Abstract
Protease production by fourteen M. anisopliae isolates differing in geographical origin and host insect were investigated. Highest protease activity was observed during 4–8 days of culture incubation. Pr1 and Pr2 activity was investigated in various media containing different carbon and nitrogen source to evaluate the induction mechanism of these enzymes. Basal levels of Pr1 and Pr2 activity were observed in minimal medium suggesting constitutive production. Casein (1%) as an exogenous protein supplement was not able to induce significant release of Pr1 and Pr2 enzymes, whereas high levels of activity were observed in the medium containing colloidal chitin (2%) as sole carbon and nitrogen source. The pH, ammonia and oxalic acid production in in vitro conditions was also investigated and the alteration in pH for protease production was not significant in the different media used except for the medium containing casein (1%) as a supplement.
Keywords: M. anisopliae, Pr1, Pr2, Protease, pH
Introduction
The concern for the development of hyphomycete fungi as suitable biocontrol agent of insect pests leads to the isolation of various insect pathogenic fungi. Amongst them, one of the most studied entomopathogenic fungus is Metarhizium anisopliae and mycoinsecticides have been developed and registered worldwide for control of agricultural pests [1]. The major barrier against the insect infection is the complex nature of the host cuticle which is primarily composed of proteins and chitin [2]. The secretion of proteases is believed to be an important pathogenic factor for fungal attachment on cuticle [3]. The best understood model of a fungal determinant of entomopathogenicity is based on the subtilisin like endoprotease, designated Pr1 [4]. The role of the Pr2 protease in insect parasitism is not yet elucidated completely, although St. Leger et al. [5] reported that Pr2 as well the other cuticle-degrading proteases may complement each other in the splitting of peptide bonds in the insect cuticle. St. Leger et al. [6] observed the regulation mechanism of Pr1 and Pr2 from M. anisopliae and reported the catabolic repression of the enzymes.
Regulation of gene expression by pH is observed in M. anisopliae that includes the cuticle-degrading subtilisin proteases and other putative virulence factors [7]. Subtilisins like proteases from M. anisopliae are only produced under alkaline conditions [8]. Ammonia production by M. anisopliae is tightly regulated by amino acids which as protein degradation products could serve as signals for the presence of proteinaceous nutrients in the environment [7]. Given that ammonium is linked with repression of several enzymes related with nitrogen metabolism, mutations in particular genes resulted in the derepression of the same including extracellular protease enzyme in some fungi. Although there are presumably additional regulatory mechanisms for instance oxidative deamination of the amino acids leading to ammonia accumulation in culture medium during fungal growth and subsequent increase in the pH favoring the protease production at basic pH [7, 9, 10]. In this study the variation in the proteolytic activity of fourteen M. anisopliae isolates was investigated for a period of 10 days to study the effect of incubation time on protease production. Subtilisin type Pr1 and trypsin like Pr2 activity of the high protease producing isolates was studied in media containing different carbon and nitrogen sources to elucidate the induction mechanism of these two enzymes. At the same time studies on pH, ammonia and oxalic acid release into the media and their effect on the Pr1 and Pr2 activity was also investigated.
Materials and Methods
Fungal Isolates and Culture Conditions
A total of 13 isolates were obtained from ARSEF (USDA-ARS Plant Protection Unit, Ithaca, NY) and one isolate was from India (Table 1). The isolates were routinely subcultured on Sabouraud Dextrose Agar (SDA) slants at 28°C and maintained at 4°C.
Table 1.
