h at room temperature, the membrane was incubated with primary antibodies [Phospho AMPK (Thr172), total AMPK, phospho-AKT (Ser473), total AKT, BCL-XL, MCL1 (from Cell Signaling Technology, Inc., Danvers, MA, USA) and P53, HIF1α, β-actin (from Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA)] for the appropriate time, washed extensively with TBS-T and then incubated with HRP-conjugated secondary antibody (1:2000 dilution). Protein bands were detected using super signal west pico chemi-luminescent substrate kit (Pierce, Thermo Fisher Scientific, Rockford, IL, USA). Densitometric analyses were done using Image J software.
MMP and ATP measurements
MMP was determined using the cationic fluorescent redistribution dye TMRM (Invitrogen). Approximately 1 × 106 cells (four replicates for each line) were seeded in each well of six-well plates. Next day, a total of 200 nm TMRM and 10 μg/ml of nuclear staining dye Hoechst-33342 (Sigma-Aldrich, St Louis, MO, USA) were added and incubated the cells for 15–20 min in a 37°C CO2 incubator. After washing, the cells were scraped and collected by centrifuging. Pellet was re-suspended in phosphate buffered saline and transferred to an opaque black 96-well microtiter plate. Fluorescence was recorded for TMRM (Ex 540 nm and Em 575 nm) and Hoechst-33342 (Ex 350 nm and Em 461 nm) in HT-BioTek fluorescence plate reader (BioTek Instrument Inc., Winooski, VT, USA). TMRM fluorescence was normalized with Hoechst reading. ATP measurements were done as described previously (10). Mean ± SE values were calculated from three independent experiments.
Cell viability measurements
Multiple identical samples of 1–5 × 105 cells were placed on six-well plates in 2 ml of the medium and cells were cultured at 37°C for 24 h. The medium was changed the next day with either regular growth media or with galactose media (containing 0.9 mg of galactose/ml and 0.5 mg of pyruvate/l, with 10% dialyzed FBS). Cell counting and viability was performed using cell viability analyzer (Beckman Coulter, Inc., Fullerton, CA, USA) based on the trypan blue exclusion method. Live cell images were taken using inverted microscope (Nikon Eclipse TE2000, Nikon Instruments Inc., Melville, NY, USA).
Soft agar assay and cell migration assay
Soft agar colony-forming assay was performed as described previously (10). For cell migration analysis, 5000 cells were seeded in 8 μm pore-sized cell culture inserts (BD Labware, Franklin lakes, NJ, USA) in media without serum. These inserts were placed in 12-well plates containing complete media and incubated for 12–16 h at 37°C and 5% CO2 incubator. The cells migrated to the lower part of insert were stained with Hema 3 staining system (Thermo Fisher Scientific, Inc.) and imaged under microscope (Nikon Eclipse TE2000). Stained cells were counted using Image J software (five random fields for each insert). All results for these experiments are the mean ± SE of at least three separate experiments.
Measurement of NAD/NADH, lactate and mitochondrial ROS measurements
The redox molecules NADH and NAD were measured using fluorescent NAD/NADH detection kit (Cell Technology Inc., Mountain View, CA, USA). The extracellular lactate level was determined as described previously (10). Mitochondrial ROS measurements were carried out using Mitosox assay (48). Briefly, after treatment, equal number of cells were re-suspended in Hank's buffered salt solution buffer with 5 μm Mitosox (Molecular Probes, Eugene, OR, USA) and Hoechst-33342 10 μg/ml, incubated at 37°C for 15 min. Cells were then washed, and the fluorescence (Mitosox: Ex 492 nm and Em 595 nm) was measured and recorded on Perkin Elmer 1000 multiwell fluorescence plate reader with temperature maintained at 37°C. Relative values were calculated after normalizing either with Hoechst fluorescence or for 1 × 106 cells. The H2O2 levels in isolated mitochondria were measured using the HRP-linked fluorometric assay (Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit; Molecular Probes). Mitochondria were isolated using mitochondrial isolation kit (Pierce) and protein was estimated using BCA kit (Pierce). Mitochondria (up to 10–20 μg in 50 μl volume) were added to 96-well plate in triplicates with a total reaction volume of 100 μl of reaction buffer containing 0.1 U/ml HRP, 50 μm Amplex Red reagents. Fluorescence (Ex 530 nm and Em 595 nm) was measured using fluorescence plate reader with temperature maintained at 37°C. The H2O2 concentrations were calculated from standard curve and normalized with per mg protein used per assay. All experiments were performed in triplicates and repeated at least three times. Values are mean ± SE of at least three separate experiments.
