Abstract
A potentially novel antimicrobial compound producing Pediococcus acidilactici LAB 5 was isolated from vacuum-packed fermented meat product. This compound was found active against some species of Enterococcus, Leuconostoc, Staphylococcus and Listeria, many of which are associated with food spoilage and food related health hazards. The strain was found to be a paired cocci which can utilize a broad range of carbohydrates and produce acid identical to the P. acidilactici and P. pentoseus. Since the antimicrobial agent was sensitive to proteolytic enzymes but quite resistant to heat, it was identified as a bacteriocin and was designated as Pediocin NV 5. The molecular weight of the bacteriocin was 10.3 kDa and the bacterium possessed a 5 kbp plasmid responsible for bacteriocin production and also for vancomycin resistance phenotype.
Keywords: Antimicrobial spectrum, Bacteriocin, Paired cocci, Plasmid, Pediocin
Introduction
Naturally fermented foods are a great source of different types of fermenting microbes specially lactic acid bacteria group. Lactic acid bacteria produce a variety of compounds during sugar fermentation such as organic acids, hydrogen peroxide, and bacteriocins etc. [1]. These compounds are either bactericidal or bacteriostatic. Among these, bacteriocins have some specialty in that they are ribosomally synthesized proteinaceous compounds that are inhibitory to a wide variety of organisms, mostly closely related to the producer organisms [2–4]. For these properties the use of bacteriocins to enhance shelf-life of fermented foods and feeds is now being exploited [5–7]. Bacteriocins from different strains of Pediococcus acidilactici H [8], P. acidilactici PAC 1.0 [9] and Pediococcus pentosaceus [10] have been characterized for effective use in foods. Pediocins, produced by several species of Pediococcus, have some common properties that they mostly are anti-listerial and plasmid-borne [9, 11].
In this study, we report on isolation and characterization of a new bacteriocin producing lactic acid bacterium, P. acidilactici LAB 5, from vacuum-packed fermented meat product (sausage), which produces a potentially novel plasmid borne bacteriocin, Pediocin NV 5.
Materials and Methods
Bacterial Strains and Media
A strain of lactic acid bacterium was isolated from vacuum packed fermented meat product (sausage) and was screened for bacteriocin production against two bacteriocin sensitive indicator strains Enterococcus faecalis MB1 and Leuconostoc mesenteroides Ly following previously described method [12]. All the producer and indicator strains were cultured in Lactobacillus MRS medium [13] (Hi Media) and were maintained in 10% glycerol-skimmed milk at −4°C. The bacteria against which the antimicrobial spectrum of Pediocin NV 5 was studied are enlisted in Table 1. All the bacteria were procured from MTCC, IMTECH, Chandigarh, India. The non-lactic acid bacteria were cultured and maintained in their respective media.
Table 1.
Spectrum of antimicrobial activity of Pediocin NV 5
| Bacterial strains | Growth medium | Deferred method | Spot-on-lawn method |
|---|---|---|---|
| B. megaterium1684a | NB | − | − |
| B. subtilis | NB | − | − |
| E. faecalis MB 1b | MRS | + | + |
| Escherichia coli | NB | − | − |
| Lactococcus lactis subsp. l 440 | BHI | − | − |
| Lactococcus lactis subsp. la 3038 | MRS | + | + |
| Lactococcus lactis subsp. ch 3042 | MRS | + | + |
| L. mesenteroides Lyb | MRS | + | + |
| L. inocua | BHI | + | + |
| L. monocytogenes 657 | TSB | + | + |
| Pantoea ananetis 2307 | NB | − | − |
| Pseudomonas aeruginosa | NB | − | − |
| Pseudomonas fluorescens | NB | − | − |
| Salmonella typhimurium 98 | NB | − | − |
| S. aureus 96 | TSB | + | + |
| S. aureus subsp. aureus 3103 | TSB | − | − |
| S. aureus subsp. aureus 737 | TSB | − | − |
| S. aureus subsp. aureus 902 | TSB | − | − |
| S. aureus subsp. aureus 1430 | TSB | − | − |
| S. aureus subsp. aureus 1144 | TSB | − | − |
| S. epidermidis 2639 | TSB | − | − |
| S. epidermidis 3086 | TSB | − | − |
| Streptococcus sp. | NB | + | + |
| Xanthomonas campestris 2286 | – | − | − |
(+) and (−) symbols indicate the presence and absence of growth inhibition zone respectively around the spot
NB Nutriet Broth (Hi Media); MRS de Man, Rogosa and Sharpe (Hi Media); BHI Brain Heart Infusion (Hi Media); TSB Tripticase Soya Broth (Hi Media)
aNumber at the end of strain name indicates MTCC strain number
bMarked strains provided by Prof. Bibek Ray, USA
Isolation of the Producer Strain
Vacuum-packed ready to eat fermented meat products were purchased from local market. Each bag was punctured with a sterile needle and liquid drops from the packet were collected from where the producer strain was isolated on MRS agar plate. Plates containing 50 colonies or less (approx.) were overlaid with melted MRS soft agar (0.7%) seeded with E. faecalis MB1 and L. mesenteroides Ly indicator organisms, and were incubated at 28°C for 24 h to observe the zone of growth inhibitions. Colonies producing zones greater than 5 mm were flipped out and subsequently transferred in MRS agar plates for pure culture. The isolated strains were examined for morphology using light and scanning electron microscope and for Gram nature using crystal violet dye followed by ethanol washing; negative stain for detection of capsule formation was done by Nelsen solution, and endospore staining using malachite green stain following standard protocol. NaCl tolerance was studied by incubating the isolate in different concentration of NaCl amended MRS broth at 28°C for 24 h and observed for cell turbidity.