Source of M. anisopliae isolates
| Isolates | Code no./accession no. ARSEF | Host insect | Geographical location |
|---|---|---|---|
| UM1 | 1745 | Nilaparvata lugens | India |
| UM2 | 2735 | Spodoptera sp. | Philippines |
| UM3 | 2153 | Nephotettix virescens | Indonesia |
| UM4 | 2424 | Lepidpotera larva | Indonesia |
| UM5 | 3210 | Coleoptera | India |
| UM6 | 2596 | Pyrausta machaeralis | India |
| UM7 | 1080 | Helicoverpa zea | U.S.A |
| UM8 | 1724 | Nilaparvata lugens | India |
| UM9 | 1727 | Nilaparvata lugens | India |
| UM10 | 3295 | Anticarsia gemmatalis | Mexico |
| UM11 | 1729 | Nilaparvata lugens | India |
| UM12 | 1744 | Nilaparvata lugens | India |
| UM13 | 1823 | Nilaparvata lugens | India |
| AR1 | Local | Unknown | India |
Seven day old SDA slants were used for the preparation of conidial suspension (1 × 106 conidia/ml) to inoculate SDY broth (4% Dextrose, 1% Peptone, and 1% Yeast extract) and incubated at 28°C and 180 rpm for 3 days. The harvested mycelium was washed twice with sterilized distilled water and inoculated into induction medium containing casein (1%) (Sigma) in basal salt medium (0.1% KH2PO4, 0.05% MgSO4, 0.05% NaCl) at 20% v/v based on the final volume (50 ml) of the culture for the determination of proteolytic activity. The pH of the culture media was adjusted to 8.0 and the cultures were incubated at 28°C and 180 rpm for a period of 10 days. The mycelia were harvested by centrifugation and the supernatant was assayed for proteolytic activity on alternate days of growth from second day till 10th day of culture.
For the determination of subtilisin (Pr1) activity and trypsin (Pr2) activity three different media, minimal media, minimal media supplemented with casein (1%) and colloidal chitin (2%) were used respectively and the pH was maintained at 7.0. For the enzyme production, conidia at a concentration of 1 × 106 conidia/ml were inoculated into each media (three replicates were maintained) and incubated as shake cultures at 180 rpm, 28°C for 72 h. The mycelia were harvested by centrifugation and the supernatants assayed for enzymatic activity (Pr1 and Pr2).
Enzyme Assays and Protein Determination
Proteolytic Activity
The protease assay was done by the method of Kunitz [11]. Casein substrate was prepared for enzyme assay by dissolving 2% casein (Sigma) in 0.01 M Tris HCl (pH 8.0) containing 10 mM CaCl2 (pH 8.0). A 400 μl of casein substrate was added to the 200 μl of culture supernatant in 0.01 M Tris HCl pH 8, 10 mM CaCl2. The reaction mixture was incubated at 35°C for 10 min and the reaction terminated by the addition of 1.2 M TCA. The reaction mixture was centrifuged at 4000 rpm for 5 min and the supernatant was taken and the absorbance at 280 nm (A280) was observed against water as blank. One unit of protease activity was defined as the amount of enzyme that produced 1 mM of Tyrosine per minute under the above conditions.
Subtilisin (Pr1) and Trypsin (Pr2) Activity
The subtilisin (Pr1) activity and trypsin (Pr2) activity was assayed by the method of St. Leger et al. [12]. The Pr1 and Pr2 activity was assayed using specific synthetic substrates, N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide and Benzoyl-phenylalanine-valine-arginine-p-nitroanilide (BAPNA) respectively. Each substrate (50 μl) was mixed with 0.85 ml of Tris–HCl buffer (pH 8.5) and 100 μl of culture supernatant. The reaction mixture was incubated for 1 h at 28°C and the reaction was terminated using 0.25 ml of 30% acetic acid. Absorbance at 410 nm (A410) was observed and concentration of para-nitro aniline was determined. One unit of protease activity was defined as the amount of enzyme that produced 1 μM of para-nitro aniline per minute under the above conditions.
Protein content was measured by the method of Bradford [13] using Bovine Serum Albumin as a standard.
Quantification of Ammonia
Ammonia concentration was measured by the colorimetric method of Chaney and Marbach [14]. An uninoculated broth was used as a negative control. For this study two solutions were prepared, solution I (phenol 10 g/l, sodium nitroprusside 0.05 g/l) and solution II (NaOH 5 g/l, NaCl 0.42 g/l) and added in equal volumes to 1 ml of culture supernatant. Absorbance at 625 nm (A625) was determined after incubating the solution at room temperature for 30 min.
