Abstract
A new algorithm is presented which permits the display of nucleic acid secondary structure by computer. This algorithm circumvents the problem of overlapping portions of the molecule which is inherent in some other drawing programs. The results from this algorithm may also be used as input to the drawing algorithm previously reported in this journal [1] to untangle most of a drawing. The algorithm also represents the molecule in a form which makes visual comparisons for similarity quite easy since it guarantees that comparable features will reside in the same relative position in the drawings when the drawings are normalized.
Full text
PDF

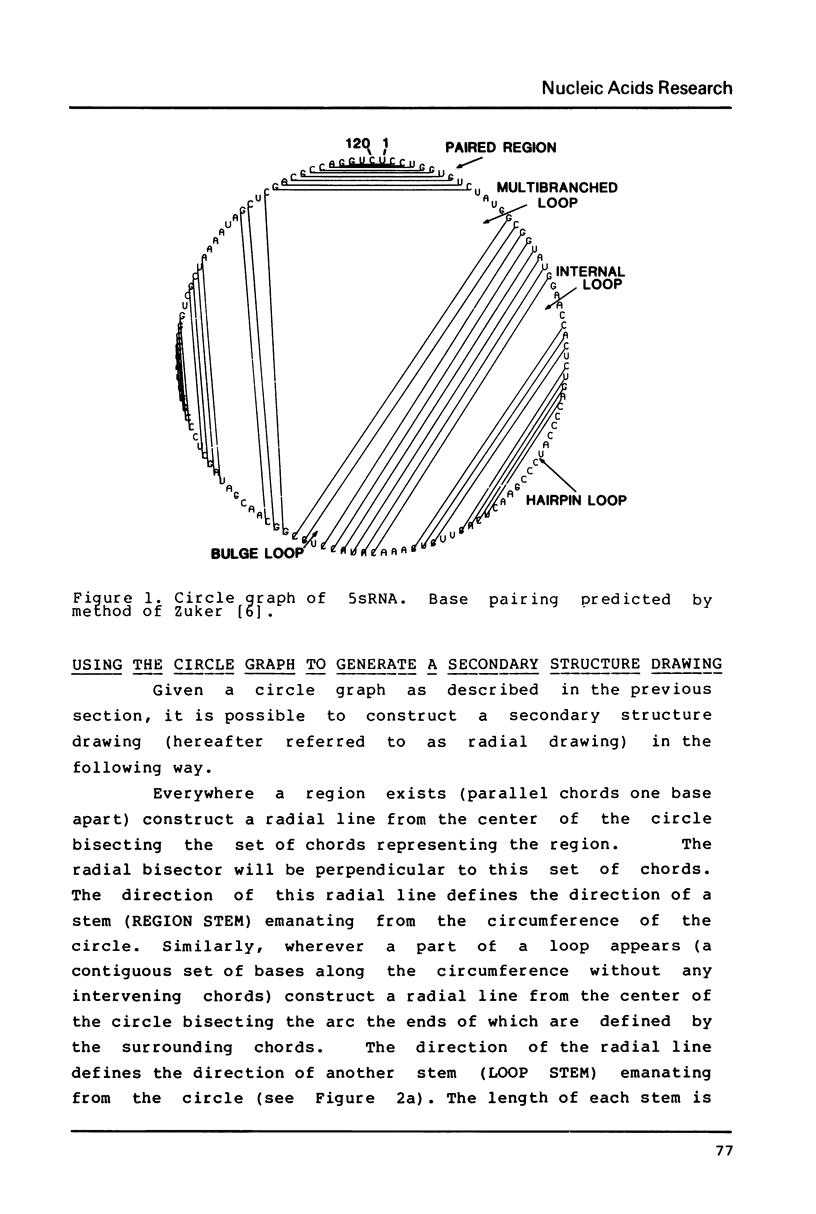
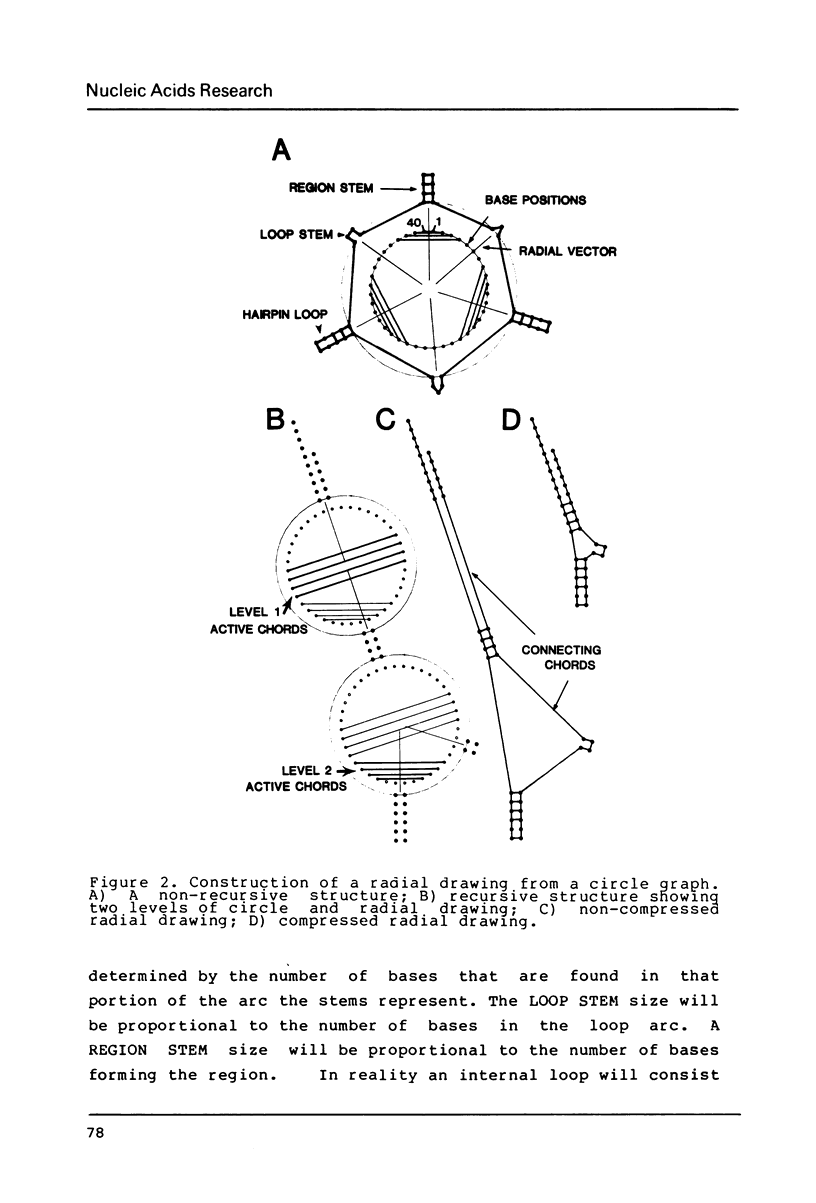


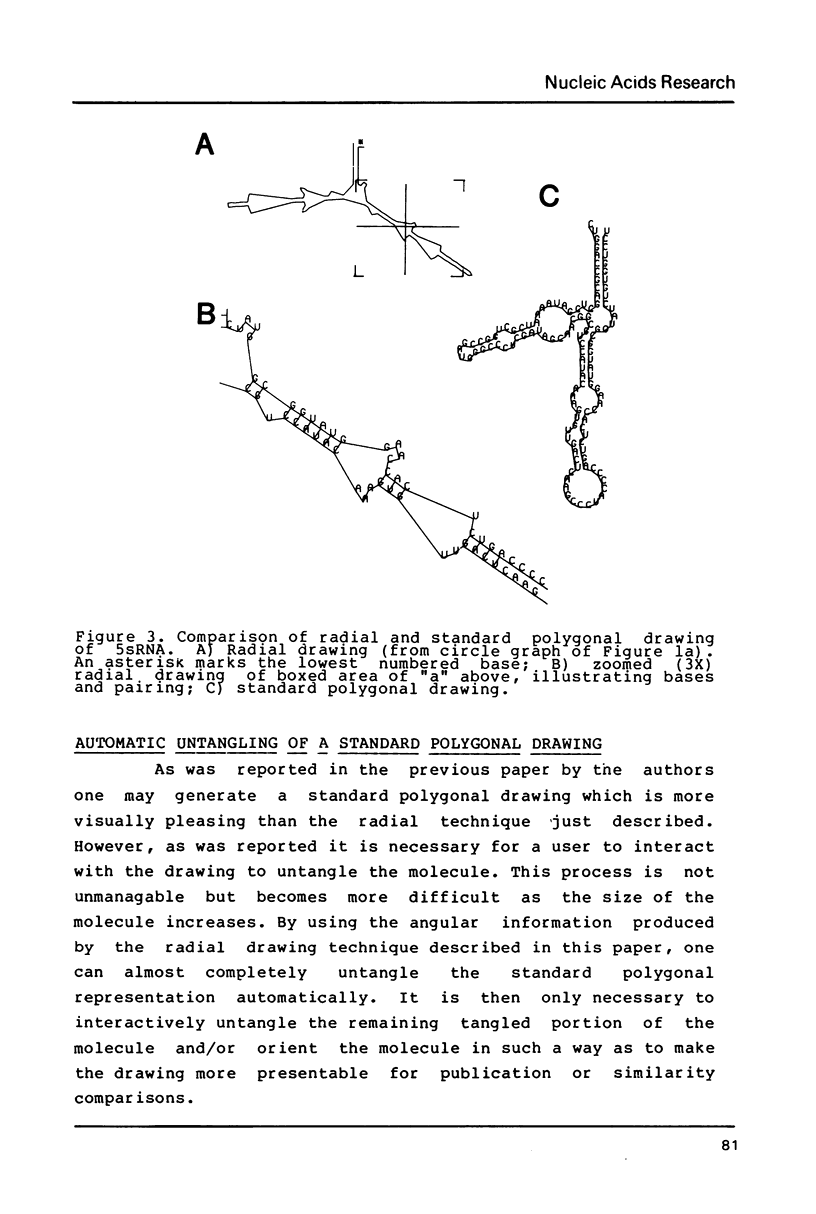



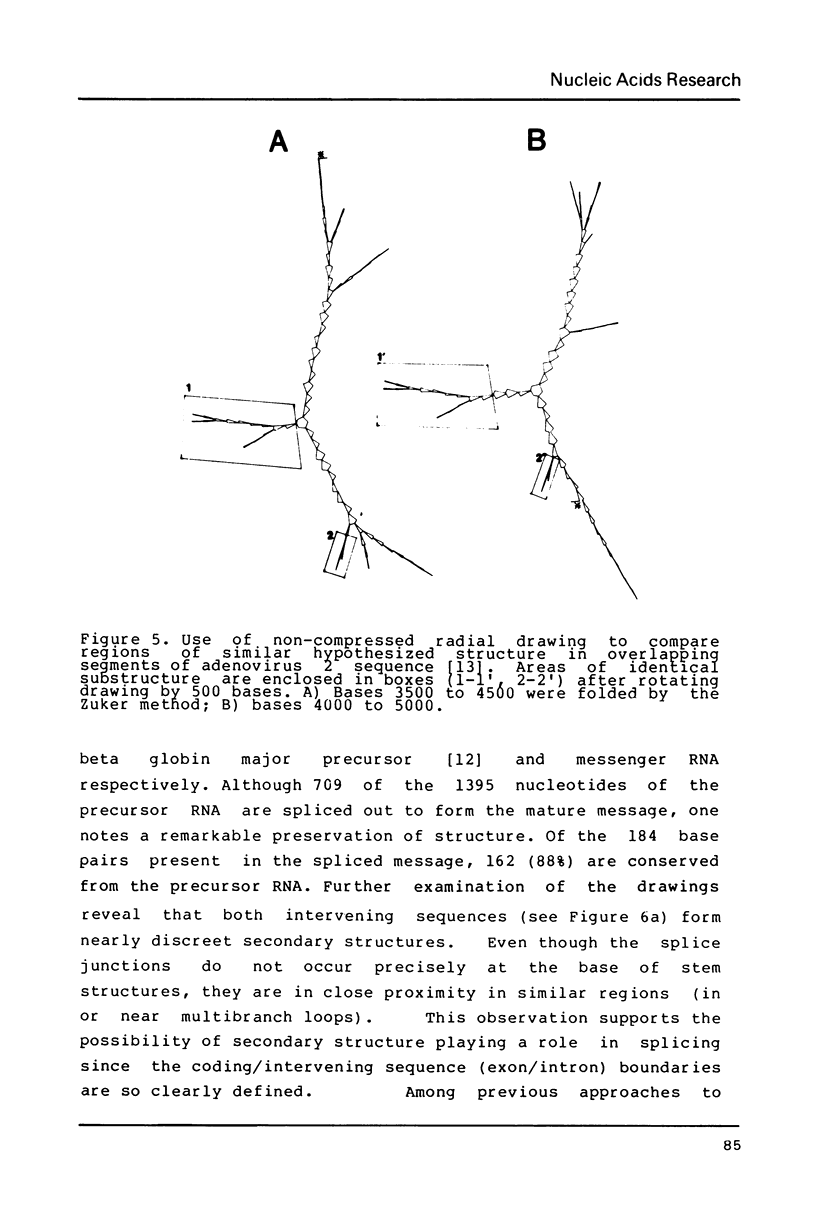