Identification and Characterization of the Isolated Strains
The isolated strain was tested for biochemical tests like catalase production, gelatin and nitrate reduction tests. Catalase test was done by flooding 3% H2O2 on slides containing the strain and observed for any bubble formation. Catalase activity and gas production from glucose were determined by the methods of Kozaki et al. [14]. Gelatin and nitrate reduction tests were done following standard protocol. The carbohydrate fermentation profile was done using API-CH 50 identification test kit (bioMerieux) and further characterized by partial 16S rDNA sequencing and subsequent phylogenetic analysis to identify the strain. 16S rDNA sequencing was done using 5′–3′ 357 forward primer and 5′–3′ 685 reverse primer and phylogenetic tree was constructed by neighbor joining method [15].
Assay of Activity and Determination of Antimicrobial Spectrum
Antimicrobial substance (bacteriocin) produced by the strain was detected by deferred method [12] and confirmed by spot-on-lawn method [12]. For deferred method, the producer strain was cultured overnight (O/N) and serially diluted and spreaded onto the surface of MRS agar plates. The plates were incubated for 24 h at 28°C to allow the colonies to appear and then overlaid with 5 ml of melted MRS soft agar (0.7%) seeded with overnight grown 6 × 107 cfu/ml of E. faecalis MB1. The plates were incubated at 28°C for 24 h and observed for clear zone of growth inhibition. In spot-on-lawn method, pre-poured MRS agar plates were overlaid with 5 ml melted soft agar (0.7%) inoculated with 50 μl of O/N grown 6 × 107 cfu/ml of indicator strain. Colony as well as the boiled and un-boiled cell-free aliquot (5 μl) of the producer strain were spotted on the lawn and incubated at 28°C for 24 h and observed for growth inhibition.
The amount of bacteriocin production was calculated as arbitrary Activity Units (AU). One AU is defined as the reciprocal of the highest serial two-fold dilution showing a clear zone of growth inhibition of the indicator strain [3]. The antimicrobial spectrum was determined against a number of pathogenic bacteria (Table 1) belonging to both Gram-positive and Gram-negative category using heat killed cell free culture aliquot by spot-on-lawn method.
Partial Purification and Characterization of the Antimicrobial Substance
Partial purification of the bacteriocin was done following the adsorption–desorption method of Yang et al. [16]. The chemical nature of the antimicrobial substance was tested against pH, temperature, organic solvents and enzymes [17]. Partially purified bacteriocin was evaluated in tricine SDS-PAGE under denaturing condition [18].
Plasmid Curing and Restriction Endonuclease Analysis
Plasmid was cured using non-hazardous chemical following the protocol of Ramesh et al. [19]. To cure plasmid, membrane sterilized ascorbic acid was introduced in sterile MRS broth at a concentration of 0.5–5.0 mM, pH of which was adjusted to 7.2 by addition of sodium bicarbonate. The producer strain was inoculated as 1% and incubated at 37°C and sub-cultured at every 18 h for two or three times in ascorbic acid added MRS broth. At every subculture the percent of viability and plasmid loss was enumerated using deferred method. The plasmid was purified following the protocol of Anderson and Mc Kay [20] and agarose gel electrophoresis was done following the procedure published by Sambrook and Russell [21]. Plasmid preparations for restriction endonuclease analyses were done in RE buffer (10 mM Tris, 4 mM NaCl, 0.1 mM EDTA, pH 8.0). Plasmid DNA was digested with EcoRI and Hind III restriction enzymes (Sigma) according to the instructions of the manufacturer. The restriction enzyme digest plasmid DNA was analysed to evaluate the plasmid size following the protocol of Sambrook and Russell [21].