Quantification of Oxalic Acid
Oxalic acid concentration was determined by the method of Yan et al. [15]. To the culture supernatant (1 ml), 1 M H2SO4, 400 μl of 0.03 M K2Cr2O7 and 8 ml of dd H2O was added. After a gentle mix, 400 μl of 1 × 104 M Victoria blue B was added and the total volume of the solution were made to 10 ml by dd H2O. The solution was incubated for 9 min at 60°C. Mixture was cooled to quench the reaction with tap water for 2 min and absorbance at 610 nm (A610) of the solution was determined.
Protease Activity Gel (GAGE)
Molecular weight of the native enzyme was observed using gelatin acrylamide gel electrophoresis. SDS-PAGE was performed on 12.5% gels containing 0.3% gelatin as a copolymerized substrate under non-reducing conditions. 2.5% Triton X-100 solution was used to remove SDS and renaturate the enzymes. Gels are then incubated in 0.01 M Tris–HCl (pH 8)–10 mM CaCl2, buffer at 37°C for 5–8 h. The gels were fixed and stained with Coomassie brilliant blue R-250. Clear proteolytic zones in the gel were visible against dark blue background.
Statistical Analysis
Statistical analyses were performed by SPSS software. Test of significance were carried out using Tukey’s test.
Results and Discussion
The approach in the present study was to explore the proteolytic activities of several isolates of entomopathogenic fungus, Metarhizium anisopliae. The studies on various factors like pH, ammonia, oxalic acid concentration and Pr1 and Pr2 activities of the isolates screened based on the proteolytic activity was also carried out.
Proteolytic Activity
Variation in the time for maximum protease production was observed with different isolates. Most of the isolates showed high protease production during 6–8 day of culture. On the second day of incubation isolate UM6 showed highest enzymatic activity of 0.62 U/ml followed by isolate UM7 (0.57 U/ml), UM2 and UM9 (0.53 U/ml) and UM5 (0.51 U/ml). Isolates UM10, UM1 and UM2 showed a specific activity of 4.65, 4.14 and 4.12 mU/mg respectively (Table 2). On the fourth day, isolate UM6 showed a maximum enzymatic activity of 1.19 U/ml and the specific activity on fourth day was high for isolates UM10 (7.16 mU/mg) and UM13 (5.08 mU/mg). Isolates UM6 (1.82 U/ml) and UM4 (0.96 U/ml) showed maximum enzyme activity on day six. Specific activity was high for isolate UM2 (9.81 mU/mg). On the eighth day of incubation isolate UM6 (1.04 U/ml) showed the maximum enzyme activity followed by isolates UM4 (0.96 U/ml) and the specific activity was high for the isolate UM2 (5.28 mU/mg). Few isolates showed high proteolytic activity on 10th day. Caseinolytic and elastolytic activity of an M. anisopliae isolate was observed to be high during 4–6 days of culture and then a steep decrease up to 16th day of culture and this probably may be due to increasing degree of autolysis of the aging culture [16]. Among the M. anisopliae isolates studied, isolate UM6 showed highest enzyme activity on all the days of culture incubation. High specific activities were observed on day six for most of the isolates studied.
Table 2.
