Abstract
Madhya Pradesh is the major soybean contributor in India. The taxonomy of nitrogen fixing bacteria forming symbiotic associations with leguminous plants has been deeply changed in recent years. The use of very sensitive and accurate molecular methods has enabled the detection of large rhizobial diversity. Molecular biotyping and characterization the Bradyrhizobium, isolates from eleven varieties of soybean from agricultural field of Sehore district of Madhya Pradesh is done using 16S rDNA typing. Bradyrhizobia were identified genetically by determining the %Guanine plus Cytosine content of the whole genome, followed by 16S rDNA-RFLP analysis % Guanine plus Cytosine content of all the Bradyrhizobium isolates reflects similarity at generic level among all Bradyrhizobial isolates. Restriction Fragment Length Polymorphism (RFLP) further showed a considerable level of genetic diversity among the Bradyrhizobial isolates. PCR-RFLP of 16S rDNA supported existence of two divergent groups among indigenous Bradyrhizobial isolates, at similarity level of 66, and 75 and 74% of similarity within the group. The technique used was helpful in characterizing Bradyrhizobium isolates to be used as inoculants for improving productivity of agricultural land of Madhya Pradesh (India).
Keywords: % Guanine plus Cytosine content (% G + C), 16S rDNA, Bradyrhizobium sp, DNA fingerprinting, Genetic diversity, Soybean
Introduction
Rhizobia are symbiotic bacteria capable of eliciting and invading root or stem nodules on leguminous plants, and are efficient to differentiate into Nitrogen fixing bacteroides [1]. Rhizobia (Rhizobium, Bradyrhizobium, Sinorhizobium, Mesorhizobium, Allorhizobium), contributes 65% of total atmospheric nitrogen fixed in an environment [2]. Prokaryotic Guanine plus Cytosine content is highly variable ranging from 25 to almost 80%. Despite such a wide range of variation, Guanine plus Cytosine content of microbial strains within a particular species is constant however, in case of variation within % G + C content the strains differs indicating variability at molecular level among themselves [3, 4]. Guanine plus Cytosine content appears to be useful in characterizing bacterial genera since variation within a genus is usually less then 10% even though in few cases the content may vary greatly between genera [5]. Based on their 16S rDNA sequences Bradyrhizobia, the nodule endosymbionts, constituting a polyphyletic assemblage of bacteria are currently grouped in 13 major phylogenetic branches of α-2 subclass of Protobacteria including the genera Allorhizobium,Mesorhizobium, Rhizobium, and Sinorhizobium. Resulting rhizobial clades and Azorhizobium, Bradyrhizobium, and Methylobacterium were found on a different and well-resolved phylogenetic branch [6, 7]. 16S rDNA regions reflects much greater diversity analysis than previously recognized molecular techniques [7–9]. However, such observations helps to enumerate systematics of Bradyrhizobia in relation to new genera and species [8, 10, 11]. In current study, agricultural land of Madhya Pradesh is selected. The state is known for highest production of Glycine max. The Rhizobium diversity has great implication in agriculture in order to define nitrogen rich biofertilizer which helps to improve fertility of soil. A single genus of host can be infected with different strains of microorganism of same species. The variation among those strains may or may not be visualized at expression level of gene, but by conserved sequences of the bacterial genome i.e. 16S rDNA [12, 13]. Genetic relatedness is known to established more strongly using 16S rDNA. These repeated ribosomal sequences of 9–65 base pair provides high polymorphism for distinguishing varying strains within a species and among different species. Presently, the use of restriction enzymes during RFLP over amplified 16S rDNA confirms location of genes indicating genetic variation in number. Recently diversity analysis of Rhizobia nodulating another host i.e. Arachis hypogea L. is being studied using RFLP analysis of 16S rDNA [14]. It has been established that cross infection is possible by efficient strains of Bradyrhizobia in different legumes hosts on the basis of 16S rDNA-RFLP assay [15]. In current study 25 isolates of Bradyrhizobium from 11 varieties of soybean have been isolated and characterized at molecular level on the basis of genetic study including % Guanine plus Cytosine content and 16S rDNA-RFLP, with banding pattern analysis using NTSYS software [16].