Antibiotic Sensitivity
The native and the plasmid-cured strains were tested for antibiotic sensitivity using antibiotic octodiscs (HiMedia product code OD 004, 023 and 026) to assess the antibiotic marker genes. Both the cured and native strains were grown in MRS broth for 15 h and inoculated in melted MRS soft agar as 1% and overlaid on MRS agar plate, octodiscs were placed on it and incubated at 28°C for 24 h and observed for zone of growth inhibition. Sensitivity and resistance were inferred by comparing the standard chart.
Results and Discussion
Isolation and Characterization of Bacteriocin Producing Strain
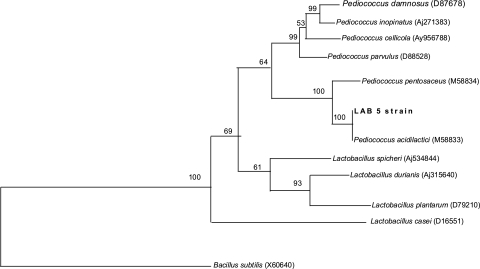
Bacteriocin producing strain of lactic acid bacteria was isolated on MRS agar plate against the sensitive indicators. The plates producing inhibition zone >5 mm around the colonies were picked up as antimicrobial compound producing organisms. Inhibition zone below that were ignored as these zones may be produced due to high amount of organic acid produced by the colonies. After screening a large number of colonies only one was selected as a potent antimicrobial-substance producer based on its inhibition zone size, which was greater than 9 mm. The isolated strain produced small white colonies on MRS agar medium. Broth cultures usually had uniform turbidity. The strain could grow at pH 7.0 and at 35°C, microaerophilic. It tolerated up to 10% of NaCl salt concentration. Scanning electron microscopic observation showed typical cocci in paired condition (Fig. 1) without any motile structure. Single cells or cells in chain were not found. Gram staining showed its positive nature and negative nature in malachite green staining showed its non-spore formation. Negative staining for slime was also found negative suggested for non-capsular structure. Hydrogen peroxide test produced no bubble formation hence it is inferred as catalase negative. Acid but no gas produced from glucose, fructose and mannose. Gelatin was not liquefied and nitrates not reduced to nitrites. It required complex medium having growth factors and amino acids for growth. From these observations it was inferred that the isolate is a species of Pediococcus [22]. Carbohydrate fermentation pattern of the isolate showed that it could ferment l-arabinose, ribose, galactose, glucose, fructose, N-acetyl glucosamine (nag), esculine, cellobiose, gentibiose, tagatose, trehalose, rhamnose, arbutin, salicin and d-xylose but unable to ferment amygdalin, raffinose, maltose, lactose, melibiose, saccharose and inositol (Table 2). It did not produce gas (CO2) during glucose fermentation, so the isolate was defined as homo-fermentative. The fermentation profile was compared with the standard fermentation chart of other lactic acid bacteria. From this fermentation profile, it is inferred that the isolated strain is P. acidilactici [23, 24]. A similar fermentation profile was found reported for P. pentosaceus which has the ability to ferment amygdalin, raffinose, maltose, lactose, melibiose, saccharose and inulin. The strain was designated as LAB 5. The partial 16S rDNA sequences of the strain were compared to sequences from type LAB strains held in GenBank. Phylogenetic tree analysis by the neighbor-joining method [15] confirmed that this bacteriocin producing organism was P. acidilactici as it showed 100% sequence similarity with P.acidilactici (M 58833) (Fig. 2). Bacillus subtilis (X60646) was used as an out-group organism. The isolated strain was designated as P. acidilactici LAB 5 (GenBank accession GQ240304) and the antimicrobial compound (bacteriocin) produced by this strain as Pediocin NV 5. Similar type of pediocin producer strains were also reported from other food sources like cucumber brines [10], sorghum beer [25], meat [26] and sausage [27].
Fig. 1.
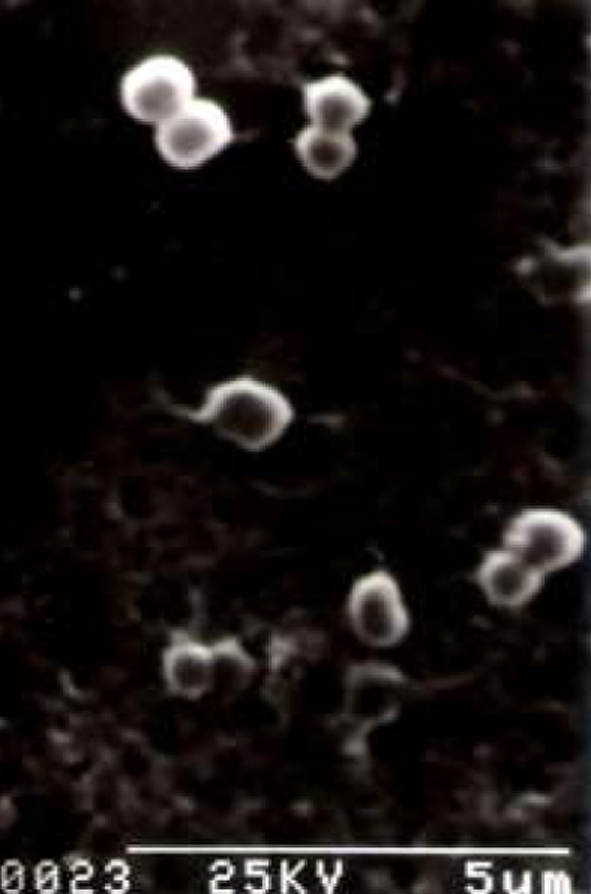
Scanning electron micrograph of the producer organism, P. acidilactici LAB 5, magnification 5k
Table 2.