Enzymatic and specific activity of M. anisopliae isolates
| Isolates | Enzyme and specific activity as on | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Day 2 | Day 4 | Day 6 | Day 8 | Day 10 | ||||||
| Enzymatic activity (U/ml) | Sp. activity (mU/mg) | Enzymatic activity (U/ml) | Sp. activity (mU/mg) | Enzymatic activity (U/ml) | Sp. activity (mU/mg) | Enzymatic activity (U/ml) | Sp. activity (mU/mg) | Enzymatic activity (mU/ml) | Sp. activity (mU/mg) | |
| UM1 | 0.4054c | 4.14a | 0.4360e | 3.74d | 0.4457e | 5.79d | 0.5118e | 3.20c | 0.4011e | 4.77b |
| UM2 | 0.5398b | 4.12a | 0.5750d | 4.76c | 0.8122c | 9.81a | 0.4274f | 5.28a | 0.3037f | 2.92d |
| UM3 | 0.3312d | 3.21b | 0.4380e | 4.33c | 0.4245e | 7.26b | 0.4382f | 4.77b | 0.3296f | 3.73c |
| UM4 | 0.2701e | 1.87d | 0.8548b | 3.78d | 0.9676b | 5.46d | 0.9617ab | 4.61b | 0.2987f | 1.14e |
| UM5 | 0.5194b | 2.76c | 0.7520c | 3.33d | 0.7859c | 4.32e | 0.7838c | 2.53d | 0.8781a | 3.84c |
| UM6 | 0.6266a | 2.97c | 1.1905a | 3.26d | 1.8241a | 6.07c | 1.0479a | 3.76c | 0.9522a | 3.61c |
| UM7 | 0.5737b | 3.15b | 0.8550b | 4.36c | 0.8639bc | 6.12c | 0.8681b | 3.96c | 0.7068b | 4.38b |
| UM8 | 0.2179f | 0.447e | 0.2811g | 3.44d | 0.3471f | 3.40f | 0.5825e | 5.71a | 0.6046c | 3.99c |
| UM9 | 0.5378b | 3.89b | 0.5467d | 1.05e | 0.5423d | 5.02d | 0.6408d | 4.25b | 0.6534c | 4.44b |
| UM10 | 0.4797c | 4.65a | 0.4929e | 7.16a | 0.5692d | 4.66e | 0.6632d | 4.73b | 0.6639c | 5.35a |
| UM11 | 0.2156f | 0.442e | 0.2330g | 4.37c | 0.4048e | 3.13f | 0.6082d | 3.74c | 0.5837d | 3.47c |
| UM12 | 0.1711 g | 0.345e | 0.2053g | 4.54c | 0.2639g | 3.35f | 0.6401d | 3.81c | 0.5907d | 3.68c |
| UM13 | 0.3227d | 2.43c | 0.3352f | 5.08b | 0.3943e | 5.89d | 0.5812e | 4.37b | 0.4211e | 3.05c |
| AR1 | 0.2755e | 0.742e | 0.3925f | 4.06c | 0.3840ef | 1.42g | 0.5794e | 2.29d | 0.5295d | 2.44d |
Values followed by same lower case alphabets in the column are statistically equivalent according to Tukey’s test
An immense variation in the protease activity was observed among the fourteen M. anisopliae isolates. This variation might be attributed to the geographical origin of the isolates as four among the five high protease producing isolates viz. UM2, UM4, UM5 and UM6 were from Asian tropical regions (Philippines, Indonesia and India), although variation can not be correlated with the insect host. The insect host of isolates UM2, UM4 and UM6 belongs to the order Lepidoptera and isolate UM5 belongs to the order Coleoptera respectively. Interestingly isolates with a common insect host viz., Nilaparvata lugens, showed low to moderate protease activity. The time for maximum protease production does not necessarily depend upon the media constituents and Kucera [17] observed high amounts of protease release 3 days post inoculation in the media with different nitrogen sources. The caseinolytic activity can be attributed to the presence of Pr1 and Pr2 activity in the culture filtrate [12]. The high protease producing isolates were screened out based on the enzyme activity on sixth day and further Pr1 and Pr2 activity was determined along with pH, ammonia and oxalic acid production. The Pr1 and Pr2 activity in three different media with different carbon and nitrogen sources was investigated for the selected seven isolates.
pH, Ammonia, Oxalic Acid, Pr1 and Pr2 Activity in Minimal Medium
The use of minimal medium for protease production could possibly emulate the enzyme activity in the initial stages of host infection when readily metabolizable compounds are extremely low. All the isolates showed low levels of Pr1 and Pr2 activity in the minimal medium indicating that the production of this enzyme was constitutive. Similar results were obtained by St. Leger [6] where a rapid increase in Pr1 and Pr2 activity during carbon and nitrogen depression for an isolate of M. anisopliae was recorded. Constitutive production of protease was also found in Aspergillus sp. although an exception was observed for a B. bassiana isolate which showed decreased protease production in the absence of exogenous protein as inducer [18, 19]. High Pr1 activity was observed for isolate UM10 (1.16 U/ml) followed by isolate UM12 (0.530 U/ml). Specific activity was high for the isolate UM13 (0.36 U/ml), UM12 (0.34 U/ml) and UM10 (0.33 U/ml). No significant difference was observed in Pr2 activities for the seven isolates studied (Table 3). Highest Pr2 activity was observed for isolate UM12 (0.18 U/ml), UM6 (0.17 U/ml) and UM13 (0.16 U/ml). The specific activity was high for the isolates UM13 and UM12.
