Abstract
Congenital myasthenic syndromes (CMS) are inherited diseases affecting the neuromuscular junction (NMJ). Mutations in AGRIN (AGRN) and other genes in the AGRIN signaling pathway cause CMS, and gene targeting studies in mice confirm the importance of this pathway for NMJ formation. However, these mouse mutations are complete loss-of-function alleles that result in an embryonic failure of NMJ formation, and homozygous mice do not survive postpartum. Therefore, mouse models of AGRIN-related CMS that would allow preclinical testing or studies of postnatal disease progression are lacking. Using chemical mutagenesis in mice, we identified a point mutation in Agrn that results in a partial loss-of-function allele, creating a valid model of CMS. The mutation changes phenylalanine 1061 to serine in the SEA domain of AGRIN, a poorly characterized motif shared by other extracellular proteoglycans. NMJs in homozygous mice progressively degrade postnataly. Severity differs with genetic background, in different muscles, and in different regions within a muscle in a pattern matching mouse models of motor neuron disease. Mutant NMJs have decreased acetylcholine receptor density and an increased subsynaptic reticulum, evident by electron microscopy. Synapses eventually denervate and the muscles atrophy. Molecularly, several factors contribute to the partial loss of AGRIN's function. The mutant protein is found at NMJs, but is processed differently than wild-type, with decreased glycosylation, changes in sensitivity to the protease neurotrypsin and other proteolysis, and less efficient externalization and secretion. Therefore, the Agrn point mutation is a model for CMS caused by Agrn mutations and potentially other related neuromuscular diseases.
INTRODUCTION
Congenital myasthenic syndromes (CMS) are inherited diseases of the neuromuscular junction (NMJ). These diseases result from mutations that cause dysfunction in proteins associated with the presynaptic motor nerve terminal, the postsynaptic specialization of the muscle or the extracellular matrix of the synaptic cleft (1). Though rare, CMS can be very debilitating for patients, causing weakness, fatigue and sometimes impairing neuromuscular function so severely that they are lethal. Several CMS variants are caused by mutations in AGRIN or other genes encoding proteins in the AGRIN signaling pathway, an essential trans-synaptic cascade that is critical for the formation and maintenance of the NMJ.
AGRIN is a heparan sulfate proteoglycan associated with the basal lamina of the NMJ. It is most studied for its role in the embryonic development of the NMJ (2,3). AGRIN is secreted from ingrowing motor nerve terminals and stabilizes nascent sites of postsynaptic differentiation that arise in the end plate band of the muscle, although AGRIN can also induce de novo clusters of acetylcholine receptors (AChRs) in cultured myotubes and in transgenic mice (4–6). This activity of AGRIN depends entirely on the inclusion of two alternatively spliced exons (Z-exons) found only in neuronal agrin transcripts (7–9). Transcripts including one, the other, or both of these exons (encoding 8, 11 or 19 amino acids) induce the activation of the receptor tyrosine kinase, muscle-specific kinase (MuSK), in concert with its co-receptor lipoprotein related protein 4 (LRP4) (10–12). In addition to the Z alternative splice site near the 3′ end of the transcripts, AGRIN also has two alternative N-termini, arising from different transcriptional and translational start sites. The short N-terminal form (SN) is the predominant form in the brain and functions as a type II transmembrane protein (13,14). The longer N-terminal form (LN) has a signal peptide for secretion and is associated with the extracellular matrix in many tissues, including the NMJ (15).
Genetic targeting studies in mice demonstrated that eliminating either the NMJ-associated LN isoforms, the alternatively spliced Z-exons required for MuSK activation, or the bulk of the Agrin coding sequence, all result in an almost complete failure to maintain NMJs (15–18). As a result, mice homozygous for these mutations die at birth and are unable to move or breathe independently. Mouse mutations in Musk, Lrp4 or their intracellular effectors have a similar phenotype (19,20). These mouse mutations demonstrate the central and essential role of the AGRIN signaling pathway in NMJ development.
The activation of MuSK/LRP4 in muscle by LN-Z+ isoforms of AGRIN secreted from the motor nerve terminal promotes AChR clustering and postsynaptic differentiation through a signaling cascade that involves the MuSK-associated adaptor protein DOK7 and the intracellular scaffolding protein RAPSYN. Consistent with their importance at the mouse NMJ, mutations in the genes encoding MSK, RAPSYN and DOK7 cause inherited CMS in humans (21–23). MuSK is also a target for autoimmune serum-negative myasthenia gravis (24). Recently, a point mutation in AGRN (Gly1709Arg) was also reported to cause a CMS (25). As anticipated from the mouse studies, this mutation is proposed to be a partial-, not complete, loss of function resulting from a single amino acid change, and the mutant protein is still able to stimulate MuSK phosphorylation and promote AChR clustering in cultured myotubes. Application of recombinant G1709R protein disrupted existing NMJs in rats, suggesting some pathological function of the mutant protein itself. Patients with the AGRNG1709R mutation presented clinically with weakness and muscle atrophy, a predominance of type 1 myosin-positive muscle fibers, and fatigue with 3 Hz nerve stimulation. In examination of muscle biopsies, NMJ morphology was abnormal, and remodeling and denervated junctions were seen. In electron microscopy, enlarged subsynaptic folds were seen at junctions that were still innervated, although no decrease in AGRIN or MuSK immunostaining intensities was seen at the NMJ.
Although the use of gene targeting in mice has demonstrated the importance of AGRIN at the NMJ, existing alleles result in a complete loss of activity at the NMJ and make studies of postnatal myasthenic phenotypes impossible. Furthermore, a loxP-flanked conditional allele of Agrin exists, but the allele deletes inefficiently in response to CRE and determining if an NMJ has been deleted for motor neuron-derived AGRIN is complicated by protein contributed by the muscle and Schwann cells (26). Here we report an N-ethyl N-nitrosourea (ENU) induced point mutation in the mouse Agrin (nmf380-F1061S) that results in a partial loss-of-function allele, thus creating a model of CMS related to AGRIN signaling.
RESULTS
Genetic identification of a new allele of Agrn
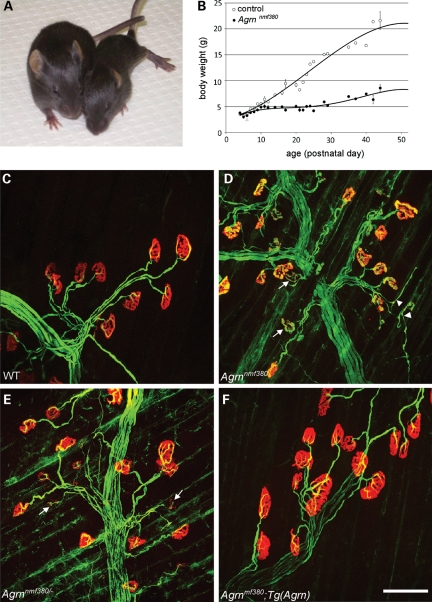
Random chemical mutagenesis screens in mice have proven to be an effective method to identify new genes involved in the development of the NMJ and to generate animal models for inherited neuromuscular diseases (19,27–29). In a third-generation recessive screen performed at The Jackson Laboratory, we identified a deviant strain, designated nmf380, which bred with the characteristics of a single recessive locus (two unaffected parents produced ∼25% affected offspring). The affected (presumed homozygous) mice were smaller than unaffected littermates (Fig. 1A and B), and displayed symptoms of neuromuscular dysfunction such as poor hindlimb motor control and atrophy. Affected mice typically died at a few weeks to a few months of age, with factors such as competition from littermates contributing to the variability. In addition to overt phenotypes, affected nmf380 mice had abnormal NMJs, as visualized in whole-mount muscle preparations from postnatal day 15 (P15) mice. In both wild-type and affected mice, motor axons, stained with antibodies to neurofilament and SV2, reached the muscle and innervated postsynaptic sites, labeled by α-bungarotoxin (BTX) staining of AChRs. In the affected mice, however, the postsynaptic sites were smaller and did not attain their normal, complicated, ‘pretzel-like’ adult morphology. In addition, the staining intensity of the BTX was reduced, and presynaptic terminals often sprouted beyond the boundaries of the postsynaptic specializations (Fig. 1C and D). Therefore, the nmf380 locus was suspected of being involved in neuromuscular development, and specifically in NMJ formation and stability.
Figure 1.
Overt phenotype and NMJ disaggregation in a new Agrn allele. (A) By P15, affected mice (right) are significantly smaller than their WT littermates, display altered locomotion and signs of muscle stiffness and atrophy. (B) Growth curves of mutant and unaffected littermates. Although the bodyweight is the same at birth, significant differences are observed by 2 weeks of age and by P21, affected mice weigh approximately half as much as controls. NMJs in control (C) and mutant (D) mice at P15. In control mice, the motor nerve terminal (green, labeled with anti-neurofilament and anti-SV2) completely overlaps the postsynaptic AChRs (red, labeled with α-BTX) on the muscle and the synapse is assuming its mature morphology. In mutant mice, the synapse still resembles an immature plaque, AChR staining intensity is reduced, AChR clusters begin to disaggregate (arrows) and motor nerve terminals are beginning to sprout beyond the endplates (arrowheads). (E) In genetic complementation tests, the nmf380 allele failed to complement Agrn knockout alleles, with similar overt and NMJ phenotypes (arrows) to nmf380 homozygotes. (F) The phenotype was completely rescued by an Agrn genomic transgene. Scale bar in (F) is 100 µm.
In the mutagenesis, C57BL/6J mice were dosed with ENU and bred to generate third-generation offspring to be examined for recessive phenotypes (see Materials and Methods). Unless otherwise specified, all analyses and data presented are from mice in an inbred C57BL/6J genetic background.
We used standard genetic mapping techniques with BALB/cByJ as a mapping partner to determine that the mutation causing neuromuscular dysfunction was on telomeric chromosome 4, closely linked to D4Mit344 at 153.5 Mb. The gene encoding AGRIN (Agrn) is known to be essential for normal NMJ formation and lies at 155.5 Mb on mouse chromosome 4 (syntenic to human 1p36.33). Given the NMJ defects in the nmf380 mice and the map location, we performed a complementation test with Agrn knockout mice to determine whether nmf380 was indeed an allele of Agrn. The Agrn knockout allele dies at birth as a homozygote with a nearly complete failure of NMJ development (16). However, the stock used for complementation testing was homozygous for the Agrn knockout on chromosome 4, but was rescued by a full-length fusion of AGRIN and cyan fluorescent protein (AGRIN-CFP) encoded by a modified Agrn bacterial artificial chromosome transgene construct on Chr. 8 [Tg2R9, (30)]. These mice were bred to heterozygous (unaffected, known carrier) nmf380 mice. As anticipated for an allele of Agrn, 11/53 offspring were affected with neuromuscular dysfunction and NMJ dysmorphology similar to that of affected nmf380 mice (Fig. 1E). In addition, none of the affected mice carried the Agrin-CFP transgene (29/53 mice were transgene positive), indicating that nmf380 fails to complement the Agrn knockout allele, but was rescued by the Agrin-CFP transgene. The failure to complement was confirmed in crosses with another Agrn knockout allele lacking the active Z exons (18), and the rescue was confirmed with a second Agrin-CFP transgene insertion, Tg6R16, with lower expression levels than the first (30). NMJs in rescued mice had normal pre- and postsynaptic morphology (Fig. 1F), indicating the transgene can fully rescue the nmf380 mutation like the knockout allele, and arguing against a dominant negative effect of the mutant protein. We therefore conclude that nmf380 is a new allele of Agrn in mice.