Carbohydrate fermentation profile by Pediococcus acidilactici LAB 5 after 48 h incubation at 37°C
| Sugars used | Fermentation |
|---|---|
| Glycerol | − |
| Erythritol | − |
| d-Arabinose | − |
| l-Arabinose | + |
| Ribose | + |
| d-Xylose | + |
| l-Xylose | − |
| Adonitol | − |
| β-methyl xyloside | − |
| Galactose | + |
| d-Glucose | + |
| d-Fructose | + |
| d-Mannose | + |
| l-Sorbose | − |
| Rhamnose | + |
| Dulcitol | − |
| Inositol | − |
| Mannitol | − |
| Sorbitol | − |
| α Methyl-d-manoside | − |
| α Methyl-d-glucoside | − |
| N-Acetyl glucosamine | + |
| Amygdaline | − |
| Arbutine | + |
| Esculine | + |
| Salicine | + |
| Cellobiose | + |
| Maltose | − |
| Lactose | − |
| Melibiose | − |
| Saccharose | − |
| Trehalose | + |
| Inuline | + |
| Melezitose | + |
| d-Raffinose | − |
| Amidon | − |
| Glycogene | − |
| Xylitol | − |
| β Gentibiose | + |
| d-Turanose | − |
| d-Lyxose | − |
| d- Tagatose | + |
| d-Fuctose | − |
| l-Fucose | − |
| d-Arabitol | − |
| l-Arabitol | − |
| Gluconate | − |
| 2 Ceto-gluconate | − |
| 5 Ceto-gluconate | − |
+ able to, − unable to perform the functions
Fig. 2.
Phylogenetic tree showing the relative position of our strain LAB 5 as inferred by the neighbour-joining method of partial 16S rDNA sequences. Bootstrap values for a total of 100 replicates are shown at the nodes of the tree. Type strains used for comparison are given along with their accession number in brackets. B. subtilis is used as an out-group
Antimicrobial Assay and Spectrum
Following spot-on-lawn and deferred methods the antimicrobial activity produced by the organism was assayed against a number of pathogenic and non-pathogenic bacteria (Fig. 3) that are associated with food and feed systems or otherwise pathogenic to human beings. The antimicrobial compound, Pediocin NV 5 was found to inhibit strongly to the species of Enterococcus, Leuconostoc, Listeria and Staphylococcus (Table 1) but failed to inhibit other Gram-positive and Gram-negative bacteria. Although deviating from the properties of the bacteriocin of lactic acid bacteria, there are reports that bacteriocin from some lactic acid bacteria can inhibit Gram-negative bacteria [17] too, the present isolate produced bacteriocin which could inhibit only related Gram-positive bacteria. Partial purification of the bacteriocin following the adsorption–desorption method [16] showed quite satisfactory in terms of cost, time and recovery of the product. The bacteriocin activity enhanced in each step of purification from the starting activity 2,400 AU/ml to a final activity of 20,000 AU/ml [17].
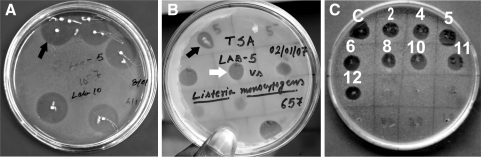
Fig. 3.