Table 3.
pH, ammonia, oxalic acid concentration, Pr1 and Pr2 activity of M. anisopliae isolates in minimal medium (MM)
| Isolates | pH | Ammonia concentration (μg/ml) | Oxalic acid concentration (μg/ml) | Pr1 | Pr2 | ||
|---|---|---|---|---|---|---|---|
| Enzyme activity (U/ml) | Sp. activity (U/mg) | Enzyme activity (U/ml) | Sp. activity (U/mg) | ||||
| UM4 | 7.04 | – | 3.5 | 0.32c | 0.16b | 0.13ab | 0.06bc |
| UM6 | 7.05 | – | 1.5 | 0.39c | 0.19b | 0.17ab | 0.09bc |
| UM7 | 7.06 | – | 3.5 | 0.37c | 0.17b | 0.14ab | 0.07bc |
| UM10 | 7.06 | – | 4.5 | 1.16a | 0.33a | 0.11b | 0.03c |
| UM11 | 7.04 | – | 21.5 | 0.21d | 0.09c | 0.13ab | 0.05c |
| UM12 | 7.05 | – | 42.5 | 0.53b | 0.34a | 0.18a | 0.12a |
| UM13 | 7.06 | – | 39 | 0.41c | 0.36a | 0.16ab | 0.14a |
Values followed by same lower case alphabets in the column are statistically equivalent according to Tukey’s test
Genes that are involved in virulence and pathogenesis are often regulated by environmental signals like pH [20]. In this investigation, no correlation was observed in relation to the pH of the culture medium and subtilisin type protease production (Pr1) in minimal medium with the seven M. anisopliae isolates. The pH of the unbuffered media increased diminutively from the initial pH 7.0 and the low protease production could also be attributed to the production of oxalic acid in the media. Oxalic acid concentration was high in isolates UM12 (42.5 μg/ml), UM13 (39.05 μg/ml). Ammonia production was not detected in any of the isolates studied probably due to the presence of oxalic acid.
pH, Ammonia, Oxalic Acid, Pr1 and Pr2 Activity in Minimal Medium Supplemented with Casein (1%)
High Pr1 enzymatic activity was observed for isolate UM10 (2.44 U/ml) and specific activity was also high at 7.43 U/mg. The specific activity was high for isolate UM12 (1.40 U/mg) which was much lower than isolate UM10 (Table 4). Isolates UM4 and UM6 showed a very low Pr1 enzymatic and specific activity. Pr2 activity was high for the isolate UM10 (2.48 U/ml) higher than Pr1 activity. The specific activity was also high for the isolate UM10 when compared to other isolates. Isolate UM6 showed the lowest Pr2 enzymatic activity followed by isolates UM7 and UM4. Low levels of Pr1 and Pr2 activity were observed in medium II containing casein (1%) as sole source of carbon and nitrogen except for isolate UM10 which showed high levels of both Pr1 and Pr2 activity (2.44 U/ml and 2.48 U/mg) respectively. Astoundingly specific activities of all the seven isolates were high for both Pr1 and Pr2 enzyme and this perhaps might be due to the exogenous protein supplement which was acting as an inducer for the enzyme production. Similar results of protease induction by protein were observed by Drucker [21] in Neurospora crassa. St. Leger et al. [6] reported that modifying the concentrations of carbon and nitrogen independently could repress protease production even in the absence of other repressor molecule. In this medium supplemented with casein (1%), low levels of ammonia were detected and oxalic acid was not produced unlike the minimal media. A significant change in the pH was noticed in all the isolates from the initial 7.0 and isolate UM10 which showed highest Pr1 and Pr2 activity showed an increase to 8.21 from the initial 7.0. The appearance of ammonia could possibly explain the increase in the pH of the medium as ammonia production by isolate UM10 (0.2238) was also highest compared to other isolates.
