Abstract
A combination of cultivation-based methods with a molecular biological approach was employed to investigate whether bacteria with identical 16S rRNA gene sequences can represent distinct eco- and genotypes. A set of eight bacterial strains wherein three were Pseudomonas putida and rest were Acinetobacter calcoaceticus, were isolated from casing soils community by conventional plating. These strains had identical 16S rRNA gene sequences and represented the dominant phylotype in the plateable fraction. Each strain utilized a specific combination of 154 carbon substrates, and the niche overlap indices were low, suggesting that each strain occupied a different ecological niche. Our results have implications for assessment of the diversity and biogeography of bacteria and increase the perception of natural diversity beyond the level of 16S rRNA gene sequences. It is worthwhile approach to explore prokaryotic diversity in different ecological niches.
Keywords: Microdiversity, Ecophysiology, 16S rRNA sequencing, Pseudomonas putida, Acinetobacter calcoaceticus
Introduction
Mumbo-Jumbo, that is how Thomas Brock [1] described the study of diversity in his pithy and prescient essay on the state of microbial ecology in 1987. Brock argued that measures of diversity were pointless because the dynamic nature of the microbial world meant that communities did not have a characteristic diversity but changed as the environment changed. Wilson [2] reinforced the futility of the study of microbial diversity when he observed that microbial diversity is ‘beyond practical calculation’. The rationale for low prokaryotic diversity is that microbial organisms are so abundant, that free-living representatives can be easily globally dispersed. Consequently allopatric speciation is discouraged in microbial communities by very high invasion rates [3]. Curtis et al. [4] developed a rough and ready universal prokaryotic diversity estimator. They used published data on rRNA gene based clone libraries and found that there could be 7,000 species in a gram of soil. Thus, a ton of soil could theoretically hold 3 million different taxa. The relationship between structure and function in a community can only be understood, predicted and engineered through an understanding of the source of diversity from which the community is drawn [5].
Analysis of 16S rRNA (ribosomal RNA) gene sequences has become the primary approach for studying the natural occurrence and distribution of bacteria in a culture independent manner [6]. The vertical and seasonal distributions of distinct 16S rRNA gene sequences (phylotypes) within one ecosystem have been used to infer the ecological niches of bacteria [7, 8]. This approach is especially valuable if the physiology of bacteria that have not been cultured yet is to be elucidated.
In many cases phylogenetically closely related bacteria (whose 16S rRNA sequences differ by between 2.7 and 0.3%) have been detected in the same fresh water, marine, or soil habitat [8]. According to macroecological principles of competitive exclusion, physiologically similar micro organisms should not co-occur in nutrient poor systems which are dominated by physical and chemical fluctuations [7]. Accordingly, phylogenetically closely related bacteria coexisting in the same habitat occupy distinct ecological niches which make up the microdiversity [8–11]. For pathogenic bacteria it is well established that even phylogenetically identical strains or species can exhibit distinct ecophysiological properties. Certain serovars of Mycobacterium intracellulare [12], serovars of Ochrobactrum anthropi [13], strains of Yersinia pestis, Yerinia pseudo tuberculosis [14], or strains of Bacillus anthracis, Bacillus cereus [15], contain identical 16S rRNA gene sequences. These phylogenetically identical organisms are also genetically highly similar based on DNA-DNA hybridization data but clearly represent different ecotypes based on their virulence properties or host ranges. Often, phenotypic differences can be traced back to the presence of plasmids, as in B. anthracis, in which the major virulence determinants are encoded by the 181-kb plasmid pX01 and the 95-kb plasmid pX02 not present in B. cereus [16].
In the present study, eight bacterial strains were selected wherein three were Pseudomonas putida and rest five strains were Acinetobacter calcoaceticus with identical 16S rRNA gene sequences. All bacterial strains were isolated from mushroom casing soil (an integrated part of mushroom compost ecosystem and used for cultivation of button mushroom). The strains were subsequently analyzed with respect to their genomic and physiological diversities, and their ecological niches.
Materials and Methods
Recovery of Bacterial Strains
Casing sample (10 g) was suspended in 90 ml of 0.85% normal saline (pH 7.0) and shaken vigorously at 150 rpm at 18°C for 1 h. The resulting slurry was serially diluted (100 μl) to 900 μl of 0.85% normal saline in each Eppendorf tube and appropriate dilution (10−4) of this suspension (100 μl) was spread plated in triplicate, on King’s B medium [17]. Cultures were incubated at 20°C ± 2 for 2 days. After incubation they were restreaked until pure cultures were obtained. The purity of all strains isolated was examined microscopically. For experimental use, isolates were transferred when needed to King’s B medium that was stored at 4°C.