Growth inhibition by Pediocin NV 5 of P. acidilactici LAB 5 a against E. faecalis MB1 by overlay method on MRS agar plate, b against L. monocytogenes MTCC 657 by spot-on-lawn method and c inhibition of L. mesenteroides Ly by different dilutions of the boiled culture aliquot of P. acidilactici LAB 5 in spot-on-lawn method, the numbers indicate the fold of dilution
Partial Purification and Characterization of the Antimicrobial Substance
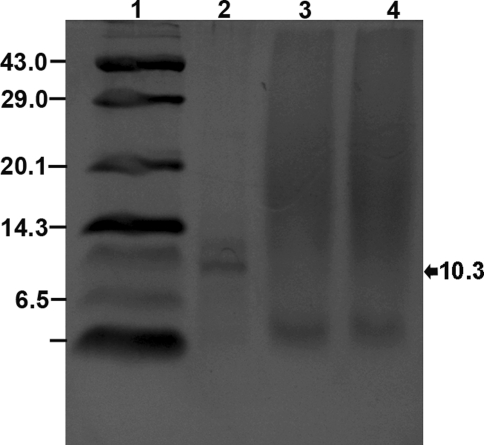
The antimicrobial compound produced by the organism was stable at a pH range from 2 to 8 with better activity in the range of 2 to 5. The loss of activity in basic pH might be due to the degradation of the molecule. Similar type of observation was reported for Pediocin PA-1 [28] and Nisin [29, 30] where probably the secondary structure is distorted at alkaline pH. It also retained its antimicrobial activity in autoclaving temperature, 121°C for 20 min [17]. Enzyme treatment of the antimicrobial compound showed loss of activity when treated with proteolytic enzymes, proteinase K but quite resistant to other enzymes like α-amylase and lysozyme. So this antimicrobial activity by the stain was due to secretion of an extra cellular proteinaceous compound as it lost its activity when treated with proteolytic enzymes. This compound was found to be almost pure protein as it had retained its activity in α-amylase, lysozyme and organic solvent treatment [17]. Failure to retain antimicrobial activity when treated with β-mercaptoethanol, a reducing agent, suggested that it contains cystine residues which are responsible for its secondary configuration [31, 32] with antimicrobial activity. So, from the above point of findings it can be concluded that the antimicrobial compound produced by the strain is bacteriocin and was designated as Pediocin NV 5. Tricine SDS-PAGE analysis of partially purified bacteriocin showed its approximate molecular weight of 10.3 kDa (Fig. 4).
Fig. 4.
4 18% Tricine SDS-PAGE of partially purified bacteriocin Pediocin NV 5; Lane 1 Molecular wt. marker (kDa) of low range (Genei, India) 43: Ovalbumin; 29: Carbonic anhydrase; 20.1: Soyabean trypsin inhibitor; 14.3: Lysozyme; 6.5: Aprotinin; 3: Insulin; Lane 2 lyophilized bacteriocin pediocin NV 5; Lane 3: culture aliquot of TGE + Tween 80 + buffer; Lane 4 blank media
Plasmid Curing and Antibiotic Markers
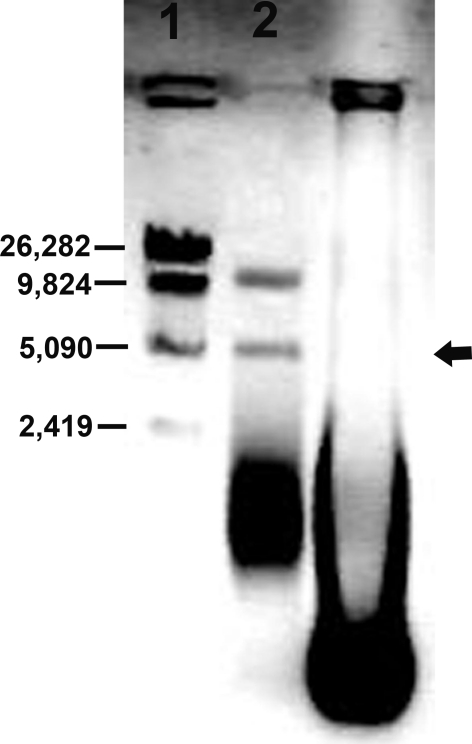
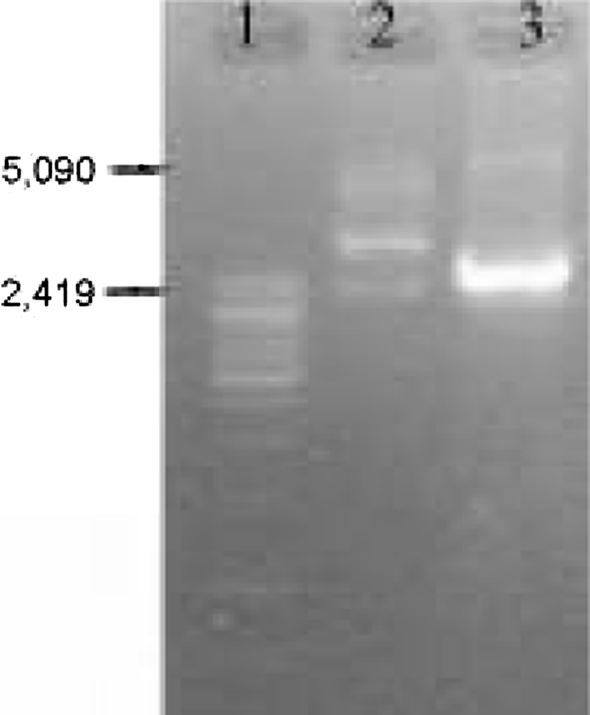
Plasmid curing with ascorbic acid resulted in producing bacteriocin negative mutant (Fig. 5). It was a good plasmid curing agent when sub-cultured for one or two times but not for repeated i.e. six to seven times. More sub-culturing with ascorbic acid decreased the percent of cured strain. The ascorbic acid concentration at 0.5–1.0 mM generated maximum number of mutants. It was found that the cured strain produced colonies with small size whereas the native strain having