Table 4.
pH, ammonia, oxalic acid concentration, Pr1 and Pr2 activity of M. anisopliae isolates in MM supplemented with casein (1%)
| Isolates | pH | Ammonia concentration (μg/ml) | Oxalic acid concentration (μg/ml) | Pr1 | Pr2 | ||
|---|---|---|---|---|---|---|---|
| Enzyme activity (U/ml) | Sp. activity (U/mg) | Enzyme activity (U/ml) | Sp. activity (U/mg) | ||||
| UM4 | 7.68 | 0.0277 | – | 0.17c | 0.52c | 0.35c | 1.07d |
| UM6 | 7.32 | 0.0354 | – | 0.18c | 0.56c | 0.19d | 0.59e |
| UM7 | 7.72 | 0.0323 | – | 0.38b | 1.18b | 0.32c | 0.99d |
| UM10 | 8.21 | 0.2238 | – | 2.44a | 7.43a | 2.48a | 7.55a |
| UM11 | 7.61 | 0.0538 | – | 0.42b | 1.26b | 0.43b | 1.29c |
| UM12 | 7.59 | 0.0531 | – | 0.46b | 1.40b | 0.41b | 1.25c |
| UM13 | 7.69 | 0.0261 | – | 0.35b | 1.07b | 0.49b | 1.49b |
Values followed by same lower case alphabets in the column are statistically equivalent according to Tukey’s test
pH, Ammonia, Oxalic Acid, Pr1 and Pr2 Activity in Minimal Medium Supplemented with Colloidal Chitin (2%)
High levels of Pr1 production was observed in medium containing colloidal chitin (2%) as a sole carbon and nitrogen source. Chitin, the main constituent of insect cuticle enhances the Pr1 activity either by inducing the protease or possibly due to the inability to produce catabolite repression. The response of Pr2 activity was not the same for all the isolates as for Pr1 activity. Low to moderate Pr2 activity was observed among the different isolates. Isolate UM7 and UM13 showed highest Pr2 activity of 1.87 and 1.22 U/ml respectively on contrary a basal level of Pr2 production was observed for isolate UM6 (0.10 U/ml) and perhaps Pr2 induction in M. anisopliae is an isolate dependent event. Pr1 activity was highest for the isolate UM11 (4.52 U/ml) followed by isolate UM13 (4.46 U/ml). The other isolates showed a very close range in the Pr1 activity. The specific activity was high for the isolate UM4 (0.76 U/ml). The Pr2 activity was high for the isolate UM7 (1.87 U/ml) followed by isolate UM13 (1.22 U/ml). The specific activity was high for the isolate UM4 (0.18 U/mg) though the enzyme activity was low (Table 5). The rest of the isolates showed very low specific activity for Pr2. A marginal change in the pH from initial 7.0 was observed with a maximum increase to 7.22 for isolate UM13 which showed highest Pr1 activity. Interestingly, in this medium supplemented with colloidal chitin (2%), low levels of ammonia were detected but there was no production of oxalic acid like the earlier medium supplemented with casein (1%). The pH and ammonia concentration obtained indicated no such correlation with the protease production. Entomopathogenic fungus B. bassiana was observed to produce moderate amounts of protease at pH 7 in unbuffered medium after 72 h of incubation [22].