DNA Extraction
For total genomic DNA extraction [18] 1.5 ml culture was centrifuged at 12,000 rpm for 10 min at 4°C; the pellet was washed with 1.5 ml of TRIS–Cl (0.1 M, pH 6.8) twice and centrifuged. Cells were lysed with a combination of 0.5% SDS and 0.001% proteinase K followed by treatment with 1% CTAB and washed with phenol: chloroform (1:1) and chloroform: isoamyl alcohol (24:1). DNA was precipitated with absolute alcohol at −20°C overnight followed by pelleting and washing with 70% ethanol. After RNAse treatment, pellet was checked for presence of DNA on 0.8% agarose gel run in TBE buffer at 70 V for 45 min. Gel was stained with ethidium bromide and visualized under UV on a Gel Doc Mega System, Biosystematica.
Quantification and Detection of Purity of Extracted DNA
Extracted genomic DNA was run in 0.8% agarose gel at 80 V for 45 min with quantitative marker in one lane (Low DNA Mass Ladder, MBI Fermentas). Gel was visualized under UV transilluminator.DNA was quantified spectrophotometrically, by measuring OD at 260 and 280 nm. Purity of DNA was checked measuring the extinction at A260/A280, on a DU 640 B Beckman spectrophotometer. The concentration of DNA was calculated as: [DNA] = A260 × 100 × Dilution factor (μg ml−1).
PCR Amplification of 16S rDNA
The amplified 16S rRNA gene was obtained from each bacterial isolate by PCR amplification employing the eubacterial universal primers [19] fDI (5′-AGAGTTTGATCCTGG-3′) and rP2 (5′-TACCTTGTTACGACTT-3′) which were targeted at universally conserved regions and permitted amplification of approximately 1,500-bp fragment. PCR amplification was carried out in a PTC-100 thermocycler (M.J. Research). Reaction tubes contained 25 ng (5 μl) of DNA extract, 1 U of Taq polymerase (Genei), 1× buffer (10 mM Tris–Chloride [pH 9.0], 1.5 mM MgCl2, 500 mM KCl) (Genei), 10 mM dNTPs (Genei) and 0.25 mM of each primer (Genei). Initial DNA-denaturation and enzyme activation steps were performed at 95°C for 7 min, followed by 25 cycles of denaturation at 94°C for 1 min, annealing at 51°C for 1 min and extension at 72°C for 1 min, and a final extraction at 72°C for 10 min. The presence and yield of specific PCR product (16S rRNA gene) was monitored on 0.8% agarose (wt/vol.) (Life Technologies Inc.); gel electrophoresis was carried out at 100 V for 30 min in 1× Tris–acetate–EDTA buffer and visualization by ethidium bromide staining and viewing on a UV transilluminator (Biosystematica).
Partial Sequencing of the 16S rDNA
PCR products obtained from bacterial strains were purified with an EXO-SAP. Components were supplemented with gold buffer (Applied Biosystem) and sequenced on an Applied Biosystem 310 Genetic analyzer, using big dye terminator cycle sequencing Ready Kit (Lab India). The partial sequences amplified by the fDI primer were used to determine the similarities.
Nucleotide Sequence Accession Numbers
The partial 16S rRNA gene sequences of strains determined in this study have been deposited in the GenBank database of NCBI under accession numbers (parentheses): UVC 2 (AY 961043), CVC 2 (AY 967724), USC 31 (DQ 074752), UVC 4 (AY 961045), UVC 3 (AY 961044), UVC 8 (AY 961047), USC 29 (AY 961061) and USC 30 (DQ 074751).
Sole Source Carbon Utilization (SSCU)
The bacterial strains were tested for their ability to catabolize 154 different compounds (see Table 1) as sole carbon sources at a concentration of 5 mM each. Bacterial strains were cultured on KB for 18 h at 20°C. Bacterial cells were scraped from the plate and suspended in phosphate buffer (0.01 M, pH 7.0). The cell suspensions were adjusted to optical density at 560 nm, 0.12. For growth tests, each microtitre well received triplicate 180 μl of minimal medium (M9) and was inoculated with 15 μl of a bacterial suspension, and 10 μl of redox dye, triphenyl-tetrazolium chloride (TTC). Control well was devoid of bacterial suspension. The plates were incubated for 5 days at 20°C, and growth was monitored by determining the colour change of dye from colourless to red.
Table 1.