Molecular definition of the Agrnnmf380 mutation
Given that Agrn knockout mice die at birth, and that the nmf380 mice were generated in an ENU mutagenesis, which typically causes A to G point mutations in DNA, we anticipated that the nmf380 mutation would be a point mutation resulting in a partial loss-of-function allele. To sequence the Agrn gene, cDNA was prepared from the brains of Agrnnmf380/nmf380 and C57BL/6J control mice by standard techniques and the entire open reading frame (ORF) was sequenced. A single base pair change was found in the cDNA encoded by exon 18 of Agrnnmf380/nmf380 mice. This change is an A to G conversion, consistent with ENU, and results in the change of phenylalanine 1061 to serine (F1061S, numbered according to the SN form of the protein) in the SEA domain of AGRIN (Fig. 2A). SEA domains are poorly characterized motifs found in extracellular matrix-associated glycoproteins. AGRIN's SEA domain is found in the middle of the protein, flanked by serine/threonine-rich repeats (Fig. 2B). The acronym stands for Sea urchin sperm protein, Enterokinase, and Agrin, thus AGRIN is a defining protein for these domains (31). Evolutionarily, the mutated phenylalanine is highly conserved in Agrin genes as distantly as they can be identified by sequence similarity in cartilaginous fishes (Fig. 2C). The phenylalanine is also conserved in other SEA domains such as those in mucins 1 and 13, and enterokinase, but is not found in all SEA domains, such as those in other mucins or perlecan. The mutated phenylalanine is predicted to be at the base of an alpha-helix by a structural prediction model extrapolated from the structure of the Mucin16 SEA domain (Fig. 2D) (32).
Figure 2.
Molecular characterization of the new Agrn mutation. (A) Sequencing of cDNA from Agrnnmf380 homozygotes reveals an A to G change in exon 18 of Agrn, resulting in a change of phenylalanine 1061 to serine (F1061S) in the protein. (B) F1061 (*) is in the SEA module of AGRIN, a poorly characterized domain in the middle of the AGRIN protein. Other domains include the type II transmembrane SN-N-terminus, the secreted, laminin-binding LN-N-terminus, folistatin repeats (F), sites of glycosaminoglycan addition (GAG), laminin domain (L), serine/threonine-rich repeats (ST), epidermal growth factor like repeats (EGF1-4), laminin-type globular domains (G1–3) and sites of alternative splicing (X, Y, Z). (C) A phenylalanine at this position in AGRIN (dark blue highlight) is conserved in rodents, human, chick, zebrafish (danre) and electric ray (disom), as well as in the SEA modules of mouse enterokinase, mucins 1, 13 and 16 (Q9D1H1). (D) A structural prediction of the SEA domain of AGRIN extrapolated from the crystal structure of Mucin 16 shows the mutation changes a phenylalanine located at the carboxy terminal end of the first alpha-helix of the SEA module (arrowhead). The autocatalytic cleavage site found in the mucins in a hinge region is also indicated (arrow). (E) Analysis of mRNA levels by northern blotting indicates that there is no reduction in Agrn transcripts or changes in transcript size in Agrnnmf380 homozygotes, as anticipated for a point mutation. Control (WT) and mutant (M) lanes are indicated, β-Actin was used as a loading control. R) Reverse-transcription PCR on brain cDNA from control (WT) and mutant (M) mice with primers in exons 31 and 34, flanking the alternative Z-splicing site (exons 32 and 33). DNA molecular weight standard is on the left side and PCRs on cloned Z0, Z8 and Z19 cDNAs with the same primers are shown on the right. Alternative splicing at the Z-splicing site is critical for AGRIN's activity and splicing at this site is not changed by the mutation.
The single nucleotide change in exon 18 of the 40-exon gene does not appear to affect transcript levels or size by quantitative polymerase chain reaction (QPCR) (data not shown) or northern analysis (Fig. 2E). Furthermore, the mutation does not affect the alternative splicing at the ‘Z’ splice site (exons 32 and 33) that is essential for AGRIN's activity in NMJ formation (Fig. 2F). The amino acid change is also not close to a potential autolytic cleavage site found in mucin SEA domains, which is in the hinge region between two beta-sheets (Fig. 2D). Therefore, it appears that the mutation causes a partial loss-of-function phenotype that impacts the protein and not the mRNA.
Postnatal NMJ defects in Agrnnmf380/nmf380 mice
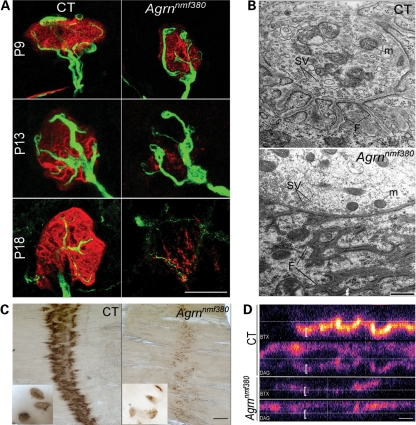
To further validate the Agrnnm380 allele as a partial loss-of-function CMS model, and to better understand the progression of the NMJ phenotype, we examined NMJs by both light and electron microscopy in different muscles and at different ages. To facilitate the visualization of motor axons in these studies, the Agrn mutant mice were bred to Thy1-YFP16, a transgene expressing yellow fluorescent protein (YFP) in all motor neurons from embryonic development onward (33). In the triangularis sterni, a thin, flat muscle on the inside of the rib cage, NMJs are of normal size and are normally innervated at P9 (Fig. 3A). However, some defects begin to arise by P13, with reduced intensity of BTX staining and abnormal presynaptic morphology. By P18, NMJs have largely degraded both pre- and postsynaptically. NMJs from the plantaris muscle in the lower leg at P20 were evaluated by transmission electron microscopy. In control muscles, motor nerve terminals rich in mitochondria and synaptic vesicles sit in depressions in the muscle plasma membrane (Fig. 3B). In the postsynaptic membrane, junctional folds are found in direct apposition to presynaptic release sites, and electron dense staining in the plasma membrane at the crests of these folds is the result of the high density of AChRs in the membrane. In the mutant NMJ, mitochondria and synaptic vesicles were still found in nerve terminals, but in some instances, the postsynaptic apparatus was abnormal, with an extensive subsynaptic reticulum that does not appear to be continuous with the plasma membrane and synaptic cleft. The presence of junctional folds indicates some maturation of postsynaptic differentiation; however, in the mutant junction shown in Figure 3B, folds extended 3.1 µm into the muscle fiber from the synaptic cleft. In other junctions with clearly identifiable junctional folds, these extended 1.1 µm and 600 nm, whereas control junctions had folds that extended 762 ± 49 nm from the synaptic cleft, thus only one mutant NMJ had ‘normal’ morphology. In other examples, such folds were found in regions of the muscle fiber extending beyond those in direct contact with the nerve terminal, suggesting that the terminal was beginning to lose contact with the muscle, resulting in partially denervated junctions (data not shown). Another frequently observed phenotype in the mutant samples was a cross-sectioned nerve terminal with 50 nm small clear vesicles and mitochondria, but no discernable postsynaptic specialization (8 identified). We presume these are sprouting terminals observed in light microscopy, and such structures were not found in the wild-type samples. Nerve terminals in the mutant mice had a normal content of vesicles and mitochondria (46.2 ± 7.8 and 1.49 ± 0.38 per sq µm, respectively) compared with controls (57.3 ± 7.5 and 1.47 ± 0.42 per sq µm, respectively, n = 11 mutant and 12 control terminal profiles measured). Therefore, axonal transport of cargoes such as synaptic vesicle components and mitochondria appears to be intact in the Agrnnmf380/nmf380 mice.
Figure 3.
The F1061S mutation causes a postnatal onset NMJ disaggregation. (A) Time course of the NMJ formation and disaggregation on the triangularis sterni muscle. Labeling with BTX (red), and Thy-1-YFP16 (green) demonstrates a progressive defects in both pre- and postsynaptic features of the NMJ. Scale bar 25 μm. (B) Transmission electron microscopy on the plantaris muscle at P20. Hallmarks of presynaptic (mitochondria, M, synaptic vesicles, SV) and postsynaptic (folds, F) differentiation are clear in control NMJs, and are present but abnormal in mutant NMJs, suggesting the synapses begin to mature, forming postsynaptic folds for example, and then disperse. Scale bar 500 nm. (C) ECM-bound AChE is also removed from the NMJs in the mutant, as demonstrated by a histochemical stain for AChE activity, although an endplate band is still discernable. Scale bar is 500 μm (50 μm for the inserts). (D) Beta-dystroglycan is also disrupted at the NMJ. Confocal stacks where the BTX (blue, top) and beta-dystroglycan (DAG, red, bottom) channels are separated, and viewed in the X–Z plane in false color. The AChR labeling colocalizes with synaptic dystroglycan (brackets) whose intensity is reduced in Agrnnmf380 homozygotes. Scale bar 5 μm.
In both wild-type and mutant electron micrographs, an extracellular basal lamina can be seen in the synaptic cleft. This extracellular matrix contains a number of specialized synaptic proteins, including acetylcholine esterase (AChE), which degrades the neurotransmitter acetylcholine (ACh), ending synaptic transmission. Consistent with a generalized loss of synaptic specialization, AChE is also lost from the mutant NMJ, as demonstrated by a histochemical reaction to detect its activity (Fig. 3C). Beta-dystroglycan (DG), the transmembrane fragment of the glycoprotein DG, is also altered in its synaptic abundance and no longer shows enrichment at sites of high BTX labeling intensity in the postsynaptic membrane of Agrnnmf380 mutant mice (Fig. 3D).
Muscle–muscle variability and fiber-type conversion in Agrnnmf380/nmf380 mice
NMJs from the triangularis sterni are shown in Figure 3; however, the severity and progression of NMJ defects varied greatly between muscles. We therefore quantified the phenotype in different muscles at different ages to assess this variability (Fig. 4A and B). NMJs in the diaphragm are affected at birth, with overgrowth of presynaptic endings and indistinct BTX staining at P1. This is more severe at P4, and by P14, virtually no intact NMJs remain. In contrast, the tibialis anterior (TA) progresses more slowly and is quite normal at P1 and P4, but NMJs are clearly disaggregating by P14 with some intact junctions persisting. As an indicator of severity, we quantified NMJ area and staining intensity of BTX in the diaphragm, soleus and TA muscle at P1, P4 and P14 (Fig. 4B). Consistent with the images in Figure 4A, the quantification revealed that NMJs in the diaphragm are affected soon after birth, the TA follows a similar progression, but the soleus is slower to disaggregate and denervate, with no differences from control at P4 but significant changes by P14. Interestingly, some other muscles, such as the extraocular muscles, are largely spared in the Agrnnmf380/nmf380 mice (Fig. 4C). The extraocular muscles are often more severely compromised in myasthenias, although this is quite variable in CMS. The extraocular muscles differ from skeletal muscles in many ways, including motor unit size, firing rate, fiber types and subunit composition of AChRs. These differences have not been characterized comprehensively in mice, although components of the AGRIN signaling pathway are found at NMJs of extraocular muscles (34). It is unclear which of the specializations of extraocular muscles may be contributing to their preservation in the Agrnnmf380/nmf380 mice; however, it is noteworthy that other motor neuron diseases and muscular dystrophies also spare these muscles in mice and people. In the gastrocnemius at P20, we saw an increase in type1 slow myosin-positive muscle fibers in Agrnnmf380/nmf380 mutant mice, consistent with motor innervation defects and with the human CMS phenotype (Fig. 4D).