Table 5.
pH, ammonia, oxalic acid concentration, Pr1 and Pr2 activity of M. anisopliae isolates in MM supplemented with colloidal chitin (2%)
| Isolates | pH | Ammonia concentration (μg/ml) | Oxalic acid concentration (μg/ml) | Pr1 | Pr2 | ||
|---|---|---|---|---|---|---|---|
| Enzyme activity (U/ml) | Sp. activity (U/mg) | Enzyme activity (U/ml) | Sp. activity (U/mg) | ||||
| UM4 | 7.01 | 0.0054 | – | 4.29b | 0.76a | 0.99c | 0.18a |
| UM6 | 7.03 | 0.0069 | – | 4.16c | 0.39c | 0.10e | 0.01d |
| UM7 | 7.17 | 0.0169 | – | 4.27b | 0.16e | 1.87a | 0.07bc |
| UM10 | 7.09 | 0.0223 | – | 4.28b | 0.49b | 0.98c | 0.11b |
| UM11 | 7.08 | 0.0300 | – | 4.52a | 0.33 cd | 0.13de | 0.01d |
| UM12 | 7.10 | 0.0300 | – | 4.21c | 0.29d | 0.19d | 0.01d |
| UM13 | 7.22 | 0.0292 | – | 4.46a | 0.15e | 1.22b | 0.04c |
Values followed by same lower case alphabets in the column are statistically equivalent according to Tukey’s test
Protease Activity gel (GAGE)
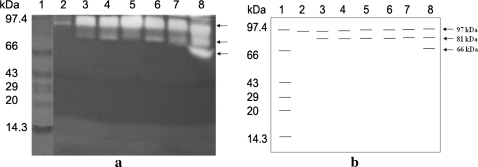
A 97 kDa band was observed for all the isolates in the activity zymogram. An additional 81 kDa protease was observed for all the isolates except isolate UM4. Isolate UM13 showed an additional 66 kDa native protease (Fig. 1a, b). Isolate UM4 showed a single band of 97 kDa and isolate UM13 showed three bands of sizes, 97, 81 and 66 kDa respectively. The rest of the isolates showed two bands (97 and 81 kDa).
Fig. 1.
a Protease activity in gelatin polyacrylamide gel electrophoresis. Lane 1 corresponds to mol wt marker after complete destaining of the gel. Lanes 2–8 correspond to isolates UM4, UM6, UM7, UM10, UM11, UM12 and UM13. b Schematic representation of (a) with the corresponding MW for each isozyme
In conclusion, the results represented in this study would increase the knowledge about the degree of protease production from various M. anisopliae isolates, the incubation time and various media. The Pr1 and Pr2 production by M. anisopliae isolates could lead to new course of understanding of the role of proteases in virulence of the entomopathogenic fungi.
Acknowledgments
We acknowledge the financial support provided by Department of Science and Technology (Project No. SR/FT/L-144) and Ministry of Human Resource and Development (Project No. F. 26-11/2004 TS V). We also thank Dr. Richard A. Humber (USDA-ARS) for providing the fungal cultures.
References
- 1.Whipps JM, Lumsden RD. Commercial use of fungi as plant disease biological control agents: status and prospectus. In: Butt TM, Jackson CW, Magan N, editors. Fungi as biocontrol agents-progress problems and potential. Wallingford: CAB International; 2001. pp. 23–69. [Google Scholar]
- 2.Anderson SO. Biochemistry of insect cuticle. Ann Rev Entomol. 1979;24:29–61. doi: 10.1146/annurev.en.24.010179.000333. [DOI] [Google Scholar]
- 3.St. Leger RJ, Cooper RM, Charnley AK. Production of cuticle-degrading enzymes by the Entomopathogen Metarhizium anisopliae during infection of cuticles from Colliphora vomitoria and Manduca sexta. J Gen Microbiol. 1987;133:1371–1382. [Google Scholar]
- 4.St. Leger RJ, Joshi L, Bidochka MJ, Roberts DW. Protein synthesis in Metarhizium anisopliae growing on host cuticle. Mycol Res. 1995;99:1034–1040. doi: 10.1016/S0953-7562(09)80769-7. [DOI] [Google Scholar]
- 5.St. Leger RJ, Bidochka MJ, Roberts DW. Isoforms of the cuticle degrading Pr1 protease and production of a metalloproteinase by Metarhizium anisopliae. Arch Biochem Biophys. 1994;313:1–7. doi: 10.1006/abbi.1994.1350. [DOI] [PubMed] [Google Scholar]
- 6.St. Leger RJ, Durrands PK, Cooper RM, Charnley AK. Regulation of production of proteolytic enzymes by the entomopathogenic fungus Metarhizium anisopliae. Arch Microbiol. 1988;150:413–416. doi: 10.1007/BF00408316. [DOI] [Google Scholar]