Materials and Methods
Isolation and Maintenance of Bradyrhizobium Isolates
Eleven varieties of Glycine max i.e. NRC-2, NRC-7, NRC-37, Punjab-1, MAUS-47, Indira soya, JS71-5, JS79-81, JS90-41, JS80-21, JS335, were obtained from agricultural field of Sehore district of Madhya Pradesh and reference strain R (RJ123) from National Research Center of soybean (NRCS), Indore, Madhya Pradesh, India, for isolation of Bradyrhizobium from the root nodule [17]. The nodule was surface sterilized and crushed to obtain the bacteria on yeast extract mannitol Agar media (YEMA) [18]. The culture was maintained on Congo red yeast extract mannitol agar media (CRYMA) [19] at 26 ± 2°C for 3–4 days. Isolates were further confirmed by biochemical tests [19] (Table 1) compared with reference strain R (RJ123) of Rhizobium japonicum.
Table 1.
Designation and properties of Bradyrhizobial isolates and reference strain used in the study
| Strain Name | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N1 | 63.928 | NRC2 | 96 | +/+ | 4.4 | S | − | − | − | G −ve | −ve | + |
| N2 | 63.196 | NRC2 | 96 | +/+ | 4.1 | S | − | − | − | G −ve | −ve | + |
| N3 | 63.928 | NRC7 | 96 | +/+ | 4.1 | S | − | − | − | G −ve | −ve | + |
| N4 | 63.928 | NRC37 | 72 | +/+ | 5.4 | M | − | − | − | G −ve | −ve | + |
| N5 | 61.976 | NRC37 | 72 | +/+ | 5.3 | M | − | − | − | G −ve | −ve | + |
| N6 | 64.172 | NRC37 | 72 | +/+ | 5.2 | M | − | − | − | G −ve | −ve | + |
| N7 | 63.928 | Punjab 1 | 96 | +/+ | 4.3 | S | − | − | − | G −ve | −ve | + |
| N8 | 63.196 | Indira Soy | 96 | +/+ | 3.9 | S | − | − | − | G −ve | −ve | + |
| N9 | 64.416 | MAUS-47 | 96 | +/+ | 3.8 | S | − | − | − | G −ve | −ve | + |
| N10 | 61.976 | MAUS-47 | 96 | +/+ | 4.2 | S | − | − | − | G −ve | −ve | + |
| N11 | 61.976 | MAUS-47 | 96 | +/+ | 3.9 | S | − | − | − | G −ve | −ve | + |
| N12 | 64.172 | MAUS-81 | 96 | +/+ | 4.3 | S | − | − | − | G −ve | −ve | + |
| N13 | 61.976 | MAUS-81 | 96 | +/+ | 4.4 | S | − | − | − | G −ve | −ve | + |
| N14 | 63.196 | JS71-5 | 72 | +/+ | 6.5 | M | − | − | − | G −ve | −ve | + |
| N15 | 64.416 | JS71-5 | 72 | +/+ | 5.7 | M | − | − | − | G −ve | −ve | + |
| N16 | 63.928 | JS71-5 | 72 | +/+ | 6.1 | M | − | − | − | G −ve | −ve | + |
| N17 | 61.00 | JS79-81 | 72 | +/+ | 5.6 | M | − | − | − | G −ve | −ve | + |
| N18 | 63.928 | JS79-81 | 72 | +/+ | 5.5 | M | − | − | − | G −ve | −ve | + |
| N19 | 63.196 | JS80-21 | 96 | +/+ | 4.2 | S | − | − | − | G −ve | −ve | + |
| N20 | 61.976 | JS80-21 | 96 | +/+ | 3.9 | S | − | − | − | G −ve | −ve | + |
| N21 | 63.928 | JS80-21 | 96 | +/+ | 4.2 | S | − | − | − | G −ve | −ve | + |
| N22 | 61.976 | JS90-41 | 96 | +/+ | 4.2 | S | − | − | − | G −ve | −ve | + |
| N23 | 61.00 | JS90-41 | 96 | +/+ | 4.1 | S | − | − | − | G −ve | −ve | + |
| N24 | 61.976 | JS335 | 72 | +/+ | 6.1 | M | − | − | − | G −ve | −ve | + |
| N25 | 64.172 | JS335 | 72 | +/+ | 6.3 | M | − | − | − | G −ve | −ve | + |
| R | 64.928 | Soybean | 96 | +/+ | 3.8 | S | − | − | − | G −ve | −ve | + |
1 %G + C = 2.44 (Tm−69.3); 2 Plant Variety; 3 Incubation at 25° ± 1°C for hours; 4 Growth on YEMA/CRYMA media; 5 Max. Colony Size after 6th day of incubation (mm); 6 Slow (S)/Moderate (M)/Fast (F) growing sp.; 7 Hofer’s alkaline medium pH (11) (Growth = +ve No growth = −ve test); 8 Glucose peptone agar medium (Growth = + ve test/No growth = −ve test); 9 Ketolactose test (Production of yellow colour = +ve test/No colour change = −ve test); 10 Gram Staining (G +ve/G −ve); 11 Liquefaction of Gelatin (liquefaction = + ve test;/no liquefaction = − ve test; 12 Action on Milk (curdling = +ve test/no curdling = −ve test)