Figure 4.
Variability of the NMJ phenotype between muscles and transition to a slow-fiber phenotype. (A) Comparison of the NMJ disaggregation in the diaphragm (Dia), a muscle affected early, and the skin-facing region of the TA, a muscle affected later at postnatal days (P) 1, 4 and 14. Presynaptic overgrowth and decreased intensity of AChR staining is evident in the diaphragm at P1, but is not evident in the TA until later ages. Scale bar 100 μm. (B) Quantification of the disaggregation of the NMJs in three representative muscles, the diaphragm, TA and soleus (Sol). Bars indicate the AChR cluster area, lines indicate the intensity of BTX labeling of AChRs expressed as a percentage of control values. (C) The NMJs of extraocular muscles are preserved at P30, an age where NMJs are severely affected in all other muscles. Scale bar 100 μm. (D) Type I, slow-myosin staining (green, BTX in red) in cross-sections of the lateral gastrocnemius at P20 demonstrates an increase in slow fibers in the Argnnmf380 mutant mice. Scale bar 50 μm.
Agrnnmf380/nmf380 shares features with motor neuron disease models
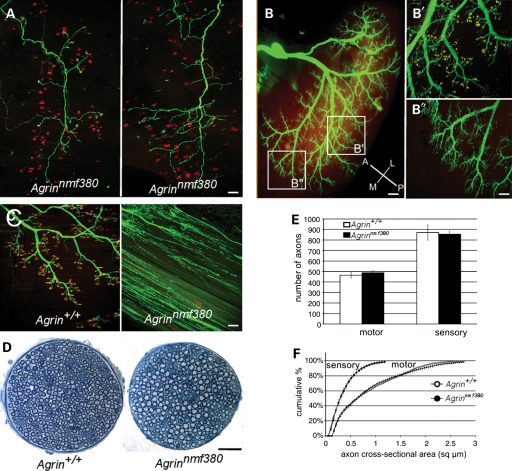
To further examine commonalities of the Agrnnmf380/nmf380 phenotype with other neuromuscular disease models, we analyzed the pattern of NMJ disaggregation and denervation at the level of single motor units and regionally within muscles. In mutant triangularis sterni muscles labeled using the transgenic Thy1-YFPH strain [which typically labels a single motor unit in this muscle (33)], we noted that the severity of NMJ defects was synchronous within a motor unit (Fig. 5A). This would be expected if the phenotype resulted from deficiencies in the motor neuron, but would not necessarily be the case if the defects were in the muscle fibers or individual NMJs. Regional differences within a given muscle have been noted previously, and these correspond to differing sensitivities to degeneration in models such as the transgenic SOD1G93A model of familial amyotrophic lateral sclerosis (ALS) (35–37). We examined regional differences in the gastrocnemius and determined that NMJs are lost in the same pattern in Agrnnmf380/nmf380 mice. NMJs in the intermediate region of the muscle are still intact, whereas synapses in the medial portion of the muscle are largely disaggregated (Fig. 5B). Similar results were found in the TA, where the pattern of NMJ disaggregation again matched the pattern reported for SOD1G93A transgenic mice (data not shown). It is interesting that the pattern of denervation in Agrn mutant muscles matches that of a motor neuron disease model. In the SOD1G93A mice, defects are seen at the NMJ before any locomotor phenotype or axon loss is evident (38), but the nerve then loses contact with the muscle and dies back. Similarly, in mouse models for spinal muscular atrophy and human patients, NMJ abnormalities are observed before the onset of axon retraction (39). However, unlike die-back neuropathies, the motor axons in the Agrnnmf380/nmf380 mice sprout throughout the muscle (Fig. 5C). This phenotype resembles the embryonic phenotype seen in complete loss-of-function alleles of Agrn. Despite the loss of NMJ integrity, the axons of peripheral nerves are surprisingly unaffected and do not retract or degenerate, with no changes in morphology, axon number or axon size in either the motor or sensory branches of the femoral nerve (Fig. 5D–F).
Figure 5.
Agrnnmf380 shares features with motor-neuron disease models, but is not a die-back neuropathy. (A) Two individual motor units in the triangularis sterni at P10, visualized with the Thy1.1-YFP-H transgenic strain in Agrnnmf380/nmf380 mice (green). Left, all the terminals of the motor unit are still connected to their NMJs (α-BTX, red). Right, all the terminals of the motor unit have lost their connections and are sprouting, indicating that preservation or disaggregation of NMJs is synchronous at the level of motor units. (B) Innervation of the lateral gastrocnemius of P10 mutants visualized with transgenic Thy1-YFP16. The antero-posterior and medio-lateral axes are shown in the lower right corner. Higher magnification of the intermediate (B′) and medial (B″) regions is shown on the right. Most of NMJs of the intermediate region (B′) are still innervated and axon terminals facing disaggregated NMJs do not extend beyond the postsynaptic site. In the medial region (B″), most of NMJs are disaggregated and axon terminals are abundantly sprouting. (C) Medial aspect of the quadriceps in 3-month-old control and mutant mice. NMJs have disaggregated in the mutant muscle and presynaptic terminals are sprouting throughout the muscle. (D) Sections of the motor branch of the femoral nerve innervating the quadriceps in 3-month-old mice show neither a noticeable loss of axons nor a reduction in their diameter in the mutant nerves. (E and F) Quantification of the number of axons in the motor and sensory branches of the femoral nerve (E) and a cumulative histogram of their cross-sectional areas (F) at 3 months of age show no significant axon loss or atrophy, more than 2 months after the loss of most of NMJs in the quadriceps (n = 4 controls and 4 mutants). Scale bars: 100 μm in A, B′, B″ and C; 200 μm in (B) and 50 μm in (D).
Genetic background influences phenotype severity
In addition to muscle-to-muscle variability and regional differences in sensitivity within muscles, we also observed effects of genetic background on the severity of the phenotype. Little change in onset, progression or lifespan was seen when the mutation was backcrossed into FVB, DBA or BALB backgrounds in N3 to N8 generations. However, in an N3 CAST background, the severity of the phenotype was markedly decreased; mice survived over 1 year and were only nominally smaller than littermates (Fig. 6A). In sensitive genetic backgrounds such as C57BL/6 and DBA, Agrnnmf380 mutant muscles were disproportionately smaller based on the ratio of muscle weight (mg) to body weight (g), indicating atrophy. However, in the CAST background, neither overall body weight nor muscle weight:body weight ratio was different than control mice from the same cross. The mutation was still fully penetrant, with all homozygous mice being affected, and NMJs had abnormal morphology, but were not disaggregated to the extent seen in other backgrounds (Fig. 6B). Examination of the endplate band in TA muscles with α-BTX staining of NMJs demonstrated an almost complete loss of postsynaptic differentiation in young Agrnnmf380/nmf380 mice in a C57BL/6 background, whereas the endplate band was largely intact in N3 CAST mice, even at 10 months of age (Fig. 6C). Muscles examined by histology were not markedly smaller in the N3 CAST mice, and there was only mild evidence of fibrosis or neurogenic atrophy typical of long-term denervation, which is observed in Agrnnmf380 homozygotes at younger ages in other backgrounds (Fig. 6A and D). An examination of B6 × CAST F2 mice suggested that the modifier effects of the CAST background are polygenic (data not shown). We have not tested the CAST background for modifier effects in Agrn knockout strains, which die at birth and have embryonic disaggregation of NMJs. We therefore do not know if the CAST modifiers are mitigating the specific cellular and molecular defects of the F1061S mutation, or are generally compensating for impaired signaling in the AGRIN pathway, or a combination of these mechanisms. However, these results show that the Agrnnmf380 phenotype can be altered by genetic background to more closely resemble the milder human disease phenotype, and emphasize the importance of working in a defined genetic background in experiments on mouse models of neuromuscular disease.
Figure 6.
Genetic background effects in Agrn mutant mice. (A) Muscle weight (mg): body weight (g) ratios illustrate the neurogenic atrophy of the lateral and medial gastrocnemius (LGC and MGC), TA and plantaris (Pl) in P15–P30 Agrnnmf380/nmf380 mutants in a C57BL6 or DBA/2 background (pooled measures on two controls and eight mutants). In an N3.CAST background (3 mutants, 3 controls, 10 months old), the atrophy of these muscles was less severe and not significant. Student t-test, *P < 0.05, **P < 0.01. Inset, body weights are reduced in mutant animals relative to age-matched littermates in a C57BL/6 background (P20), but not in a CAST background (10 months). (B) NMJs of the TA display an abnormal shape but are still partially innervated in 10-month-old mutants in an N3 CAST background. Scale bar 10 μm. (C) The endplate band of the TA shown by BTX staining. Genotypes are indicated at the bottom of (D). C57BL/6 animals were 1 month old, N3 CAST animals were 10 months old. Scale bar 100 μm. (D) Hematoxylin and eosin staining on hindlimb cross-sections shows the developmental delay and atrophy of all mutant muscles in the C57BL/6 background, whereas the atrophy is very limited in the N3.CAST background. Inserts show high magnification of the muscle fibers. While most of the fibers have a polygonal shape and are tightly arranged in the controls (Agrn+/+), numerous rounded, smaller fibers characteristic of a neurogenic atrophy, are seen in the mutant in a C57BL/6 background. A few rounded fibers are also seen in the N3 CAST mutant (asterisk), but the atrophy is limited. C57BL/6 animals were 1 month old, N3 CAST animals were 10 months old. Scale bar is 1 mm on the top panels and 100 μm in the inserts.
Mutant AGRIN protein is stable
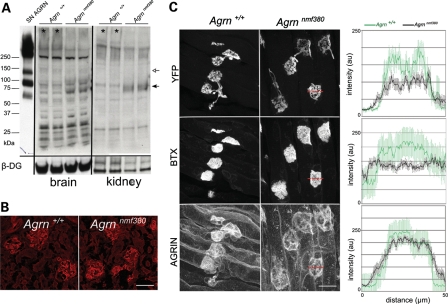
To begin to assess the cellular and molecular basis for the Agrnnmf380/nmf380 partial loss-of-function phenotype, we examined the trafficking and stability of the protein. In western blot analysis from P30 brain and kidney, a decrease in glycosylated, full-length AGRIN (>250 kDa) was observed, as well as an increase in a 75 kDa fragment detected with antibodies that recognize the C-terminal portion of the protein (Fig. 7A). The predominant form of AGRIN in the brain is the transmembrane SN-isoform, but NMJ development relies on the secreted, matrix-associated LN-isoform (15). In other tissues, LN-AGRIN is found in basement membranes, such as in the glomerulus of the kidney. In the Agrnnmf380/nmf380 mice, localization and staining in the kidney was indistinguishable from controls (Fig. 7B). Quantitative fluorescence was used to evaluate the abundance of AGRIN at the NMJs of P4 triangularis sterni muscles. Both the YFP-intensity of presynaptic motor terminals and BTX-intensity of postsynaptic AChRs were decreased in the mutant NMJs relative to controls, but the intensity of AGRIN immunoreactivity was similar using an antibody that recognizes the N-terminus of AGRIN (40) (Fig. 7C), suggesting that AGRIN is trafficked to the NMJ and is stably incorporated in the extracellular matrix. However, this analysis is complicated by the fact that terminal Schwann cells and the muscle fibers themselves are also making AGRIN at the NMJ. In addition, the western blot of AGRIN suggested that mutant AGRIN may be differentially processed, which may not be evident by immunocytochemistry.