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auron P. E., Rindone W. P., Vary C. P., Celentano J. J., Vournakis J. N. Computer-aided prediction of RNA secondary structures. Nucleic Acids Res. 1982 Jan 11;10(1):403–419. doi: 10.1093/nar/10.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craik C. S., Rutter W. J., Fletterick R. Splice junctions: association with variation in protein structure. Science. 1983 Jun 10;220(4602):1125–1129. doi: 10.1126/science.6344214. [DOI] [PubMed] [Google Scholar]
- Nussinov R., Jacobson A. B. Fast algorithm for predicting the secondary structure of single-stranded RNA. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6309–6313. doi: 10.1073/pnas.77.11.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipas J. M., McMahon J. E. Method for predicting RNA secondary structure. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2017–2021. doi: 10.1073/pnas.72.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro B. A., Lipkin L. E., Maizel J. An interactive technique for the display of nucleic acid secondary structure. Nucleic Acids Res. 1982 Nov 11;10(21):7041–7052. doi: 10.1093/nar/10.21.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studnicka G. M., Rahn G. M., Cummings I. W., Salser W. A. Computer method for predicting the secondary structure of single-stranded RNA. Nucleic Acids Res. 1978 Sep;5(9):3365–3387. doi: 10.1093/nar/5.9.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilghman S. M., Curtis P. J., Tiemeier D. C., Leder P., Weissmann C. The intervening sequence of a mouse beta-globin gene is transcribed within the 15S beta-globin mRNA precursor. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1309–1313. doi: 10.1073/pnas.75.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]