DNA Isolation
All the 25 isolates were inoculated in yeast extract mannitol broth (YEMB) for 4 days on shaking incubator at 26 ± 2°C. The total genomic DNA was isolated using standard phenol–chloroform extraction method [21]. The pellets were washed with 70% ethanol, dried and redissolved in 150 μl of TE buffer. Concentration and purity of DNA were estimated spectrophotometrically at 260 and 280 nm, respectively for further assay.
% Guanine Plus Cytosine Content
The purified DNA was then dissolved in 0.15 M NaCl, 0.015 M citrate solution and divided into four parts, incubated at 60, 70, 80, and 90°C for 30 min and change in absorption was determined spectrophotometrically at 260 nm. The value obtained was used to calculate the Tm (temperature of melting). The % Guanine plus Cytosine was then calculated using the formula % Guanine plus Cytosine = 2.44 (Tm−69.3) [20].
16S rDNA-RFLP Analysis
The universal primer rD1 (5′-ACGGCTACCTTGTTACGACTT-3′) and fD1 (5′-AGAGTTTGATCCTGGCTCAG-3′) were used for PCR amplification of 16S rDNA [21] in 25 μl of volume, having 20 mM/l tris–HCl (pH 8), 50 Mm/l KCl, 2.0 Mm/l MgCl2, 200 μmol/l of dNTP’s, 1 μmol/l of each primer, 50 ng of genomic DNA and 1.5 U of Taq DNA polymerase (Bangalore Genei, India) followed by the following touchdown programme starting with initial denaturation at 94°C for 5 min, 40 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 40 s, extension at 72°C for 1 min, followed by final extension at 72°C for 10 min. The PCR product was digested with restriction enzymes, HinfI, MspI, CfoI, NdeII, DdeI, RsaI MboI, HpaII, TaqI. The digested were separated by electrophoresis on pre-caste agarose gel of 2% in submarine electrophoresis system (Bangalore Genei) for 2.5 h at 70 V/cm at room temperature. The restriction pattern was visualized under UV illuminator after staining with 10 mg/ml ethidium bromide (EtBr) solution and saved as TIFF (tag image file format) image for data analysis.
Data Analysis
All restriction patterns were coded in binary form and analysed using NTSYS software [16]. A simple matching coefficient was calculated to construct a similarity matrix and the UPGMA algorithm was used to perform hierarchical cluster analysis and to construct a dendrogram.
Results
Twenty-five soybean isolates of Bradyrhizobium were subjected to biochemical analysis and compared with reference strain R (RJ 123) and were found to be Gram negative, capsulated, motile, non-sporing rods (Table 1). On the basis of biochemical confirmatory records the isolated Bradyrhizobium strains from agricultural fields were subjected to genetic diversity analysis. The Guanine plus Cytosine content data of isolated strains shows slight variation ranging between 61.0 and 64.0%. In the current study, restriction enzyme HinfI, MspI, CfoI, NdeII, DdeI, RsaI, MboI, HpaII, TaqI were used to restrict the amplified 16S rDNA segments separately. All the enzymes resulted different banding patterns showing differences among the Bradyrhizobium isolates. Observations with MboI, TaqI was highly discriminatory and was used to generate dendrograms.