Figure 7.
AGRIN abundance and deposition in tissues. (A) Western blot on brain and kidney protein preparations. Using an antibody that recognizes the C-terminus of AGRIN, glycosylated, full-length AGRIN (>250 kDa, *) is reduced in P30 mutant samples, and a shorter peptide (75 kDa, black arrow) is increased. Recombinant SN-AGRIN was run in parallel on the same gel and shows the full-length protein, the 110 kDa, NT-dependent cleavage product (white arrow) and the 75 kDa product. Beta-dystroglycan was used as a loading control. (B) LN-AGRIN deposition in P10 kidney glomeruli is normal in the mutants, as detected with an LN-specific N-terminal antibody. Scale bar 10 mm. (C) Quantitative immunofluorescence for agrin at the NMJs on P4 triangularis sterni. Representative images of the three separate channels (presynaptic Thy1-YFP, BTX and AGRIN) are shown. Measures of staining intensity (0–250) were made along a line across the NMJ profile (red lines) and averaged (right graphs, mean ± SEM). Scale bar 10 mm.
Proteolytic processing, membrane targeting and secretion of AGRIN
To better address questions of glycosylation, trafficking, stability and processing, we turned to expression of recombinant AGRIN constructs in heterologous cells (Fig. 8A). AGRIN is processed by the protease neurotrypsin (NT), which cleaves the protein immediately upstream of the SEA domain and at a second site between the fourth EGF repeat and third G-domain. AGRIN is the only known substrate of NT, and the processing of AGRIN by the protease has been shown to occur at central nervous system synapses and at the NMJ in transgenic mice expressing NT in motor neurons (41–43). To examine glycosylation and processing quantitatively, HEK293 cells were transfected with full-length AGRIN with a C-terminal MYC tag in either wild-type or F1061S forms, as well as NT with a hemagglutinin (HA) tag. HEK293 cells express endogenous NT, which was not detected with the HA antibody. Cell membrane fractions and conditioned media were prepared and used for western blotting (Fig. 8B). Both wild-type and F1061S AGRIN were found in the membrane fractions as full-length proteins of ∼240 kDa. The cells transfected with wild-type AGRIN also show larger bands indicative of glycosylated proteins, and these are reduced in abundance in cells co-transfected with NT and in cells transfected with F1061S AGRIN, with or without NT. This indicates that glycosylated full-length AGRIN is sensitive to NT cleavage and that the F1061S protein is less glycosylated than wild-type AGRIN. Consistent with proteolytic cleavage, C-terminal AGRIN fragments were found in the conditioned media of transfected cells. Transfection with wild-type AGRIN produced fragments of 110 and 29 kDa, consistent with NT cleavage products, as well as fragment of ∼75 kDa. Cotransfection with NT and wild-type AGRIN resulted in the elimination of larger glycosylated proteins and also the elimination of the 75 kDa fragment, which presumably was reduced to the 29 kDa NT cleavage product. Interestingly, the F1061S protein was cleaved by NT to produce the 29 kDa C-terminal fragment, but no evidence of cleavage at the upstream site near the mutant SEA domain was found. The reduced level of AGRIN fragments in the media may be the result of impaired externalization of the F1061S AGRIN protein, since NT is membrane associated and active on the cell surface.
Figure 8.
Glycosylation, NT cleavage and plasma membrane targeting are affected by the F1061S mutation. (A) Schematic of the expression constructs used. Full-length SN-AGRIN with C-terminal myc tag, C110 AGRIN with vector signal peptide and C-terminal myc tag, AGRIN-SEA domain fused to alkaline phosphatase with signal peptide and C-terminal myc tag, and full-length NT with HA tag. The NT cleavage sites in AGRIN, upstream of the SEA domain and upstream of the third G-domain are indicated on the SN-AGRIN schematic. (B) Western blot of transfected HEK293 cell lysates and their conditioned media. The western blot membranes were probed with either an anti-myc antibody to visualize the carboxy terminal myc tag on AGRIN, an anti-HA antibody to detect NT and anti-tubulin as a loading control. In cell membranes, full-length AGRIN is present in both wild-type and F1061S transfected cells, but the larger, glycosylated forms are reduced in the mutant protein. In conditioned media, cells transfected with wild-type AGRIN produce 110 and 29 kDa C-terminal fragments as a result of NT cleavage, as well as a non-NT-dependent 75 kDa product. In cells transfected with F1061S constructs, less NT cleavage product is seen without NT cotransfection, and only the C-terminal 29 kDa fragment is detected when NT is cotransfected, suggesting the N-terminal site near the SEA domain is insensitive to NT cleavage. (C) Live cell staining without fixation or permeabilization for the C-terminal myc tag on full-length, type II transmembrane AGRIN. Cells were co-transfected with a mitochondrial COX8-YFP fusion (green, to identify individual transfected cells), and WT or mutant AGRIN, and without or with NT. Anti-myc (red) detects externalized AGRIN in both wild-type and F1061S transfected cells, and this cell surface staining is lost with cotransfection with NT, indicating both proteins are sensitive to NT cleavage. (D) Western blots on transfected CHO cells and their conditioned media. Cells were transfected with the full-length, C110 or SEA-alkaline phosphatase fusion DNAs, with or without the F1061S mutation. Blots of cell lysates and media were probed for the C-terminal myc tag. Full-length transmembrane AGRIN remains cell associated, whereas C110 AGRIN is secreted into the media in both wild-type and F1061S constructs. The F1061S SEA domain causes almost complete failure of secretion of the alkaline phosphatase fusion construct.
To examine externalization of AGRIN on the plasma membrane, Chinese hamster ovary (CHO) cells (which do not express endogenous NT) were transfected with the full-length SN-AGRIN–C-MYC construct, with and without NT-HA. The cells were co-transfected with a mitochondrial Cox8–YFP construct to mark transfected cells. Live cells were stained with the MYC antibody without permeabilization to detect AGRIN in the plasma membrane with the C-terminal epitope exposed extracellularly by the type II transmembrane protein (Fig. 8C). The intensity of MYC staining on the cell surface was reduced in cells transfected with F1061S AGRIN compared with wild-type AGRIN. Cotransfection with NT resulted in an almost complete elimination of MYC staining on the membrane for both wild-type and mutant AGRIN, consistent with the proteolytic activity of NT releasing the C-terminal epitope, and with mutant AGRIN still being sensitive to NT cleavage. Therefore, live cell staining also suggests a decrease in AGRIN with the correct topology associated with the plasma membrane in cells transfected with the F1061S form, but wild-type and F1061S AGRIN were found in similar abundance in membrane fractions (which were not necessarily purely plasma membranes) and both were similarly sensitive to NT-mediated cleavage at the C-terminal site, although the F1061S protein appears resistant to NT cleavage at the site upstream of the SEA domain.
The persistence of AGRIN immunoreactivity at the NMJ in vivo and the targeting of AGRIN to the plasma membrane in vitro suggest that the F1061S protein is both stable and properly trafficking through the cellular protein synthesis pathway (Figs 7C and 8B and C). However, the reduced levels of the protein detected on the cell surface and in conditioned media suggest some impediment in secretion. Consistent with this, defects in secretion were observed in constructs carrying the mutant SEA domain (Fig. 8D). Full-length SN-AGRIN was found in cell lysates of transfected CHO cells and not in the media, as anticipated. However, a truncated C-terminal construct (C110-C-MYC), beginning just upstream of the SEA domain and using a signal peptide for secretion provided by the expression vector, was less efficiently secreted. Comparable levels of protein were found in cell lysates, but the protein was reduced in abundance in the media. This effect was most pronounced when AGRIN's SEA domain was fused to alkaline phosphatase, again with a signal peptide for secretion provided by the vector. In these fusions, the mutant SEA domain resulted in an almost complete intracellular retention of the fusion protein. The fusion protein was detectable in cell lysates with similar abundance to wild-type–SEA fusions, indicating it was expressed, but it was undetectable in the media, indicating it was not secreted. These results indicate that the mutant SEA domain can have very deleterious effects on protein folding and trafficking, resulting in intracellular retention; however, these effects are more pronounced in the context of an isolated SEA domain and are reduced in severity in the context of additional flanking AGRIN sequence.
DISCUSSION
The Agrin point mutation described here (F1061S) creates a genetically and phenotypically valid mouse model of human CMS. It is therefore a useful research tool for the study of AGRIN's role at the postnatal NMJ, and for testing therapeutic strategies for CMS caused by mutations in AGRIN and downstream components of it signaling cascade. Previous mouse mutants generated through gene targeting of Agrin, or downstream genes such as Musk, Lrp4, Rapsyn or Dok7, all resulted in complete loss of function and neonatal lethality (17,19,20,44,45). These results confirm the importance of this signaling pathway for NMJ development, but these previous mutants do not provide accurate animal models for human CMS.
The nmf380-F1061S allele of Agrin is a recessive, partial loss-of-function mutation with no indication of a dominant phenotype in heterozygous mice that would suggest a pathological function of the mutant protein. The phenotype closely resembles human CMS pathology (25): early postnatal onset, impaired neuromuscular performance, weakness, fatigue, a preponderance of slow muscle fibers, a disaggregation of NMJs resulting in remodeling, functional denervation, enlarged subsynaptic folds and premature lethality at a few weeks to months of age. The shortening of lifespan represents a more severe condition than reported for the G1709R patients, but is dependent on genetic background in the mice. The NMJ defects occur at the level of motor units and affect nerve terminals and postsynaptic specializations, but more proximal peripheral axons are unaffected. Muscle groups innervated by motor neuron pools that are resistant to NMJ defects, including the extraocular muscles, are also preserved in the Agrinnmf380 mice, and the pattern of sensitivity within muscles matches that of the ALS model mice (37). The basis for this differential sensitivity remains largely unknown (36), but this model provides an additional tool for exploring the mechanisms underlying the resistance of these motor units to synaptic phenotypes.