Data analysis
Data in graphs are reported as ±SE and depict the average of at least three independent experiments. Morphological images are representative of at least three independent experiments with similar results. Statistical analysis was performed using GraphPad software (GraphPad Software, La Jolla, CA, USA) with Student's paired t-test or a one-way analysis of variance followed by a Dunnett's multiple comparison test when appropriate.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work is supported by grants from National Institute of Health (1R21 NS072777), Wendy Will Case Cancer Fund and Chinese National Science Foundation (31070765/C050605).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Takao Yagi (The Scripps Research Institute, La Jola, CA, USA) for providing anti-NDI1 antibodies and Dr Giovanni Manfredi (Weill Cornell Medical College, NY, USA) for providing COX1 mutant cell lines. The SS31 peptide was a kind gift from Dr P. Hemachandra Reddy (OHSU, Portland, OR, USA). We thank Dr C.S. Jonathan Liu (SoftGenetics, LLC State College, PA, USA) for his help in the usage of Mutation Surveyor software program. We also thank Peiqing Hu for their technical assistance.
REFERENCES
- 1.Wallace D.C. Mitochondria as chi. Genetics. 2008;179:727–735. doi: 10.1534/genetics.104.91769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 3.Sharma L.K., Lu J., Bai Y. Mitochondrial respiratory complex I: structure, function and implication in human diseases. Curr. Med. Chem. 2009;16:1266–1277. doi: 10.2174/092986709787846578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu J., Sharma L.K., Bai Y. Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell Res. 2009;19:802–815. doi: 10.1038/cr.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai Y., Hu P., Park J.S., Deng J.H., Song X., Chomyn A., Yagi T., Attardi G. Genetic and functional analysis of mitochondrial DNA-encoded complex I genes. Ann. N. Y. Acad. Sci. 2004;1011:272–283. doi: 10.1007/978-3-662-41088-2_26. [DOI] [PubMed] [Google Scholar]
- 6.Bai Y., Attardi G. The mtDNA-encoded ND6 subunit of mitochondrial NADH dehydrogenase is essential for the assembly of the membrane arm and the respiratory function of the enzyme. EMBO J. 1998;17:4848–4858. doi: 10.1093/emboj/17.16.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai Y., Shakeley R.M., Attardi G. Tight control of respiration by NADH dehydrogenase ND5 subunit gene expression in mouse mitochondria. Mol. Cell Biol. 2000;20:805–815. doi: 10.1128/mcb.20.3.805-815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofhaus G., Attardi G. Efficient selection and characterization of mutants of a human cell line which are defective in mitochondrial DNA-encoded subunits of respiratory NADH dehydrogenase. Mol. Cell Biol. 1995;15:964–974. doi: 10.1128/mcb.15.2.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polyak K., Li Y., Zhu H., Lengauer C., Willson J.K., Markowitz S.D., Trush M.A., Kinzler K.W., Vogelstein B. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat. Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 10.Park J.S., Sharma L.K., Li H., Xiang R., Holstein D., Wu J., Lechleiter J., Naylor S.L., Deng J.J., Lu J., et al. A heteroplasmic, not homoplasmic, mitochondrial DNA mutation promotes tumorigenesis via alteration in reactive oxygen species generation and apoptosis. Hum. Mol. Genet. 2009;18:1578–1589. doi: 10.1093/hmg/ddp069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yagi T., Seo B.B., Di Bernardo S., Nakamaru-Ogiso E., Kao M.C., Matsuno-Yagi A. NADH dehydrogenases: from basic science to biomedicine. J. Bioenerg. Biomembr. 2001;33:233–242. doi: 10.1023/a:1010787004053. [DOI] [PubMed] [Google Scholar]