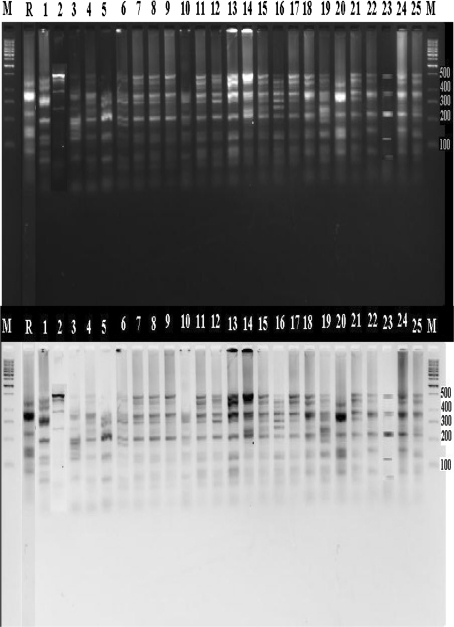
16S rDNA study with TaqI digestion formed common band of 200 bp in case of all Bradyrhizobium isolates except N2. 500 bp band was seen in Bradyrhizobium strains N2, N4, N7, N8, N9, N11, N12, N13, N14, N15, N16, N17, N18, N19, N21, N22, N23, N24 and N25 (Fig. 1). Computation of DNA profiles using NTSYS, based on Unweighted Pair Group Method with Arithmetic averaging (UPGMA) over TaqI digested 16S rDNA, demonstrated two major groups, one having R, N1, N2 and N25 with 65% similarity among them (Fig. 2). Other group included Bradyrhizobium strains N3, N4, N5, N6, N7, N8, N11, N12, N13, N14, N15, N16, N17, N18, N19, N21, N22 and N20. 100% similarity was further revealed by N6, N14, N15, N16 and N19. Bradyrhizobium group including N7, N8, N9, N11, N12, N13, N17, N18, N21 and N22 also represented 100% similarity within themselves. More than 80% similarity was recorded in N3 and N5 (83%); N4, N6, N14, N15, N19 (88%) and N7, N8, N9, N11, N12, N13, N17, N18 N21 and N22 (86%). Thus, present observations revealed that Bradyrhizobium isolates are more related to same species except strains N2, N10 and N23 were not similar to rest of Bradyrhizobial isolates.
Fig. 1.
Restriction enzyme TaqI treated 16S rDNA segments of isolated strains of Bradyhizobium sp. on 2% agarose gel
Fig. 2.
Dendogram based on RFLP of 16S rDNA segment of Bradyrhizobium isolates using TaqI restriction enzyme
The electrophoresis picture obtained following MboI restriction enzyme digested 16S r DNA, showed existence of common band (100 bp) in N1, N2, N4, N5, N6, N7, N8, N9, N10, N11, N13, N16, N17, N21 and N24 strains (Fig. 3). Another group with common band of 400 bp (N1, N2, N4, N5 N6, N8, N10, N11, N12, N13, N14, N17, N18, N24 and N25) was also observed. Strains N1, N2, N4, N6, N8, N10, N11, N12, N13, N14, N17 and N24 showed both bands of 100 and 400 bp bands. Overall analysis of 16S rDNA segment, with the help of restriction enzyme digestion revealed a remarkable dissimilarity among all the Bradyrhizobium isolates. MboI restriction enzyme on amplification with 16S rDNA segment also resulted into two groups, one having similarity of 60% (N2, N3, N5, N6, N8, N9, N10, N11and N12), however, another with 63% similarity (N1, N7, N13, N15, N17, N18, N19, N25 and R) (Fig. 4). Only four strains N13, N15, N2 and N3 were found to be 100% similar as per the availability of restriction site on the 16S rDNA. Binary analysis with NTSYS grouped the isolated strains into three groups with similarity of 34% (N23, N24 and N25); 42% (N13, N14, N15, N16, N18, N17, N19, N20 and N22) and 33% (N1, N2, N3, N5, N6, N7, N8, N9, N10, N11 and N12), reflecting similar gene pool. Current study with 16S rDNA was carried out with restriction enzymes MboI, TaqI, and HpaII only tells the presence and absence of restriction site on 16S rDNA segments of Bradyrhizobium isolates. The analysis showed all the strains to be different from each other with some degree of similarity amongst themselves. The dendrogram generated from data obtained by HinfI, MspI, CfoI, NdeII, DdeI, RsaI, MboI, HpaII, TaqI digestion confirms two groups, one at 74% (N1, N14, N10, N11, N6, N7, N8, N9, N13, N5, N2, N3, N4, N24, N25 and other at 75% (N12, N17, N18, N19, N15, N16, N20, N21, N22, N23) similarity. More then 85% similarity was reflected in N1 and N14; N10 and N11 (88%); N8 and N9 (94%); N22 and N23 (87%) and strains N12, N17, N18, and N19 (86%). These observations revealed that the Bradyrhizobium isolates are directly related to each other although differ with respect to place of isolation.