The importance of the SEA domain for AGRIN function had not been previously explored. Many recombinant AGRIN expression constructs used in previous studies did not include the SEA domain and instead initiated immediately downstream to include the C-terminal half of the protein. The isoforms of AGRIN including the Z-exon encoded inserts between the EGF4 and G3 domains have been well studied, and C-terminal constructs including these sequences are sufficient for MuSK activation and AChR clustering. Indeed, the SEA domain is clearly dispensable for this critical activity of AGRIN. Recombinant proteins including the C-terminal half of AGRIN without an SEA domain are equally potent in inducing AChR clustering in vitro as full-length constructs (46). An SEA domain could conceivably be provided in these assays by the Z0 AGRIN expressed by the myotubes themselves. However, treatment of primary myotube cultures derived from Agrin knockout animals revealed the same AChR clustering activity of C-terminal AGRIN constructs as observed in myotube cultures derived from control mice (R.W.B., unpublished data). In these assays, there is no SEA domain provided by either the truncated, recombinant Z+ AGRIN protein or the Z0 AGRIN that would normally be secreted by the muscle. Furthermore, knockout mice are rescued by motor neuron-specific expression of an Agrin ‘mini-gene,’ consisting of the N-terminal laminin-binding portion of LN AGRIN (Nta), and the C-terminal EGF repeats, G-domains and Z-inserts, omitting the SEA domain (47). These mice also die prematurely, with central nervous system defects and also possible neuromuscular dysfunction. Any NMJ defects are likely to be the result of a failure of the motor neuron-specific HB9 promoter construct to sustain transgene expression in adults. While this transgenic rescue model similarly demonstrates the postnatal requirement of AGRIN in maintaining NMJs, it is not as valid as the F1061S allele for mechanistic and therapeutic studies of CMS because the pattern and progression of NMJ loss is determined by insufficiency of the HB9 promoter, and not by mutant AGRIN protein expressed under the control of its endogenous regulatory elements. However, these mice do provide an interesting comparison. The age of onset of the F1061S mice corresponds to a postnatal reduction in Agrin gene expression and may reflect the point at which reduced expression combined with impaired secretion, glycosylation and processing results in AGRIN levels at the NMJ that are below a critical threshold for maintaining NMJ integrity.
SEA domains are found in other extracellular proteoglycans including the MUCINs and PERLECAN (31). Structurally, DG is also proposed to contain an SEA domain, although there is little primary sequence homology (48). In the MUCINs and DG, the SEA domains may serve as a site of autolytic breakage of the full-length protein into two fragments (49–51). It is unclear if this is a universal function of SEA domains, and whether AGRIN undergoes such processing, although the presence of smaller ‘degradation products’ on AGRIN western blots is a consistent observation. Instead, AGRIN's SEA domain may be more like that of perlecan, which is N-terminal, but also flanked by serine/threonine-rich repeats and also modulates glycosylation (52).
AGRIN is also proteolytically processed by the protease NT in the brain and possibly the NMJ, and by matrix metalloprotease 3 during synaptic remodeling after muscle reinnervation (41–43,53–55). The pattern of AGRIN fragments detected on western blots in Agrnnmf380 homozygotes is not identical to wild-type AGRIN; however, the basis for these differences remains unclear. The NT cleavage site near the SEA domain appears to be resistant to NT in the F1061S protein, but the physiological role of NT processing of AGRIN in NMJ development remains unclear, and NT knockout mice do not have an NMJ phenotype, indicating that NT-mediated cleavage of AGRIN is not essential at the NMJ (43). Therefore, while the change in NT sensitivity in the F1061S protein demonstrates that the mutation can alter proteolytic processing by a known AGRIN protease, it is unlikely that this change underlies the NMJ disaggregation phenotype.
The F1061S mutation is predicted to fall in a helical region of the SEA domain. The deleterious effects of the amino acid change on SEA domain conformation is demonstrated by the almost complete failure of secretion of constructs consisting of just the SEA domain with a signal peptide provided by the vector and a C-terminal fusion with alkaline phosphatase. The SEA domain is also flanked by heavily glycosylated serine/threonine-rich repeats in AGRIN and changes in AGRIN peptide sequence nearer the C-terminus can alter the glycosylation of serine/threonine-rich regions near the SEA domain (56). Furthermore, the high molecular weight glycosylated forms of full-length AGRIN are reduced both in tissue and in transfected cells. Therefore, altered glycosylation may also contribute to the differences in trafficking, processing or activity of AGRIN that we have observed in the F1061S mutant mice.
In summary, the Agrinnmf380 mice create a valid model of human CMS associated with AGRIN mutations and possibly other genes in the AGRIN signaling pathway. The mutation results in a partial loss of function allele with a phenotype that closely resembles human CMS. The mutation is in the SEA domain of AGRIN, a region that had not been previously implicated as important for AGRIN function. The defect appears to be impaired conformation and secretion of the protein. Cleavage by the known processing protease, NT, is not lost at a C-terminal site, but is impaired at an upstream site close to the SEA domain. Other mechanisms such as altered autolytic cleavage or glycosylation may also contribute. However, altered activation of MuSK in postsynaptic muscle is unlikely since the SEA domain is entirely dispensable for this activity and it is also not a factor in the human G1709R mutation. This model will be useful for studies of postnatal myasthenic phenotypes and for testing therapies, and may also help explain mechanisms underlying other neuromuscular and motor neuron diseases.
MATERIALS AND METHODS
Mice
All mice were housed in the research animal facility at The Jackson Laboratory under standard conditions, including PIV caging and ad libitum food and water. All procedures were approved by the Institutional Animal Care and Use Committee of The Jackson Laboratory in accordance with policies outlined in The Guide to the Care and Use of Laboratory Animals. The Thy1-YFPH and Thy1-YFP16 strains were obtained from The Jackson Laboratory and are officially designated B6.Cg-Tg(Thy1-YFPH)2Jrs/J and B6.Cg-Tg(Thy1-YFP16)2Jrs/J, stock numbers 3782 and 3709, respectively. The YFP16 strain labels virtually all motor axons brightly, even during embryonic development. The YFPH strain labels a sparse, random subset of motor axons beginning at a few weeks of age.
Mutagenesis and mapping
The nmf380 allele was generated in a mutagenesis screen for recessive neurological mutations performed at The Jackson Laboratory. Male C57BL/6J (B6) mice were mutagenized using ENU, allowed to recover fertility, and bred to B6 females. Male offspring in generation 1 (G1), each carrying a unique assortment of mutations, were mated to B6 females to generate G2 females, which were bred back to the G1 male ancestor, producing homozygous recessive mutations in G3. G3 offspring were screened for neurological phenotypes using a battery of behavioral, clinical and histological analyses. The nmf380 strain was initially identified based on overt neuromuscular dysfunction and hind limb wasting, followed by histological and immunofluorescence evaluation of NMJs.
Once the nmf380 strain was ascertained, an affected female (presumed homozygote) was used for ovary transplantation and bred to a BALB/cByJ male to generate obligate heterozygous F1 mice for genetic mapping in the F2 generation. A genome-wide scan using simple sequence length polymorphisms (Mit markers) between C57BL/6J and BABL/cByJ was performed on pooled DNA from 10 affected versus 17 unaffected littermate F2 mice. A suggestive bias for C57BL/6J markers was found on distal chromosome 4 (D4Mit332, D4Mit54, D4Mit334), and individual mice were genotyped with these markers. The best association was found with the distal-most marker, D4Mit344 at 153.5 Mb on chromosome 4 (NCBIm37), with 9/10 affected mice being homozygous C57BL/6J and 15/17 unaffected mice being heterozygous or homozygous BALB/cByJ at this marker. These results suggested that the locus was on chromosome 4 and was distal to the D4Mit344 marker. No other regions of the genome showed a bias for C57BL/6J markers in the F2 genome scan.
Sequencing
Trizol (Invitrogen) was used according to manufacturer's instructions to prepare total RNA from the brains of a C57BL/6 control and an affected (presumed homozygote) nmf380 mouse. Five microgram of total RNA was reverse transcribed to first-strand cDNA using Superscript III reverse transcriptase (Invitrogen) and a mix of oligo-dT and random primers. The Agrn coding sequence was amplified by PCR in overlapping reactions of ∼800 bp each and the products sequenced directly on both strands and compared with sequence from the mouse genome project (www.Ensembl.org) and from isogenic control mice using the Sequencher analysis program. The SN-specific sequence was amplified from cDNA. The alternatively spliced Z-exons were sequenced from genomic DNA. Only the single base A to G substitution in exon 18 was found in the mutant sequence.
NMJ and transfected cell imaging
NMJ imaging was performed as described (57). In brief, muscles were dissected and fixed in cold 2% buffered paraformaldehyde (prepared fresh) overnight. P1 to P4 muscles were then rinsed, blocked and permeabilized in phosphate buffered saline (PBS) supplemented with 1% bovine serum albumin and 1% Triton X-100, and compressed between two microscope slides under pressure from a binder clip for 30 min. Beyond P4, muscles were embedded in 3% agarose in PBS and sectioned (100 μm thick sections) with a vibratome. Samples were then incubated overnight in primary antibody (a cocktail of mouse monoclonal anti-neurofilament 2H3 and anti-SV2 from the Developmental Studies Hybridoma bank), washed the following day and incubated overnight with AlexaFluor 488 conjugated anti-mouse IgG1 and AlexaFluor 594 conjugated α-BTX (Invitrogen). Muscles were mounted in 80% glycerol and imaged on a Leica SP5 laser confocal microscope. In cases where YFP16 transgenic mice were used to visualize motor neurons, samples were prepared identically, but were only stained with AlexaFluor 594 BTX.
Live cell staining was performed by adding primary antibodies to the culture medium for 1h. Cells were then washed twice with ice-cold PBS, fixed for 10 min on ice in 4% buffered paraformaldehyde, washed and incubated for an hour in the secondary antibody in 0.3% Triton X-100, 1% BSA, PBS.
Antibodies
The following antibodies were used in these studies: rabbit anti-C-terminal AGRIN [generously provided by Dr Markus Ruegg, Biozentrum, University of Basel, Switzerland (58)], sheep anti-N-terminal LN-AGRIN [GR-14, generously provided by Dr Berden, Radboud University, Nijmegen, Netherlands, (40)], Anti-MYC (9E 10), Anti-FLAG M2 (Sigma), Anti-BETA-DYSTROGLYCAN (MANDAG2, clone 7D11) and Anti-SLOW MYOSIN (Sigma, M8421). Species-specific secondary antibodies were Alexa Fluor® 488 and 594 goat anti-mouse IgG1 (γ1), Alexa Fluor® 594 goat anti-rabbit IgG (Invitrogen), Cy3-AffiniPure Donkey Anti-Sheep IgG (H+L) (Jackson Immunoresearch), HRP-labeled goat anti-mouse and anti-rabbit IgG (Perkin Elmer). The monoclonal antibodies 2H3, SV2, 9E10, MANDAG2 developed by T.M. Jessell and J. Dodd, K.M. Buckley and J.M. Bishop, and G.E. Morris were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242, USA.
Quantifications of NMJ area and intensity, and immunofluorescence intensity
For semi-quantitative imaging, control and mutant tissues were processed for immunofluorescence in parallel and images were acquired by confocal microscopy with the equivalent settings (laser power, gain, magnification, thickness of the stacks). Image analysis was performed with ImageJ. For NMJ morphometry, images were collected with a ×20 objective. Individual AChR clusters facing an axon terminal were delineated either with the Analyse Particles Macro (P1) or by hand (P4 and older) on the Z projection (maximum intensity) of each confocal stack. An average of 500 NMJs (P1) and 50 NMJs (P4 and older) per muscle were analyzed, on at least three muscles per genotype and age group. Areas in pixels were converted into square microns based on the confocal settings and mean intensities in the BTX channel (8 bits) expressed for each NMJ as a percentage of the average control intensity. Data for each condition (muscle, genotype and age) were pooled, averaged and the Student t-test was used to find statistical differences. For fluorescence quantification, images were collected with a ×63 objective; the intensity profiles in each channel (green for YFP, blue for BTX, red for AGRIN, 8 bits each) were measured with the Plot Profile Macro along a 50 µm line crossing the NMJs in their larger diameter. Profiles of 5 control and 16 mutant NMJs from the triangularis sterni muscles of 2 and 3 P4 pups were averaged.