- 7.St. Leger RJ, Nelson JO, Screen SE. The entomopathogenic fungus Metarhizium anisopliae alters ambient pH, allowing extracellular protease production and activity. Microbiology. 1999;145:2691–2699. doi: 10.1099/00221287-145-10-2691. [DOI] [PubMed] [Google Scholar]
- 8.St. Leger RJ, Joshi L, Bidochka MJ, Rizzo NW. Biochemical characterization and ultrastructural localization of two extracellular trypsins produced by Metarhizium anisopliae in infected insect cuticles. Appl Environ Microbiol. 1996;62:1257–1264. doi: 10.1128/aem.62.4.1257-1264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arst HN, Cove DJ. Methyl ammonium resistance in Aspergillus nidulans. J Bacteriol. 1969;98:1284–1293. doi: 10.1128/jb.98.3.1284-1293.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen BL. Ammonium repression of extracellular protease in Aspergillus nidulans. J Gen Microbiol. 1972;71:293–299. [Google Scholar]
- 11.Kunitz M. Crystalline soybean trypsin inhibitor. II. General properties. J Gen Physiol. 1947;30:291–310. doi: 10.1085/jgp.30.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.St. Leger RJ, Charnley AK, Cooper RM. Characterization of cuticle-degrading proteases produced by the entomopathogen Metarhizium anisopliae. Arch Biochem Biophys. 1987;253:221–232. doi: 10.1016/0003-9861(87)90655-2. [DOI] [PubMed] [Google Scholar]
- 13.Bradford MM. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 14.Chaney AL, Marbach EP. Modified reagents for the determination of urea and ammonia. Clin Chem. 1962;8:130–132. [PubMed] [Google Scholar]
- 15.Yan ZY, Xing GM, Li ZX. Quantitative determination of oxalic acid using victoria blue B based on a catalytic kinetic spectrophotometric method. Microchim Acta. 2004;144:199–205. doi: 10.1007/s00604-003-0085-2. [DOI] [Google Scholar]
- 16.Braga GUL, Destefano RHR, Messias CL. Protease production during growth and autolysis of submerged Metarhizium anisopliae cultures. Rev Microbiol. 1999;30:107–113. doi: 10.1590/S0001-37141999000200004. [DOI] [Google Scholar]
- 17.Kucera M. Toxins of the entomophagous fungus Beauveria bassiana. II. Effect of nitrogen sources on formation of the toxic protease in submerged culture. J Invertebr Pathol. 1971;17:211–215. doi: 10.1016/0022-2011(71)90093-0. [DOI] [PubMed] [Google Scholar]
- 18.Cohen BL, Drucker M. Regulation of exocellular protease in Neurospora crassa: induction and repression under conditions of nitrogen starvation. Arch Biochem Biophys. 1977;182:601–613. doi: 10.1016/0003-9861(77)90541-0. [DOI] [PubMed] [Google Scholar]
- 19.Bidochka MJ, Khachatourians GG. Regulation of extracellular protease in the entomopathogenic fungus Beauveria bassiana. Exp Mycol. 1988;13:161–168. doi: 10.1016/0147-5975(88)90005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernardis F, Muhlschlegel FA, Cassone A, Fonzi WA. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect Immun. 1998;66:3317–3325. doi: 10.1128/iai.66.7.3317-3325.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drucker H. Regulation of exocellular proteases in Neurospora crassa: induction and repression of enzyme synthesis. J Bacteriol. 1972;110:1041–1049. doi: 10.1128/jb.110.3.1041-1049.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dias BA, Neves PMOJ, Furlaneto-Maia L, Furlaneto MC. Cuticle degrading proteases produced by the entomopathogenic fungus Beauveria bassiana in the presence of coffee berry borer cuticle. Braz J Microbiol. 2008;39:301–306. doi: 10.1590/S1517-83822008000200019. [DOI] [PMC free article] [PubMed] [Google Scholar]