Fig. 3.
Restriction enzyme MboI treated 16S rDNA segments of isolated strains of Bradyrhizobium sp. on 2% agarose gel
Fig. 4.
Dendogram based on RFLP of 16S rDNA segment of Bradyrhizobium isolates using MboI restriction enzyme
Discussion
During the course of identification notable feature was recoded with the Bradyrhizobium sp. as they produce more mucilage, they utilize more glucose and were able to grow on nitrogenous compounds [18, 21]. Identical characteristics of Bradyrhizobium and growth pattern confirm the close relationship among the isolates and reference strain [22]. Diversification within a species can only be found out when genomic analysis is made as characteristics of individual is govern by expression of genetic pool which in turn supports position of individual in evolutionary hierarchy modification of genetic setups necessitates the evolution in species occupying beneficial supporting habitat.
The genetic diversity of microbial community is assessed by measuring the heterogeneity of DNA within entire microbial community. To do this, DNA was extracted from the total microbial community in a sample collected from a given habitat. The recovered DNA represents the total gene pool of the community. First and, possibly the simplest, technique to be employed is the determination of DNA base composition. Genetic content of organism reveals its position in biological diversity in general. The bacteria including nitrogen-fixing nodules also occupies bacterial diversity directly influenced by geographical and biological factors.
Guanine plus Cytosine content can be chemically ascertained after hydrolysis of DNA and separation of its bases. Physical methods are easier and more often used. The Guanine plus Cytosine content often determined from the melting temperature (Tm) of DNA. Despite such a wide range of variation, the Guanine plus Cytosine content of strains within a particular species is constant. The Guanine plus Cytosine content data of isolated strains shows slight variation ranging between 61.0 and 64.0% in all the isolates. Guanine plus Cytosine content appears to be useful in characterizing bacterial genera since the variation within a genus is usually less then 10% even though the content may vary greatly between genera [5]. Adenine.Thymine/Guanine.Cytosine pressure changes the Guanine plus Cytosine content of various part of genome in same direction depending upon their functional importance [23], this may account for the difference in % Guanine plus Cytosine content of Bradyrhizobium isolates. The % Guanine plus Cytosine content revealed that the bacterial isolates were different from that of Gram-positive bacteria along with variation in % Guanine plus Cytosine among itself.
A novel molecular strategy for studying microbiological diversity is based on restriction digestion of 16S rDNA sequence. Separate primers are used to amplify bacterial and archeal ribosomal RNA genes using the polymerase chain reaction (PCR). Ribosomal RNA are ancient molecules, functionally constant, universally distributed and mordantly well conserved across broad phylogenetic distance. Also because the number of different possible sequences of large molecule like ribosomal RNAs is so large, similarity in two sequences. However, it is the degree of similarity in ribosomal sequences between two organisms that indicates their common evolutionary relatedness. A wide range of genes in accordance with the aim of research are used in the PCR/RFLP technique [24]. The 16S rDNA of 25 isolated strains was PCR amplified with universal primer rD1 and fD1. The PCR products were individually restricted with endonuclease, HinfI, MspI, CfoI, NdeII, DdeI, RsaI, MboI, TaqI, and HpaII. Although each restriction enzyme produces polymorphic banding patterns the most discriminative was HpaII. Bradyrhizobium isolates on amplification with universal primer, reverse and forward primer resulted in the formation of a single band of around 1500 bp in all the 25 isolates. The polymorphism occurs due to availability and