AChE histochemistry
Staining for AChE activity was performed as described (59). In brief, tissue was postfixed in 4% paraformaldehyde for 1–2 h at 4°C. The tissues were then transferred to saturated sodium sulfate and stored overnight at 4°C. The muscle was then incubated in buffer [ethopropazine HCl (Sigma) 0.2 mm, acetylthiocholine iodide (Sigma) 4 mm, glycine 10 mm, cupric sulfate 2 mm and sodium acetate 65 mm (pH = 5.5)] for 30 min. Staining for acetylcholinesterase was developed by incubating for 1.5 min in sodium sulfide (1.25%, pH 6). Tissue was rinsed extensively with water before photographing.
Transmission electron microscopy
Muscles were fixed in 2% glutaraldehyde, 2% paraformaldehyde in 0.1 m cacodylate buffer. After fixation, the muscles were cut in cross-section at the endplate band (the point at which the nerve enters the muscle) and were processed and embedded for electron microscopy using standard techniques (57). Thin sections (75 µm) were cut and mounted on grids for TEM. Sections were viewed on a Jeol 1230 electron microscope and images were captured with a Hamamatsu digital camera system.
Histology of muscle
Whole lower limbs were fixed in 4% buffered paraformaldehyde or Bouins fixative and embedded in paraffin for sectioning. Four µm sections were mounted on slides and stained with hematoxylin and eosin in a Leica automated slide processor according to standard procedures.
Genotyping and PCR
Agrnnmf380 was genotyped using the following primers: forward AAT GGC ACG GAC TAA TTT GC, reverse TAT GAG GGT TTG TGG GGT GT. These primers amplify 212 bp spanning the end of intron 17 and the beginning of exon 18, the latter harboring the nmf380 mutation. PCR reaction products were analyzed by single strand conformational polymorphism according to references (60) and (61). The Z-exons of agrin were amplified using the following primers in exons 31 and 34: CAG GGG ATA GTT GAG AAG TCA GTG G and CAC AGT GGA GCG CAG CAC AAC TGG C.
Western blots
For the western blots on tissues, cell fractionation and membranes and extracellular matrix preparation were modified from reference (62). Tissues were first homogenized with a Fisher Scientific PowerGen Model 125 followed by a Dounce Homogenizer in lysis buffer (250 mm sucrose, 50 mm Tris pH 7.4, 5 mm MgCl2, 1 mm DTT and protease inhibitors (Complete protease inhibitor cocktail, Roche), at 4°. Nuclei and mitochondria were pelleted by centrifugation at 6000g for 15 min and the supernatant was centrifuged at 100 000g for 1h. The pellet of membranes and ECM was solubilized in the extraction buffer (20 mm Tris pH 7.4, 0.4 m NaCl, 15% glycerol, 1 mm DTT, 1.5% Triton X-100). Protein concentration was measured (BCA Protein Assay, Pierce) and 50 μg was heated at 70° for 5 min in Laemmli sample buffer before being loaded on a 4–15% acrylamide gradient gel (Mini-PROTEAN® TGX™ Precast Gel, Biorad).
For western blots on transfected cells, HEK293, CHO or COS-7 cells, were lysed by sonication in lysis buffer (20 mm Tris pH 7.5, 150 mm NaCl, 1% Igepal, protease inhibitors) following the protocol of reference (63). The lysate was cleared by centrifugation at 15 000g for 20 min and 50 μg of protein was loaded on the acrylamide gel as described above. The purification of 6xHis tagged secreted proteins from the conditioned medium was performed by nickel chromatography (Ni-NTA His•Bind® Resins, Novagen) following the manufacturer's instructions.
Acrylamide gels were run at 200 V for 30 min and transferred for 1 h at 100 V on PVDF membranes. The membranes were subsequently blocked in 2% milk in TBS-0.1%Tween20 and incubated overnight at 4° with the primary antibody diluted in the blocking solution. HRP-conjugated secondary antibodies were applied for 2h at room temperature in the blocking solution and the membranes were visualized by reaction with the Western Lightning Chemiluminescence Reagent (Perkin Elmer). BioMax MS Films (Kodak) were used for maximum detection.
Expression constructs
First-strand cDNAs from wild-type or Agrnnmf380 mutant brains were used as PCR templates for the cloning of the SEA module, C110 and full-length wild-type and mutant constructs. All constructs were cloned into the APtag-5 vector [Genhunter, (64)], which contains a cloning site in 5′ of the alkaline phosphatase sequence, and a myc-epitope and 6×HIS tag in its 3′. The secretion signal peptide of APtag-5 was kept in the SEA module and C110 constructs, but removed in the AGRIN full-length construct to be replaced by AGRIN's own start codon. The alkaline phosphatase sequence was preserved in frame in the SEA module construct but removed in the C110 and full-length constructs. The C-terminal myc epitope and 6×HIS tag were retained in frame in all AGRIN constructs. The SEA module was amplified with the primers BglII-Flag-SEA F TAT AGA TCT GAT TAC AAG GAT GAC GAC GAT AAG AGC GTT GTG GTG ACC CAC and SEA-Cterm-BglII R CGC AGA TCT CAG TCG GCT CAC AGT GGT to fuse an N-terminal FLAG tag and the BglII restriction sites for a ligation in the AP5 vector upstream of the alkaline phosphatase sequence. The C110 construct was amplified with the same forward primer and Agrin-Cterm-XhoI R CCG CTC GAG GCA GGG TCT TAG CTC TGG to be ligated in APtag-5 after removal of its alkaline phosphatase sequence. This added the myc epitope and 6×HIS tag of the vector in frame at the C-terminus of AGRIN. For the cloning of the full-length SN-AGRIN, the 3 kb N-terminal half of Agrn between the start codon and a BstEII site was amplified with NheI-Flag-SN f CTG GCT AGC CAC CAT GGA TTA CAA GGA TGA CGA CGA TAA GAT GCC TCC TCT GCC ACT GGA A and Agrin-BstEII-2932 r TGT AAC AGG AAG CCC TCT CG, partially digested by NheI and BstEII and inserted 5′ of the wild-type and mutant C110 constructs in APtag-5, preserving its Kozak sequence but removing its signal peptide sequence. This strategy also fused an N-terminal FLAG tag to SN-AGRIN and kept the C-terminal myc epitope and 6×HIS tag of the C110 constructs. Each construct was sequenced to verify the correct amino acid sequence and the presence of the F1061S change in the mutant constructs.
NT was amplified from brain cDNAs and cloned in APtag-5 with the primers NheI-Prss12F CTG GCT AGC CAC CAT GGC GCT CGC CCG CTG C and XbaI-HA-Prss12R ATA TCT AGA AGC GTA GTC TGG CAC GTC GTA TGG GTA CAG ACT GGT GAC ACT TTT that fused an N-terminal NheI site and a C-terminal HA-tag and XbaI site. The ligation in APtag-5 preserved its Kozak sequence but removed the vector signal peptide sequence. The reverse primer was also designed so that a STOP codon after the XbaI site prevented the fusion of the myc epitope and the 6×HIS tag of the vector to the NT ORF.
A fusion of the cytochrome c oxidase subunit 8 (COX 8) mitochondrial targeting sequence to the YFP sequence in the N1 vector (Clontech) was used as a transfection control for imaging of the membrane targeting of AGRIN full-length constructs (65).
Lipofectamine 2000 (Invitrogen) was used for all the transfection experiments. HEK293 cells were used for the transfection of the full-length SN-agrins (with or without the HA-tagged recombinant NT) because they endogenously express NT and allowed to study the differential sensitivities of wild-type and mutant agrin to this protease. CHO cells, which express little or no endogenous NT, were used for the membrane localization and live cell staining studies. COS-7 cells also do not express NT, but express the SV40 large T antigen that allows a replication of the APtag-5 vector through its SV40 origin. They were used to achieve high levels of recombinant protein production in the intracellular retention assay. All cells were grown in the DMEM medium (Invitrogen) with 10% FBS (Atlanta Biologicals) in the absence of antibiotics.
Structural model
Among the amino acid sequences grouped in the SEA module family of the PFAM database [accession PF01390, http://pfam.sanger.ac.uk, (66)], 10 were used for the Figure 2C: AGRIN in different vertebrates (mouse, rat, human, chicken, zebrafish and electric ray), and enterokinase, mucin 1, mucin 13 and mucin 16 in the mouse. The alignment was visualized with Jalview (67), and the F1061 (position 56 in the alignment) is conserved in all these proteins. The mapping of the SEA module of AGRIN onto the three-dimensional protein structure of the SEA module of mucin 16 [Uniprot entry: Q9D1H1, PDB ID: 1ivz, (32)], visualized with the Protein Workshop Viewer (68), localizes F1061 at the C-terminal portion of the first alpha-helix. This localization was independently confirmed with the secondary structure prediction software in Jalview, Jpred (69).
FUNDING
This work was supported by funding from the National Institutes of Health (NS054154 to R.W.B.) and fellowships from the Arthur K. Watson Charitable Trust, the Myasthenia Gravis Association and the Muscular Dystrophy Association (L.P.B.).