- 12.Park J.S., Li Y.F., Bai Y. Yeast NDI1 improves oxidative phosphorylation capacity and increases protection against oxidative stress and cell death in cells carrying a Leber's hereditary optic neuropathy mutation. Biochim. Biophys. Acta. 2007;1772:533–542. doi: 10.1016/j.bbadis.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song X., Deng J.H., Liu C.J., Bai Y. Specific point mutations may not accumulate with aging in the mouse mitochondrial DNA control region. Gene. 2005;350:193–199. doi: 10.1016/j.gene.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Bai Y., Hajek P., Chomyn A., Chan E., Seo B.B., Matsuno-Yagi A., Yagi T., Attardi G. Lack of complex I activity in human cells carrying a mutation in MtDNA-encoded ND4 subunit is corrected by the Saccharomyces cerevisiae NADH-quinone oxidoreductase (NDI1) gene. J. Biol. Chem. 2001;276:38808–38813. doi: 10.1074/jbc.M106363200. [DOI] [PubMed] [Google Scholar]
- 15.Seo B.B., Kitajima-Ihara T., Chan E.K., Scheffler I.E., Matsuno-Yagi A., Yagi T. Molecular remedy of complex I defects: rotenone-insensitive internal NADH-quinone oxidoreductase of Saccharomyces cerevisiae mitochondria restores the NADH oxidase activity of complex I-deficient mammalian cells. Proc. Natl Acad. Sci. USA. 1998;95:9167–9171. doi: 10.1073/pnas.95.16.9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholls D.G. Mitochondrial membrane potential and aging. Aging Cell. 2004;3:35–40. doi: 10.1111/j.1474-9728.2003.00079.x. [DOI] [PubMed] [Google Scholar]
- 17.Macpherson I., Montagnier L. Agar suspension culture for the selective assay of cells transformed by polyoma virus. Virology. 1964;23:291–294. doi: 10.1016/0042-6822(64)90301-0. [DOI] [PubMed] [Google Scholar]
- 18.Shin S.I., Freedman V.H., Risser R., Pollack R. Tumorigenicity of virus-transformed cells in nude mice is correlated specifically with anchorage independent growth in vitro. Proc. Natl Acad. Sci. USA. 1975;72:4435–4439. doi: 10.1073/pnas.72.11.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valster A., Tran N.L., Nakada M., Berens M.E., Chan A.Y., Symons M. Cell migration and invasion assays. Methods. 2005;37:208–215. doi: 10.1016/j.ymeth.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 20.D'Aurelio M., Pallotti F., Barrientos A., Gajewski C.D., Kwong J.Q., Bruno C., Beal M.F., Manfredi G. In vivo regulation of oxidative phosphorylation in cells harboring a stop-codon mutation in mitochondrial DNA-encoded cytochrome c oxidase subunit I. J. Biol. Chem. 2001;276:46925–46932. doi: 10.1074/jbc.M106429200. [DOI] [PubMed] [Google Scholar]
- 21.Lin S.J., Guarente L. Nicotinamide adenine dinucleotide, a metabolic regulator of transcription, longevity and disease. Curr. Opin. Cell Biol. 2003;15:241–246. doi: 10.1016/s0955-0674(03)00006-1. [DOI] [PubMed] [Google Scholar]
- 22.Cuezva J.M., Krajewska M., de Heredia M.L., Krajewski S., Santamaria G., Kim H., Zapata J.M., Marusawa H., Chamorro M., Reed J.C. The bioenergetic signature of cancer: a marker of tumor progression. Cancer Res. 2002;62:6674–6681. [PubMed] [Google Scholar]
- 23.Stacpoole P.W. Lactic acidosis and other mitochondrial disorders. Metabolism. 1997;46:306–321. doi: 10.1016/s0026-0495(97)90259-6. [DOI] [PubMed] [Google Scholar]
- 24.Fleury C., Mignotte B., Vayssiere J.L. Mitochondrial reactive oxygen species in cell death signaling. Biochimie. 2002;84:131–141. doi: 10.1016/s0300-9084(02)01369-x. [DOI] [PubMed] [Google Scholar]
- 25.Weinberg F., Hamanaka R., Wheaton W.W., Weinberg S., Joseph J., Lopez M., Kalyanaraman B., Mutlu G.M., Budinger G.R., Chandel N.S. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl Acad. Sci. USA. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson K.M., Janes M.S., Pehar M., Monette J.S., Ross M.F., Hagen T.M., Murphy M.P., Beckman J.S. Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proc. Natl Acad. Sci. USA. 2006;103:15038–15043. doi: 10.1073/pnas.0601945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohanty J.G., Jaffe J.S., Schulman E.S., Raible D.G. A highly sensitive fluorescent micro-assay of H2O2 release from activated human leukocytes using a dihydroxyphenoxazine derivative. J. Immunol. Methods. 1997;202:133–141. doi: 10.1016/s0022-1759(96)00244-x. [DOI] [PubMed] [Google Scholar]
- 28.Barrientos A., Moraes C.T. Titrating the effects of mitochondrial complex I impairment in the cell physiology. J. Biol. Chem. 1999;274:16188–16197. doi: 10.1074/jbc.274.23.16188. [DOI] [PubMed] [Google Scholar]