larger one. This indicated that the plasmid bears some factors of multiplication. The size of the plasmid is approximately 5 kbp (Fig. 6) when tested on 0.6% agarose gel. From the figure it is evident that the cured strain lost the plasmid DNA of 5 kbp which is involved for bacteriocin production. Restriction enzyme digestion of plasmid DNA with EcoRI showed two bands with approx. molecular wt 2.8 and 2.2 kbp whereas HindIII digestion showed one band of approx. molecular wt 2.5 kbp (Fig. 7). The restriction enzyme analysis also showed its 5 kbp size. This plasmid DNA is quite dissimilar in size than other P. acidilactici strains like P. acidilactici CFR K7 which have 7.8 kbp plasmid [19], P. acidilactici H (7.4 MDa) [33], and P. cerevisiae FBB63 (10.5 MDa) [34] and P. pentosaceus FBB61 and L7230 possessed a 13.6-megadalton plasmid (pMD136) [11].
Fig. 5.
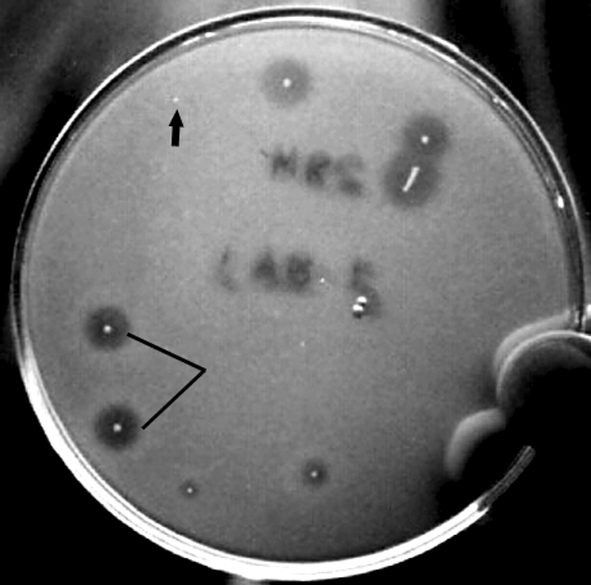
The loss of Pediocin NV 5 production of plasmid cured mutant resulting loss of inhibition zone areas against indicator E. faecalis MB1on MRS agar plate by deferred method (arrow marked is the mutant colony, dash line is the native colony)
Fig. 6.
0.6% Agarose gel of plasmid DNA; Lane 1 Marker DNA ladder of λ Mlu digest, Lane 2 RNase treated plasmid DNA of wild type (LAB 5+) P. acidilactici LAB 5, top band of nicked DNA, middle of coiled circular plasmid DNA (arrow marked) and lower band of supercoiled plasmid DNA, Lane 3 RNase untreated mutant type (LAB 5−) P. acidilactici LAB 5, thick band of total RNA
Fig. 7.
Plasmid profile of P. acidilactici LAB 5 in 1% agarose gel upon restriction digestion; Lane 1 Marker DNA ladder of λ Mlu digest, Lane 2EcoRI digest; Lane 3HindIII digest
Both the native and the cured strains are sensitive to ampicillin (10 µg), cephalothin (30 µg), cephalexin (30 µg), cephaloridine (30 µg), chloramphenicol (30 µg), clindamycin (2 µg), erythromycin (15 µg), gentamicin (10 µg), kanamycin 30 µg), lincomycin (2 µg), methicillin (5 µg), norfloxacin (10 µg), oleandomycin (15 µg), oxacillin (1 µg), penicillin (2 units), penicillin V (3 µg), penicillin G (10 units), tetracycline (10 µg) and tobramycin (10 µg) antibiotics but the mutant strain in contrast to native strain is sensitive to amoxycillin (10 µg), cloxacillin (5 µg) and vancomycin (30 µg) and both the native and cured mutant strains are resistant to co-trimoxazole (25 µg). From this observation it is inferred that amoxycillin, cloxacillin and vancomycin resistant genes are located in the plasmid DNA whereas co-trimoxazole resistant gene is located in the genomic DNA. Pediococcus spp. are generally vancomycin resistant [35]. The present isolate also showed the same character. Similarity in the vancomycin resistance predicts its common ancestry with other species of Pediococcus. These antibiotic markers will help in performing transformation experiment and the suitability of the strain to use as dietary adjunct.
