Abstract
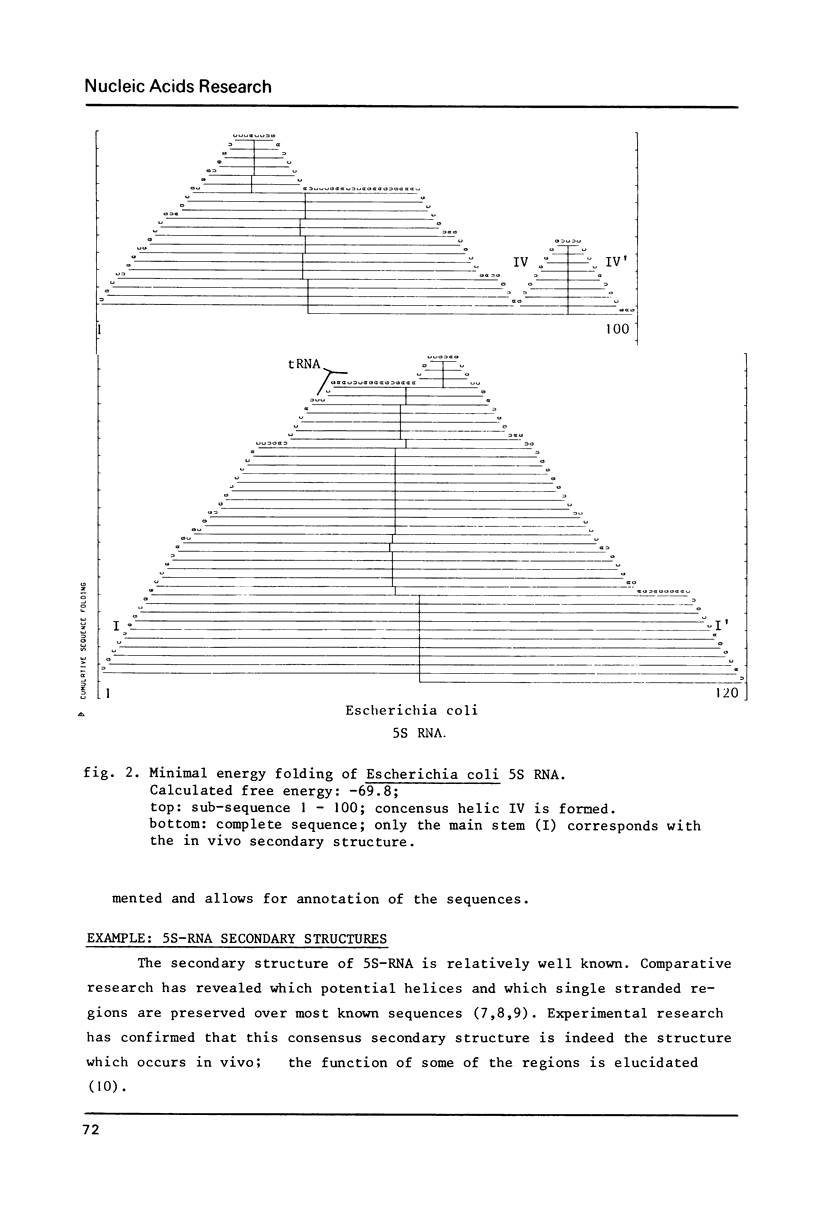
A modification of Nussinov's algorithm (1) for (planar) secondary structure generation is described. Our algorithm postpones decisions on matches involving destabiling loops until they prove to be energetically more favourable than more local matches. We present, moreover, an alternative way of representing secondary structures which avoids unwarranted suggestions on higher order neighbourhood, can be automated easily, allows for any amount of annotation of the sequences, makes comparison of alternate foldings easy and is pleasing to the eye. 5S RNA sequences are used to illustrate the methods.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Hori H. Molecular evolution of 5S RNA. Mol Gen Genet. 1976 May 7;145(2):119–123. doi: 10.1007/BF00269583. [DOI] [PubMed] [Google Scholar]
- Nussinov R., Jacobson A. B. Fast algorithm for predicting the secondary structure of single-stranded RNA. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6309–6313. doi: 10.1073/pnas.77.11.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R., Tinoco I., Jr, Jacobson A. B. Small changes in free energy assignments for unpaired bases do not affect predicted secondary structures in single stranded RNA. Nucleic Acids Res. 1982 Jan 11;10(1):341–349. doi: 10.1093/nar/10.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R., Tinoco I., Jr Sequential folding of a messenger RNA molecule. J Mol Biol. 1981 Sep 25;151(3):519–533. doi: 10.1016/0022-2836(81)90008-5. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Luehrsen K. R., Pribula C. D., Fox G. E. Sequence characterization of 5S ribosomal RNA from eight gram positive procaryotes. J Mol Evol. 1976 Aug 3;8(2):143–153. doi: 10.1007/BF01739100. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]