- 29.Cocheme H.M., Murphy M.P. Complex I is the major site of mitochondrial superoxide production by paraquat. J. Biol. Chem. 2008;283:1786–1798. doi: 10.1074/jbc.M708597200. [DOI] [PubMed] [Google Scholar]
- 30.Szeto H.H. Mitochondria-targeted peptide antioxidants: novel neuroprotective agents. AAPS J. 2006;8:E521–E531. doi: 10.1208/aapsj080362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carew J.S., Huang P. Mitochondrial defects in cancer. Mol. Cancer. 2002;1:9. doi: 10.1186/1476-4598-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bayley J.P., Devilee P. Warburg tumours and the mechanisms of mitochondrial tumour suppressor genes. Barking up the right tree? Curr. Opin. Genet. Dev. 2010;20:324–329. doi: 10.1016/j.gde.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 33.Tomlinson I.P., Alam N.A., Rowan A.J., Barclay E., Jaeger E.E., Kelsell D., Leigh I., Gorman P., Lamlum H., Rahman S., et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 34.Zhao S., Lin Y., Xu W., Jiang W., Zha Z., Wang P., Yu W., Li Z., Gong L., Peng Y., et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu P.P., Sabatini D.M. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 36.Matoba S., Kang J.G., Patino W.D., Wragg A., Boehm M., Gavrilova O., Hurley P.J., Bunz F., Hwang P.M. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 37.Bensaad K., Tsuruta A., Selak M.A., Vidal M.N., Nakano K., Bartrons R., Gottlieb E., Vousden K.H. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 38.Osthus R.C., Shim H., Kim S., Li Q., Reddy R., Mukherjee M., Xu Y., Wonsey D., Lee L.A., Dang C.V. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J. Biol. Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 39.Petros J.A., Baumann A.K., Ruiz-Pesini E., Amin M.B., Sun C.Q., Hall J., Lim S., Issa M.M., Flanders W.D., Hosseini S.H., et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc. Natl Acad. Sci. USA. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishikawa K., Takenaga K., Akimoto M., Koshikawa N., Yamaguchi A., Imanishi H., Nakada K., Honma Y., Hayashi J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 41.Zhou S., Kachhap S., Sun W., Wu G., Chuang A., Poeta L., Grumbine L., Mithani S.K., Chatterjee A., Koch W., et al. Frequency and phenotypic implications of mitochondrial DNA mutations in human squamous cell cancers of the head and neck. Proc. Natl Acad. Sci. USA. 2007;104:7540–7545. doi: 10.1073/pnas.0610818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manning B.D., Cantley L.C. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edinger A.L., Thompson C.B. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol. Biol. Cell. 2002;13:2276–2288. doi: 10.1091/mbc.01-12-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elstrom R.L., Bauer D.E., Buzzai M., Karnauskas R., Harris M.H., Plas D.R., Zhuang H., Cinalli R.M., Alavi A., Rudin C.M., et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 45.Clerkin J.S., Naughton R., Quiney C., Cotter T.G. Mechanisms of ROS modulated cell survival during carcinogenesis. Cancer Lett. 2008;266:30–36. doi: 10.1016/j.canlet.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 46.Plas D.R., Talapatra S., Edinger A.L., Rathmell J.C., Thompson C.B. Akt and Bcl-xL promote growth factor-independent survival through distinct effects on mitochondrial physiology. J. Biol. Chem. 2001;276:12041–12048. doi: 10.1074/jbc.M010551200. [DOI] [PubMed] [Google Scholar]
- 47.Pelicano H., Xu R.H., Du M., Feng L., Sasaki R., Carew J.S., Hu Y., Ramdas L., Hu L., Keating M.J., et al. Mitochondrial respiration defects in cancer cells cause activation of Akt survival pathway through a redox-mediated mechanism. J. Cell Biol. 2006;175:913–923. doi: 10.1083/jcb.200512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Degli Esposti M., McLennan H. Mitochondria and cells produce reactive oxygen species in virtual anaerobiosis: relevance to ceramide-induced apoptosis. FEBS Lett. 1998;430:338–342. doi: 10.1016/s0014-5793(98)00688-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.