ACKNOWLEDGEMENTS
We would like to thank Drs Berden and Ruegg for generously providing anti-AGRIN antibodies. We would also like to thank Drs Greg Cox and Wayne Frankel for comments on the manuscript, and the scientific services at The Jackson Laboratory, particularly the Light Microscopy and Histology services.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Engel A.G., Ohno K., Sine S.M. Chapter 66: Congenital Myasthenic Syndrome. In: Engel A.G., Franzini-Armstrong C., editors. Myology. 3rd edn. Vol. 2. New York: McGraw-Hill; 2004. pp. 1801–1854. [Google Scholar]
- 2.Sanes J.R., Lichtman J.W. Development of the vertebrate neuromuscular junction. Ann. Rev. Neurosci. 1999;22:389–442. doi: 10.1146/annurev.neuro.22.1.389. doi:10.1146/annurev.neuro.22.1.389. [DOI] [PubMed] [Google Scholar]
- 3.Burden S.J. Building the vertebrate neuromuscular synapse. J. Neurobiol. 2002;53:501–511. doi: 10.1002/neu.10137. doi:10.1002/neu.10137. [DOI] [PubMed] [Google Scholar]
- 4.Kummer T.T., Misgeld T., Sanes J.R. Assembly of the postsynaptic membrane at the neuromuscular junction: paradigm lost. Curr. Opin. Neurobiol. 2006;16:74–82. doi: 10.1016/j.conb.2005.12.003. doi:10.1016/j.conb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Nitkin R.M., Smith M.A., Magill C., Fallon J.R., Yao Y.M., Wallace B.G., McMahan U.J. Identification of agrin, a synaptic organizing protein from Torpedo electric organ. J. Cell Biol. 1987;105:2471–2478. doi: 10.1083/jcb.105.6.2471. doi:10.1083/jcb.105.6.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen I., Rimer M., Lomo T., McMahan U.J. Agrin-induced postsynaptic-like apparatus in skeletal muscle fibers in vivo. Mol. Cell Neurosci. 1997;9:237–253. doi: 10.1006/mcne.1997.0623. doi:10.1006/mcne.1997.0623. [DOI] [PubMed] [Google Scholar]
- 7.Ferns M., Hoch W., Campanelli J.T., Rupp F., Hall Z.W., Scheller R.H. RNA splicing regulates agrin-mediated acetylcholine receptor clustering activity on cultured myotubes. Neuron. 1992;8:1079–1086. doi: 10.1016/0896-6273(92)90129-2. doi:10.1016/0896-6273(92)90129-2. [DOI] [PubMed] [Google Scholar]
- 8.Hoch W., Ferns M., Campanelli J.T., Hall Z.W., Scheller R.H. Developmental regulation of highly active alternatively spliced forms of agrin. Neuron. 1993;11:479–490. doi: 10.1016/0896-6273(93)90152-h. doi:10.1016/0896-6273(93)90152-H. [DOI] [PubMed] [Google Scholar]
- 9.Ferns M.J., Campanelli J.T., Hoch W., Scheller R.H., Hall Z. The ability of agrin to cluster AChRs depends on alternative splicing and on cell surface proteoglycans. Neuron. 1993;11:491–502. doi: 10.1016/0896-6273(93)90153-i. doi:10.1016/0896-6273(93)90153-I. [DOI] [PubMed] [Google Scholar]
- 10.Kim N., Stiegler A.L., Cameron T.O., Hallock P.T., Gomez A.M., Huang J.H., Hubbard S.R., Dustin M.L., Burden S.J. Lrp4 is a receptor for Agrin and forms a complex with MuSK. Cell. 2008;135:334–342. doi: 10.1016/j.cell.2008.10.002. doi:10.1016/j.cell.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang B., Luo S., Wang Q., Suzuki T., Xiong W.C., Mei L. LRP4 serves as a coreceptor of agrin. Neuron. 2008;60:285–297. doi: 10.1016/j.neuron.2008.10.006. doi:10.1016/j.neuron.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glass D.J., Bowen D.C., Stitt T.N., Radziejewski C., Bruno J., Ryan T.E., Gies D.R., Shah S., Mattsson K., Burden S.J., et al. Agrin acts via a MuSK receptor complex. Cell. 1996;85:513–523. doi: 10.1016/s0092-8674(00)81252-0. doi:10.1016/S0092-8674(00)81252-0. [DOI] [PubMed] [Google Scholar]
- 13.Burgess R.W., Dickman D.K., Nunez L., Glass D.J., Sanes J.R. Mapping sites responsible for interactions of agrin with neurons. J. Neurochem. 2002;83:271–284. doi: 10.1046/j.1471-4159.2002.01102.x. doi:10.1046/j.1471-4159.2002.01102.x. [DOI] [PubMed] [Google Scholar]
- 14.Neumann F.R., Bittcher G., Annies M., Schumacher B., Kroger S., Ruegg M.A. An alternative amino-terminus expressed in the central nervous system converts agrin to a type II transmembrane protein. Mol. Cell Neurosci. 2001;17:208–225. doi: 10.1006/mcne.2000.0932. doi:10.1006/mcne.2000.0932. [DOI] [PubMed] [Google Scholar]
- 15.Burgess R.W., Skarnes W.C., Sanes J.R. Agrin isoforms with distinct amino termini: differential expression, localization, and function. J. Cell Biol. 2000;151:41–52. doi: 10.1083/jcb.151.1.41. doi:10.1083/jcb.151.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin W., Burgess R.W., Dominguez B., Pfaff S.L., Sanes J.R., Lee K.F. Distinct roles of nerve and muscle in postsynaptic differentiation of the neuromuscular synapse. Nature. 2001;410:1057–1064. doi: 10.1038/35074025. doi:10.1038/35074025. [DOI] [PubMed] [Google Scholar]
- 17.Gautam M., Noakes P.G., Moscoso L., Rupp F., Scheller R.H., Merlie J.P., Sanes J.R. Defective neuromuscular synaptogenesis in agrin-deficient mutant mice. Cell. 1996;85:525–535. doi: 10.1016/s0092-8674(00)81253-2. doi:10.1016/S0092-8674(00)81253-2. [DOI] [PubMed] [Google Scholar]
- 18.Burgess R.W., Nguyen Q.T., Son Y.J., Lichtman J.W., Sanes J.R. Alternatively spliced isoforms of nerve- and muscle-derived agrin: their roles at the neuromuscular junction. Neuron. 1999;23:33–44. doi: 10.1016/s0896-6273(00)80751-5. doi:10.1016/S0896-6273(00)80751-5. [DOI] [PubMed] [Google Scholar]
- 19.Weatherbee S.D., Anderson K.V., Niswander L.A. LDL-receptor-related protein 4 is crucial for formation of the neuromuscular junction. Development. 2006;133:4993–5000. doi: 10.1242/dev.02696. doi:10.1242/dev.02696. [DOI] [PubMed] [Google Scholar]
- 20.DeChiara T.M., Bowen D.C., Valenzuela D.M., Simmons M.V., Poueymirou W.T., Thomas S., Kinetz E., Compton D.L., Rojas E., Park J.S., et al. The receptor tyrosine kinase MuSK is required for neuromuscular junction formation in vivo. Cell. 1996;85:501–512. doi: 10.1016/s0092-8674(00)81251-9. doi:10.1016/S0092-8674(00)81251-9. [DOI] [PubMed] [Google Scholar]
- 21.Beeson D., Higuchi O., Palace J., Cossins J., Spearman H., Maxwell S., Newsom-Davis J., Burke G., Fawcett P., Motomura M., et al. Dok-7 mutations underlie a neuromuscular junction synaptopathy. Science. 2006;313:1975–1978. doi: 10.1126/science.1130837. doi:10.1126/science.1130837. [DOI] [PubMed] [Google Scholar]
- 22.Chevessier F., Faraut B., Ravel-Chapuis A., Richard P., Gaudon K., Bauche S., Prioleau C., Herbst R., Goillot E., Ioos C., et al. MUSK, a new target for mutations causing congenital myasthenic syndrome. Hum. Mol. Genet. 2004;13:3229–3240. doi: 10.1093/hmg/ddh333. doi:10.1093/hmg/ddh333. [DOI] [PubMed] [Google Scholar]
- 23.Ohno K., Engel A.G., Shen X.M., Selcen D., Brengman J., Harper C.M., Tsujino A., Milone M. Rapsyn mutations in humans cause endplate acetylcholine-receptor deficiency and myasthenic syndrome. Am. J. Hum. Genet. 2002;70:875–885. doi: 10.1086/339465. doi:10.1086/339465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoch W., McConville J., Helms S., Newsom-Davis J., Melms A., Vincent A. Auto-antibodies to the receptor tyrosine kinase MuSK in patients with myasthenia gravis without acetylcholine receptor antibodies. Nat. Med. 2001;7:365–368. doi: 10.1038/85520. doi:10.1038/85520. [DOI] [PubMed] [Google Scholar]
- 25.Huze C., Bauche S., Richard P., Chevessier F., Goillot E., Gaudon K., Ben Ammar A., Chaboud A., Grosjean I., Lecuyer H.A., et al. Identification of an agrin mutation that causes congenital myasthenia and affects synapse function. Am. J. Hum. Genet. 2009;85:155–167. doi: 10.1016/j.ajhg.2009.06.015. doi:10.1016/j.ajhg.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harvey S.J., Jarad G., Cunningham J., Rops A.L., van der Vlag J., Berden J.H., Moeller M.J., Holzman L.B., Burgess R.W., Miner J.H. Disruption of glomerular basement membrane charge through podocyte-specific mutation of agrin does not alter glomerular permselectivity. Am. J. Pathol. 2007;171:139–152. doi: 10.2353/ajpath.2007.061116. doi:10.2353/ajpath.2007.061116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seburn K.L., Nangle L.A., Cox G.A., Schimmel P., Burgess R.W. An active dominant mutation of glycyl-tRNA synthetase causes neuropathy in a Charcot-Marie-Tooth 2D mouse model. Neuron. 2006;51:715–726. doi: 10.1016/j.neuron.2006.08.027. doi:10.1016/j.neuron.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 28.Patton B.L., Wang B., Tarumi Y.S., Seburn K.L., Burgess R.W. A single point mutation in the LN domain of LAMA2 causes muscular dystrophy and peripheral amyelination. J. Cell Sci. 2008;121:1593–1604. doi: 10.1242/jcs.015354. doi:10.1242/jcs.015354. [DOI] [PubMed] [Google Scholar]
- 29.Achilli F., Bros-Facer V., Williams H.P., Banks G.T., AlQatari M., Chia R., Tucci V., Groves M., Nickols C.D., Seburn K.L., et al. An ENU-induced mutation in mouse glycyl-tRNA synthetase (GARS) causes peripheral sensory and motor phenotypes creating a model of Charcot-Marie-Tooth type 2D peripheral neuropathy. Dis. Model Mech. 2009;2:359–373. doi: 10.1242/dmm.002527. doi:10.1242/dmm.002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuerst P.G., Rauch S.M., Burgess R.W. Defects in eye development in transgenic mice overexpressing the heparan sulfate proteoglycan agrin. Dev. Biol. 2007;303:165–180. doi: 10.1016/j.ydbio.2006.11.033. doi:10.1016/j.ydbio.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bork P., Patthy L. The SEA module: a new extracellular domain associated with O-glycosylation. Prot. Sci. 1995;4:1421–1425. doi: 10.1002/pro.5560040716. doi:10.1002/pro.5560040716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maeda T., Inoue M., Koshiba S., Yabuki T., Aoki M., Nunokawa E., Seki E., Matsuda T., Motoda Y., Kobayashi A., et al. Solution structure of the SEA domain from the murine homologue of ovarian cancer antigen CA125 (MUC16) J. Biol. Chem. 2004;279:13174–13182. doi: 10.1074/jbc.M309417200. doi:10.1074/jbc.M309417200. [DOI] [PubMed] [Google Scholar]