Compound tested for eight strains belonged to Pseudomonas putida and Acinetobacter calcoaceticus, for their ability to supports growth of the bacterial strains on sole source carbon utilization (SSCU) when incorporated into minimal medium
| Type | Compounds |
|---|---|
| Amino acids | d-Alanine, l-alanine, l-alanylglycine, l-arginine, l-aspartic acid, l-asparagine, citrulline, l-cysteine, l-glutamic acid, l-glutamine, l-glycine, glycyl-l-aspartic acid, glycyl- l-glutamic acid, l-histidine, l-homoserine, hydroxy-l-proline, l-isoleucine, l-leucine, l-lysine, l-methionine, l-phenylalanine, l-proline, pyro-glutamic acid, d-serine, l-serine, threonine, tryptophan, valine, norvaline |
| Organic acids | Adipic acid, acetic acid, aconitic acid, anthranilic acid, α-aminobutyric acid, γ-aminobutyric acid, citraconic acid, citric acid, folic acid, formic acid, fumaric acid, galacturonic acid, galacturonic acid lactone, polygalacturonic acid, gentisic acid, gluconic acid, glucuronic acid, glucosaminic acid, glutaric acid, glyceric acid, glycolic acid, α-hydroxybutyric acid, β-hydroxybutyric acid, γ-hydroxybutyric acid, hyroxyphenylacetic acid, itaconic acid, α-ketobutyric acid, α-ketoglutaric acid, α-ketovaleric acid, lactic acid, malic acid, maleic acid, malonic acid, methylpyruvic acid, monomethylsuccinic acid, mucic acid, nicotinic acid, para-aminobenzoic acid, pantothenic acid, pimelic acid, pipe-colic acid, propionic acid, pyruvic acid, quinic acid, saccharic acid, salicylic acid, sebacic add, shikimic acid, succinic acid, bromosuccinic acid, succinamic acid, tartaric acid, urocanic acid |
| Carbohydrates | Amylose, arabinose, cellobiose, cyclodextrin, dextran, dextrin, fructose, fucose, furanose, galactose, gentiobiose, glucose, glucose 1-phosphate, glucose 6-phosphate, glycogen, lactose, lactulose, maltose, mannose, melezitose, melibiose, palatinose, psicose, raffinose, rhamnose, ribose, sorbose, starch, sucrose, trehalose, xylose |
| Sugar alcohols | Adonitol, arabitol, dulcitol, erythritol, inositol, mannitol, sorbitol, xylitol |
| Amides and amines | Acetamide, N-acetyl-d-glucosamine, N-acetyl-d-galactosamine, alaninamide, glucuronamide, phenylethylamine |
| Alcohols | 2,3-Butanediol, ethanol, 2-aminoethanol, dl-α-glycerolphosphate, glycerol, methanol |
| Fatty acids | Capric acid, caproic acid, caprylic acid, lauric acid, lauryl sulfate, levulinic acid, myristic acid |
| Miscellaneous | Betaine, dl-carnitine, choline chloride, inosine, β-methylglucoside, ornithine, putrescine, salicin, sarcosine, thymidine, Tween 40, Tween 80 |
Niche Overlap Index (NOI)
The NOI was defined in this study as the number of carbon sources utilized by both strains as a proportion of the total number of carbon sources utilized by the strain in question (Ntot): NOI = NA∩B/Ntot [20].
Physiological Similarity
The physiological similarity of the eight bacterial strains was determined by cluster analysis. A matrix with a binary code for the presence or absence of each phenotypic trait of the isolate was constructed. The Jaccard coefficient for all pairs of strains was calculated employing the SIMQUAL similarity programme for qualitative data of the NTSYS-pc numerical taxonomy computer package [21]. Cluster analysis was performed with the SAHN program of the NTSYS-pc package and by employing the unweighted pair group method with arithmetic average (UPGMA).
Results
Colour Formation in Microtiter Plates
Development of redox sensitive dyes such as TTC and incorporation of these dyes into microtiter plates has allowed for rapid profiling of sole source carbon utilization by bacterial isolates [22]. Colour formation in microplate wells in SSCU is based on the conversion of the redox-sensitive tetrazolium dye which is reduced during respiratory activity, and accumulates as insoluble formazan inside active cells. No colour development was observed in the control well.
Ecological Similarity and Co-Existence
Strains were morphotypically quite similar and they were Pseudomonas putida and Acinetobacter calcoaceticus species based on partial 16S rRNA gene sequencing. Consequently, their potential ecological niches were assessed based on their carbon substrate utilization patterns.
Each strain used a unique combination of the 154 carbon substrates. The lowest metabolic diversity was observed for strain USC 31 (P. putida) isolated from FYM + SC (3:1) whereas it was higher for two strains of P. putida viz, UVC 2 and CVC 2. The higher NOI calculated for all pairs of P. putida strains indicated that they occupy different ecological niches i.e. cannot coexist. Similarly, NOI for all pairs of A. calcoaceticus strains was calculated (Table 2).
Table 2.