unavailability of the restrictions sites on the genomic segment. Each enzyme has got a different restriction site thus generates polymorphic pattern on agarose gel. All the three enzymes generated different banding patterns showing differences among the Bradyrhizobium isolates. On combining the data generated by restriction enzyme analysis with three enzymes MboI, TaqI, and HpaII, with the help of NTSYS software, strains found to be grouped into three, with similarity of 34% (N23, N24 and N25); 42% (N13, N14, N15, N16, N18, N17, N19, N20 and N22) and 33% (N1, N2, N3, N5, N6, N7, N8, N9, N10, N11 and N12), revealing it from similar genus (Fig. 5). This study over 16S rDNA was restricted to restriction enzymes MboI, TaqI, and HpaII only tells about the presence and absence of restriction site on 16Sr DNA segments of Bradyrhizobium isolates. But the over all restriction enzyme analysis over amplified 16S rDNA segment of bradyrhizobial isolates with restriction enzymes HinfI, MspI, CfoI, NdeII, DdeI, RsaI, MboI, TaqI, and HpaII, grouped the strains in two major groups (Fig. 5) with similarity of 74% (N1,N14, N10, N11, N6, N7, N8, N9, N13, N5, N2, N3, N4, N24, N25) and 75% (N12, N17, N18, N19, N15, N16, N20, N21, N22, N23) living aside the reference strain “R” at similarity of 63% (Fig. 5), which was found to be grouped with N23, N24, N25 at 34% with combine data of restriction enzymes MboI, TaqI, and HpaII (Fig. 5). The restriction enzyme analysis though showed less % of similarity with data generated by single restriction enzyme, where as the data generated by the multiple restriction enzymes leads us towards the more accuracy over the discussion over the similarity and dissimilarity among same species of isolated strains. This analysis complements to the conclusion drawn by % Guanine plus Cytosine, strains isolated from different varieties of Glycine max, have a common range of % Guanine plus Cytosine belongs to same genus (Table 1). Similarity though, the strains showed similarity with HinfI, MspI, CfoI, NdeII, DdeI, RsaI, MboI, TaqI, and HpaII, separately, and dissimilarity on combining the whole data refers the same origin. The horizontal transfer of homologous DNA segment among closely related organism is major source of genomic diversification [25] thus the strains came out to be similar to each other with least level of dissimilarity.
Fig. 5.
Dendogram based on analysis of 16S rDNA segment of Bradyrhizobium
The significance of nodulation by Bradyrhizobium also throws several genetic factor related to establishment of symbiotic relationship. Therefore, the study of genetic variation with existing molecular techniques along with biochemical analysis helped in specifying and determining phylogeny amongst isolated Bradyrhizobium cultures.
Acknowledgments
The authors are thankful to funding agency Council of Scientific and Industrial Research (CSIR), India for JRF, SRF fellowship of Anupama Bikrol.
References
- 1.Rhijn P, Vanderleyden J. The rhizobium–plant symbiosis. Microbiol Rev. 1995;59:124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lodwig EM, Hosie AHF, Bourdes A, Findlay K, Allaway D, Karunakaran R, Downie JA, Poole PS. Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature. 2003;422(6933):722–726. doi: 10.1038/nature01527. [DOI] [PubMed] [Google Scholar]
- 3.Oshima T, Imahari K. Description of Thermus thermophilluscomb. nov., a non sporulating thermophilic bacterium from Japanese thermal spa. Int J Syst Evol Microbiol. 1974;24:102–112. [Google Scholar]
- 4.Sakai Y, Oshima T. Isolation and characterization of bacteriophage infectious to an extreme thermophile, Thermus thermophilus HB8. J Virol. 1975;15:1449–1453. doi: 10.1128/jvi.15.6.1449-1453.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prescott ML, Harley JP, Klein AD (1996) Chapter 19: Microbial taxonomy. In: Microbiology, 3rd edn. Wm C. Brown Publishers, Dubuque (IOWA), pp 390–414