- 33.Feng G., Mellor R.H., Bernstein M., Keller-Peck C., Nguyen Q.T., Wallace M., Nerbonne J.M., Lichtman J.W., Sanes J.R. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. doi:10.1016/S0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 34.Khanna S., Richmonds C.R., Kaminski H.J., Porter J.D. Molecular organization of the extraocular muscle neuromuscular junction: partial conservation of and divergence from the skeletal muscle prototype. Invest. Ophth. Vis. Sci. 2003;44:1918–1926. doi: 10.1167/iovs.02-0890. doi:10.1167/iovs.02-0890. [DOI] [PubMed] [Google Scholar]
- 35.Gatesy S.M., English A.W. Evidence for compartmental identity in the development of the rat lateral gastrocnemius muscle. Dev. Dyn. 1993;196:174–182. doi: 10.1002/aja.1001960304. doi:10.1002/aja.1001960304. [DOI] [PubMed] [Google Scholar]
- 36.Saxena S., Cabuy E., Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat. Neurosci. 2009;12:627–636. doi: 10.1038/nn.2297. doi:10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- 37.Pun S., Santos A.F., Saxena S., Xu L., Caroni P. Selective vulnerability and pruning of phasic motoneuron axons in motoneuron disease alleviated by CNTF. Nat. Neurosci. 2006;9:408–419. doi: 10.1038/nn1653. doi:10.1038/nn1653. [DOI] [PubMed] [Google Scholar]
- 38.Fischer L.R., Culver D.G., Tennant P., Davis A.A., Wang M., Castellano-Sanchez A., Khan J., Polak M.A., Glass J.D. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp. Neurol. 2004;185:232–240. doi: 10.1016/j.expneurol.2003.10.004. doi:10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Kariya S., Park G.H., Maeno-Hikichi Y., Leykekhman O., Lutz C., Arkovitz M.S., Landmesser L.T., Monani U.R. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2008;17:2552–2569. doi: 10.1093/hmg/ddn156. doi:10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raats C.J., Bakker M.A., Hoch W., Tamboer W.P., Groffen A.J., van den Heuvel L.P., Berden J.H., van den Born J. Differential expression of agrin in renal basement membranes as revealed by domain-specific antibodies. J. Biol. Chem. 1998;273:17832–17838. doi: 10.1074/jbc.273.28.17832. doi:10.1074/jbc.273.28.17832. [DOI] [PubMed] [Google Scholar]
- 41.Reif R., Sales S., Hettwer S., Dreier B., Gisler C., Wolfel J., Luscher D., Zurlinden A., Stephan A., Ahmed S., et al. Specific cleavage of agrin by neurotrypsin, a synaptic protease linked to mental retardation. FASEB J. 2007;21:3468–3478. doi: 10.1096/fj.07-8800com. doi:10.1096/fj.07-8800com. [DOI] [PubMed] [Google Scholar]
- 42.Stephan A., Mateos J.M., Kozlov S.V., Cinelli P., Kistler A.D., Hettwer S., Rulicke T., Streit P., Kunz B., Sonderegger P. Neurotrypsin cleaves agrin locally at the synapse. FASEB J. 2008;22:1861–1873. doi: 10.1096/fj.07-100008. doi:10.1096/fj.07-100008. [DOI] [PubMed] [Google Scholar]
- 43.Bolliger M.F., Zurlinden A., Luscher D., Butikofer L., Shakhova O., Francolini M., Kozlov S.V., Cinelli P., Stephan A., Kistler A.D., et al. Specific proteolytic cleavage of agrin regulates maturation of the neuromuscular junction. J. Cell Sci. 2010;123:3944–3955. doi: 10.1242/jcs.072090. doi:10.1242/jcs.072090. [DOI] [PubMed] [Google Scholar]
- 44.Gautam M., Noakes P.G., Mudd J., Nichol M., Chu G., Sanes J.A., Merlie J. Failure of postsynaptic specialization to develop at neuromuscular junctions of rapsyn-deficient mice. Nature. 1995;377:232–236. doi: 10.1038/377232a0. doi:10.1038/377232a0. [DOI] [PubMed] [Google Scholar]
- 45.Okada K., Inoue A., Okada M., Murata Y., Kakuta S., Jigami T., Kubo S., Shiraishi H., Eguchi K., Motomura M., et al. The muscle protein Dok-7 is essential for neuromuscular synaptogenesis. Science. 2006;312:1802–1805. doi: 10.1126/science.1127142. doi:10.1126/science.1127142. [DOI] [PubMed] [Google Scholar]
- 46.Gesemann M., Denzer A.J., Ruegg M.A. Acetylcholine receptor-aggregating activity of agrin isoforms and mapping of the active site. J. Cell Biol. 1995;128:625–636. doi: 10.1083/jcb.128.4.625. doi:10.1083/jcb.128.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ksiazek I., Burkhardt C., Lin S., Seddik R., Maj M., Bezakova G., Jucker M., Arber S., Caroni P., Sanes J.R., et al. Synapse loss in cortex of agrin-deficient mice after genetic rescue of perinatal death. J. Neurosci. 2007;27:7183–7195. doi: 10.1523/JNEUROSCI.1609-07.2007. doi:10.1523/JNEUROSCI.1609-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akhavan A., Crivelli S.N., Singh M., Lingappa V.R., Muschler J.L. SEA domain proteolysis determines the functional composition of dystroglycan. FASEB J. 2008;22:612–621. doi: 10.1096/fj.07-8354com. doi:10.1096/fj.07-8354com. [DOI] [PubMed] [Google Scholar]
- 49.Palmai-Pallag T., Khodabukus N., Kinarsky L., Leir S.H., Sherman S., Hollingsworth M.A., Harris A. The role of the SEA (sea urchin sperm protein, enterokinase and agrin) module in cleavage of membrane-tethered mucins. FEBS J. 2005;272:2901–2911. doi: 10.1111/j.1742-4658.2005.04711.x. doi:10.1111/j.1742-4658.2005.04711.x. [DOI] [PubMed] [Google Scholar]
- 50.Levitin F., Stern O., Weiss M., Gil-Henn C., Ziv R., Prokocimer Z., Smorodinsky N.I., Rubinstein D.B., Wreschner D.H. The MUC1 SEA module is a self-cleaving domain. J. Biol. Chem. 2005;280:33374–33386. doi: 10.1074/jbc.M506047200. doi:10.1074/jbc.M506047200. [DOI] [PubMed] [Google Scholar]
- 51.Macao B., Johansson D.G., Hansson G.C., Hard T. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat. Struct. Mol. Biol. 2006;13:71–76. doi: 10.1038/nsmb1035. doi:10.1038/nsmb1035. [DOI] [PubMed] [Google Scholar]
- 52.Dolan M., Horchar T., Rigatti B., Hassell J.R. Identification of sites in domain I of perlecan that regulate heparan sulfate synthesis. J. Biol. Chem. 1997;272:4316–4322. doi: 10.1074/jbc.272.7.4316. doi:10.1074/jbc.272.7.4316. [DOI] [PubMed] [Google Scholar]
- 53.VanSaun M., Werle M.J. Matrix metalloproteinase-3 removes agrin from synaptic basal lamina. J. Neurobiol. 2000;43:140–149. doi:10.1002/(SICI)1097-4695(200005)43:2<140::AID-NEU4>3.0.CO;2-K. [PubMed] [Google Scholar]
- 54.VanSaun M., Herrera A.A., Werle M.J. Structural alterations at the neuromuscular junctions of matrix metalloproteinase 3 null mutant mice. J. Neurocytol. 2003;32:1129–1142. doi: 10.1023/B:NEUR.0000021907.68461.9c. doi:10.1023/B:NEUR.0000021907.68461.9c. [DOI] [PubMed] [Google Scholar]
- 55.Werle M.J., VanSaun M. Activity dependent removal of agrin from synaptic basal lamina by matrix metalloproteinase 3. J. Neurocytol. 2003;32:905–913. doi: 10.1023/B:NEUR.0000020631.69804.f5. doi:10.1023/B:NEUR.0000020631.69804.f5. [DOI] [PubMed] [Google Scholar]
- 56.Xia B., Martin P.T. Modulation of agrin binding and activity by the CT and related carbohydrate antigens. Mol. Cell Neurosci. 2002;19:539–551. doi: 10.1006/mcne.2001.1095. doi:10.1006/mcne.2001.1095. [DOI] [PubMed] [Google Scholar]
- 57.Burgess R.W., Cox G.A., Seburn K.L. Neuromuscular disease models and analysis. Methods Mol. Biol. 2010;602:347–393. doi: 10.1007/978-1-60761-058-8_20. doi:10.1007/978-1-60761-058-8_20. [DOI] [PubMed] [Google Scholar]
- 58.Eusebio A., Oliveri F., Barzaghi P., Ruegg M.A. Expression of mouse agrin in normal, denervated and dystrophic muscle. Neuromuscul. Disord. 2003;13:408–415. doi: 10.1016/s0960-8966(03)00036-1. doi:10.1016/S0960-8966(03)00036-1. [DOI] [PubMed] [Google Scholar]
- 59.Enomoto H., Araki T., Jackman A., Heuckeroth R.O., Snider W.D., Johnson E.M., Jr, Milbrandt J. GFR alpha1-deficient mice have deficits in the enteric nervous system and kidneys. Neuron. 1998;21:317–324. doi: 10.1016/s0896-6273(00)80541-3. doi:10.1016/S0896-6273(00)80541-3. [DOI] [PubMed] [Google Scholar]
- 60.Hongyo T., Buzard G.S., Calvert R.J., Weghorst C.M. Cold SSCP’: a simple, rapid and non-radioactive method for optimized single-strand conformation polymorphism analyses. Nucleic Acids Res. 1993;21:3637–3642. doi: 10.1093/nar/21.16.3637. doi:10.1093/nar/21.16.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie T., Ho S.L., Ma O.C. High resolution single strand conformation polymorphism analysis using formamide and ethidium bromide staining. Mol. Pathol. 1997;50:276–278. doi: 10.1136/mp.50.5.276. doi:10.1136/mp.50.5.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kislinger T., Cox B., Kannan A., Chung C., Hu P., Ignatchenko A., Scott M.S., Gramolini A.O., Morris Q., Hallett M.T., et al. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. doi:10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 63.Lin L., McCroskery S., Ross J.M., Chak Y., Neuhuber B., Daniels M.P. Induction of filopodia-like protrusions by transmembrane agrin: role of agrin glycosaminoglycan chains and Rho-family GTPases. Exp. Cell Res. 2010;316:2260–2277. doi: 10.1016/j.yexcr.2010.05.006. doi:10.1016/j.yexcr.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Flanagan J.G., Cheng H.J., Feldheim D.A., Hattori M., Lu Q., Vanderhaeghen P. Alkaline phosphatase fusions of ligands or receptors as in situ probes for staining of cells, tissues, and embryos. Methods Enzymol. 2000;327:19–35. doi: 10.1016/s0076-6879(00)27264-9. doi:10.1016/S0076-6879(00)27264-9. [DOI] [PubMed] [Google Scholar]
- 65.Misgeld T., Kerschensteiner M., Bareyre F.M., Burgess R.W., Lichtman J.W. Imaging axonal transport of mitochondria in vivo. Nat. Methods. 2007;4:559–561. doi: 10.1038/nmeth1055. doi:10.1038/nmeth1055. [DOI] [PubMed] [Google Scholar]
- 66.Finn R.D., Mistry J., Tate J., Coggill P., Heger A., Pollington J.E., Gavin O.L., Gunasekaran P., Ceric G., Forslund K., et al. The Pfam protein families database. Nucleic Acids Res. 2009;38:D211–D222. doi: 10.1093/nar/gkp985. doi:10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. doi:10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moreland J.L., Gramada A., Buzko O.V., Zhang Q., Bourne P.E. The Molecular Biology Toolkit (MBT): a modular platform for developing molecular visualization applications. BMC Bioinformatics. 2005;6:21. doi: 10.1186/1471-2105-6-21. doi:10.1186/1471-2105-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cole C., Barber J.D., Barton G.J. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. doi:10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]