There is an increasing demand in controlling and combating the highly pathogenic and hazardous food pathogenic organisms like Listeria monocytogenes, L. mesenteroides, Enterococcus etc. associated with fermented milk and meat products with bio-preservatives. Pediocin NV 5 had proved its strong antimicrobial efficacy against L. mesenteroides, E. faecalis and L. monocytogenes. As this bacteriocin molecule is acidic pH resistant and stable at high temperature (121°C) so this Pediocin NV 5 opened the scope for its use in ready to eat fermented food products and the strain P. acidilactici LAB 5 and its antimicrobial products may also be applied in starter culture and bio-preservative in a variety of fermented foods.
Acknowledgments
We are grateful to Prof. Bibek Ray, University of Wyoming University, USA for providing the indicator strains. We also acknowledge the scientists of IMTECH, Chandigarh, India for partial 16S rDNA analysis.
Contributor Information
Vivekananda Mandal, Phone: +091-354-2254316, Phone: +091- 9434556340, Email: mandal_vivek@yahoo.co.in.
Narayan Chandra Mandal, Phone: +091-03463-262751, Phone: +091- 03463265033, Email: mandalnc@rediffmail.com.
References
- 1.Lindgren SW, Dobrogosz WJ. Antagonistic activities of lactic acid bacteria in food and feed fermentation. FEMS Microbiol Rev. 1990;87:149–154. doi: 10.1111/j.1574-6968.1990.tb04885.x. [DOI] [PubMed] [Google Scholar]
- 2.Tagg JR, Dajani AS, Wannamaker LW. Bacteriocins of Gram-positive bacteria. Bacteriol Rev. 1976;40:722–756. doi: 10.1128/br.40.3.722-756.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baerfoot SF, Klaenhammer TR. Detection and activity of lactacin B, a bacteriocin produced by Lactobacillus acidophilus. Appl Environ Microbiol. 1983;45:1808–1815. doi: 10.1128/aem.45.6.1808-1815.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brink B, Minekns M, Vander Vossen JMBM, Leer RJ, Huisin’t Veld JHJ. Antimicrobial activity of lactobacilli. J Appl Bacteriol. 1994;77:140–145. doi: 10.1111/j.1365-2672.1994.tb03057.x. [DOI] [PubMed] [Google Scholar]
- 5.Hugenholz J, Veer GJCM. Application of nisin A and Z in dairy technology. In: Jung G, Sahl HG, editors. Nisin and novel lantibiotics. Leiden: Escom Publishers; 1991. pp. 404–421. [Google Scholar]
- 6.Ray B, Miller KW. Pediocin. In: Naidu AS, editor. Natural food antimicrobial systems. Boca Raton: CRC Press Inc.; 2000. pp. 525–566. [Google Scholar]
- 7.Moliter E, Sahl HG. Application of nisin: a literature survey. In: Jing H, Sahl HG, editors. Nisin and novel lantibiotics. Leiden: Escom Publishers; 1991. pp. 434–441. [Google Scholar]
- 8.Bhunia AK, Johnson MC, Ray B. Purification, characterization and antimicrobial spectrum of a bacteriocin produced by Pediococcus acidilactici. J Appl Bacteriol. 1988;65:261–268. doi: 10.1111/j.1365-2672.1988.tb01893.x. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez CF, Kunka BS. Plasmid associated bacteriocin production and sucrose fermentation in Pediococcus acidilactici. Appl Environ Microbiol. 1987;53:2534–2538. doi: 10.1128/aem.53.10.2534-2538.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flemming HP, Etchells JL, Costilow RN. Microbial inhibition by an isolate of Pediococcus from cucumber brines. Appl Microbiol. 1975;30:1040–1042. doi: 10.1128/am.30.6.1040-1042.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daeschel M, Klaenhammer TR. Association of a 13.6 megadalton plasmid in Pediococcus pentosaceus with bacteriocin activity. Appl Environ Microbiol. 1985;50:1538–1541. doi: 10.1128/aem.50.6.1538-1541.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ko Hyun S, Ahn C. Bacteriocin production by Lactococcuslactis KCA 2386 isolated from white kimchi. Food Sci Biotechnol. 2000;9:263–269. [Google Scholar]
- 13.Man JC, Rogosa M, Sharpe ME. A medium for the cultivation of lactobacilli. J Appl Bacteriol. 1960;23:130–135. [Google Scholar]
- 14.Kozaki M, Uchimura T, Okada S. Experimental manual of lactic acid bacteria. Tokyo: Asakurasyoten; 1992. pp. 34–37. [Google Scholar]