- 6.Sy A, Giraud E, Jourand P, Garcia N, Willems A, Lajudie P, Prin Y, Neyra M, Gillis M, Boivin-Masson C, Dreyfus B. Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J Bacteriol. 2001;183:214–220. doi: 10.1128/JB.183.1.214-220.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young JPW, Downer HL, Eardly BD. Phylogeny of the phototrophic Rhizobium strain BTAi1 by polymerase chain reaction-based sequencing of a 16SrRNA gene segment. J Bacteriol. 1991;173:2271–2277. doi: 10.1128/jb.173.7.2271-2277.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martı′nez-Romero E, Caballero-Mellado J. Rhizobium phylogenies and bacterial genetic diversity. Crit Rev Plant Sci. 1996;15:113–140. [Google Scholar]
- 9.Oyaizu H, Matsumoto S, Minamisawa K, Gamou T. Distribution of rhizobia in leguminous plants surveyed by phylogenetic identification. J Gen Appl Microbiol. 1993;39:339–354. doi: 10.2323/jgam.39.339. [DOI] [Google Scholar]
- 10.Lindström K, Laguerre G, Normand P, Rasmussen U, Heulin T, Jarvis BDW, de Lajudie P, Martı′nez.Romero E, Chen WX (1998) Taxonomy and phylogeny of diazotrophs. In: C. Elmerich, A. Kondorosi, and W. E. Newton (eds) Biological nitrogen fixation for the 21st century, Kluwer Academic Publishers, Dordrecht. Proceedings of the 11th international congress on nitrogen fixation, Paris, July 1997
- 11.Young JPW, Haukka KE. Diversity and phylogeny of rhizobia. New Phytol. 1996;133:87–94. doi: 10.1111/j.1469-8137.1996.tb04344.x. [DOI] [Google Scholar]
- 12.Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. At least in 20 16SrDNA sequence record currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel JB, Leonard DGB, Pan X, Musse JM, Berman RE, Nachamkin I. Sequence-based identification of Mycobaterium species using the Micro Seq 500 16S rDNA bacterial identification system. J Clin Microbiol. 2000;38:246–251. doi: 10.1128/jcm.38.1.246-251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ngo NkotL, Krasova-Wade T, Etoa FX, Sylla SN, Nwaga D. Genetic diversity of rhizobia nodulating Arachis hypogea L in diverse land use systems of humid forest zone in Cameroon. Appl Soil Ecol. 2008;40(3):411–416. doi: 10.1016/j.apsoil.2008.06.007. [DOI] [Google Scholar]
- 15.Jiang KY, Jun CZ. Diversity and host specificity of soybean and peanut Bradyrhizobia. Biol Fertil Soils. 2008;44(6):843–851. doi: 10.1007/s00374-008-0269-3. [DOI] [Google Scholar]
- 16.Rohlf FJ. NTSYS-pc numerical taxonomy and multivariate analysis system. Version 1.60. New York: Exeter Software, Setauket; 1990. [Google Scholar]
- 17.Allan AI, Sinclair JB, Schilling PE. Laboratory and green house evaluations of four systemic fungicides. Phytopathalogy. 1969;59:169–1662. [PubMed] [Google Scholar]
- 18.Subba Rao NS. Current developments in biological nitrogen fixation. New Delhi: Oxford and IBH Publishing Co. Pvt. Ltd; 1984. p. 351. [Google Scholar]
- 19.Hahn M, Studer D. Competitive of nif−Bradyrhizobium japonicum mutant against wild type strain. FEMS Microbiol Lett. 1986;33:143. [Google Scholar]
- 20.Hofer AW. Methods for distinguishing between legumes bacteria and their most common contaminant. J Am Soc Agron. 1935;27:228–230. [Google Scholar]
- 21.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor; 1989. [Google Scholar]
- 22.Rapley R. Molecular biology and basic techniques. In: Wilson K, Walker J, editors. Practical biochemistry principles and techniques. 5. Cambridge: Cambridge University Press; 2002. pp. 80–137. [Google Scholar]
- 23.Saiki RK, Gelfland DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 24.Giongo A, Ambrosini A, Vargas LK, Freire JRJ, Bodanese-Zanettini MH, Passaglia LMP. Evaluation of genetic diversity of Bradyrhizobia strains nodulating soybean [Glycine max (L.) Merrill] isolated from South Brazilian fields. Appl Soil Ecol. 2008;38:261–269. doi: 10.1016/j.apsoil.2007.10.016. [DOI] [Google Scholar]
- 25.Flores M, Morales L, Avila A, González V, Bustos P, Gareía D, Mora Y, Guo X, Collado-vides J, Piñero D, Dávila G, Mora J, Palacios R. Diversity of DNA sequences in the symbiotic genome of Rhizobium etli. J Bacteriol. 2005;187:7185–7192. doi: 10.1128/JB.187.21.7185-7192.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]