- 15.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 16.Yang R, Johnson MC, Ray B. Novel method to extract large amount of bacteriocins from lactic acid bacteria. Appl Environ Microbiol. 1992;58:3355–3359. doi: 10.1128/aem.58.10.3355-3359.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandal V, Sen SK, Mandal NC. Optimized culture conditions for bacteriocin production by Pediococcus acidilactici LAB 5 and its characterization. Indian J Biochem Biophys. 2008;45:106–110. [PubMed] [Google Scholar]
- 18.Schägger H, Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 19.Ramesh A, Halami PM, Chandrashekar A. Ascorbic acid induced loss of a pediocin encoding plasmid in P. acidilactici CFR K7. World J Microbiol Biotechnol. 2000;16:695–697. doi: 10.1023/A:1008958517001. [DOI] [Google Scholar]
- 20.Anderson DG, McKay L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Russell D. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- 22.Pederson CS. The genus Pediococcus. Bacteriol Rev. 1949;13:225–232. doi: 10.1128/br.13.4.225-232.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buchanan RE, Gibbons NE. Bergey’s manual of determinative bacteriology. 8. Baltimore: The Williams and Wilkins Company; 1975. pp. 513–515. [Google Scholar]
- 24.Schleifer KH. Gram-positive cocci. In: Sneath PHA, Mair NS, Holt JG, editors. Bergey’s manual of systematic bacteriology. Baltimore: Williams and Wilkins; 1986. pp. 1075–1079. [Google Scholar]
- 25.Chikindas ML, Garcia-Garcera MJ, Driessen AJ, Ledeboer AM, Nissen-Meyer J, Nes IF, Abee T, Konings WN, Venema G. Pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0, forms hydrophilic pores in the cytoplasmic membrane of target cells. Appl Environ Microbiol. 1993;59:3577–3584. doi: 10.1128/aem.59.11.3577-3584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schved F, Lalazar A, Henis Y, Juven BJ. Purification, partial characterization and plasmid-linkage of pediocin SJ-1, a bacteriocin produced by Pediococcus acidilactici. J Appl Bacteriol. 1993;74:67–77. doi: 10.1111/j.1365-2672.1993.tb02998.x. [DOI] [PubMed] [Google Scholar]
- 27.Osmanagaoglu Ö, Gündüz U, Beyatli Y, Çökmüs C. Purification and characterization of Pediocin F, a bacteriocin produced by Pediococcus acidilactici F. Turkish J Biol. 1998;22:217–228. [Google Scholar]
- 28.FimLand G, Jack R, Jung G, Nes IF, Meyer JN. The bactericidal activity of Pediocin PA-1 is specifically inhibited by a 15-mer fragments that spans the bacteriocin from the center the C terminus. Appl Environ Microbiol. 1998;64:5057–5060. doi: 10.1128/aem.64.12.5057-5060.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gross E, Morell JL. The structure of nisin. J Am Chem Soc. 1971;93:4634–4635. doi: 10.1021/ja00747a073. [DOI] [PubMed] [Google Scholar]
- 30.Hurst A. Nisin. Adv Appl Microbiol. 1981;27:85–123. doi: 10.1016/S0065-2164(08)70342-3. [DOI] [Google Scholar]
- 31.Metivier A, Pilet MF, Dousset X, Sorokine O, Anglade P, Zagorec M, Piard JCH, Marion D, Cenatempo Y, Fremaux C. Divercin V 41, a new bacteriocin with two disulphide bonds produced by Carnobacterium divergens V 41: primary structure and genomic organization. Microbiology. 1998;144:2837–2844. doi: 10.1099/00221287-144-10-2837. [DOI] [PubMed] [Google Scholar]
- 32.Fimland G, Johnsen L, Axelsson L, Brurberg MB, Nes IF, Eijsink VGH, Nissen-Meyer J. A C-terminal disulfide bridge in pediocin-like bacteriocins renders bacteriocin activity less temperature dependent and is a major determinant of the antimicrobial spectrum. J Bacteriol. 2000;182:2643–2648. doi: 10.1128/JB.182.9.2643-2648.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ray SK, Kim WJ, Johnson MC, Ray B. Conjugal transfer of a plasmid encoding bacteriocin production and immunity in Pediococcus acidilactici H. J Appl Bacteriol. 1989;66:393–399. [Google Scholar]
- 34.Graham DC, Mckay LL. Plasmid DNA in strains of Pediococcus cerevisiae and Pediococcus pentosaceus. Appl Environ Microbiol. 1985;50:532–534. doi: 10.1128/aem.50.2.532-534.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Facklam R, Hollis D, Collins MD. Identification of Gram-positive coccal and coccobacillary vancomycin-resistant bacteria. J Clin Microbiol. 1989;27:724–730. doi: 10.1128/jcm.27.4.724-730.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]