NOIs for casing soil bacterial strains, derived from carbon source utilization
| Strainsa | Total compounds utilized | Amino acids | Organic acids | Carbohydrates |
|---|---|---|---|---|
| P. putida | ||||
| USC 31 | 1.000 | 0.889 | 0.870 | 1.000 |
| UVC 2 | 0.988 | 0.960 | 1.000 | 0.875 |
| CVC 2 | 0.945 | 0.942 | 0.933 | 1.000 |
| A. calcoaceticus | ||||
| UVC 3 | 0.904 | 1.000 | 1.000 | 0.962 |
| UVC 4 | 0.954 | 0.989 | 0.900 | 1.000 |
| UVC 8 | 0.908 | 1.000 | 1.000 | 0.915 |
| USC 29 | 0.877 | 1.000 | 0.983 | 1.000 |
| USC 30 | 0.965 | 0.988 | 1.000 | 1.000 |
aNOIs for pair of strains
Compositional Similarity
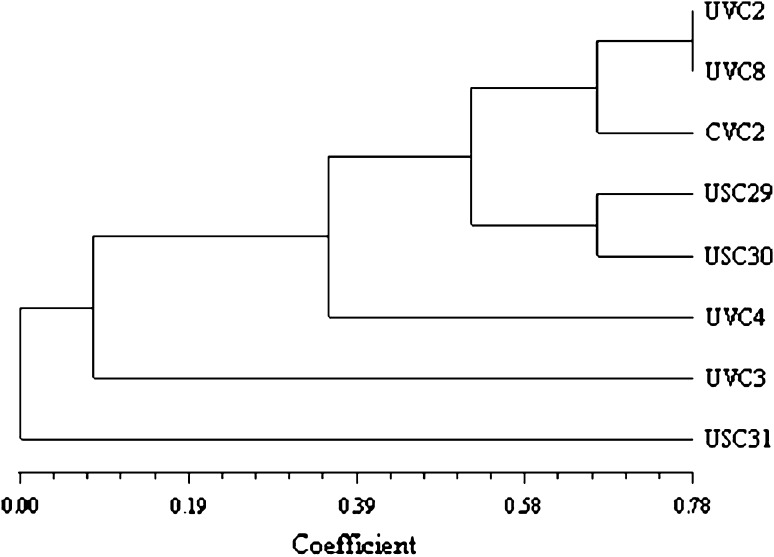
Measure of niche overlap index (NOI) does not provide information about the types of substrates that are utilized by the bacterial strains. Four strains showed identical NOI but still catabolized different substrates. Beside NOI, a consistent relationship among strains was made by cluster analysis, based on the presence or absence of substrate utilized (Fig. 1). Two strains UVC 2 and UVC 8 showed 78% similarity and they had 65% similarity with CVC 2 whereas USC 29 and USC 30 showed 65% similarity with each other. The least relationship was observed for UVC 4, UVC 3 and USC 31 with each other. Strain USC 31 was entirely different i.e. different and had least no of substrates utilization by USC 31 while UVC 3 utilized quite more no of C-sources. They showed 15% similarity with UVC 4 and UVC 4 had 40% similarity with each other.
Fig. 1.
Physiological similarity of strains of the species Pseudomonas putida and Acinetobacter calcoaceticus, assessed by cluster analysis based on the substrate utilization patterns employing Jaccard coefficient
Discussion
It is important to study microbial diversity not only for basic scientific research, but also to understand the link between diversity and community structure and function. Although methods to study diversity (numerical, taxonomic, and structural) are improving for both bacteria and fungi, there is still not a clear association between diversity and function. Even if an organism is functionally redundant in one function, chances are it is not redundant in all functions and will have different susceptibilities and tolerances to abiotic and biotic stresses. It is generally thought that a diverse population of organisms will be more resilient to stress and more capable of adapting to environmental changes [23].
The Pseudomonas and Acinetobacter sp. strains investigated in the present study were retrieved from the casing soils. Therefore, a hitherto unknown multitude of ecotypes must thrive in the same habitat. Based on our results, it has to be concluded that the extent of genomic and physiological diversity masked by identical 16S rRNA sequences is much larger than has been assumed previously and that this so-called microdiversity has ecological relevance. If a bacterial species is defined as a “monophyletic and genomically coherent cluster of individual bacteria that show a high degree of overall similarity in many independent characteristics”, then bacterial diversity may indeed exceed present estimates by several order of magnitude, as previously suggested [24, 25]. Wilson and Lindow [20] calculated niche overlap index for ice-nucleating (Ice+) P. syringae strain with respect to non-ice-nucleating (Ice−) P. syringae strain TLP2 del. It was uniformly high indicating that they were ecologically similar but had low level of coexistence. These authors reported that in the phyllosphere resource partitioning among different bacterial species with NOI values of 0.25–0.59 allowed stable coexistence, whereas catabolically identical strains (NOI 1.0), even if they belong to different species, cannot coexist.
Jaspers and Overmann [26] used a combination of cultivation based methods with nine molecular biological approaches to investigate whether planktonic bacteria with identical 16S rRNA gene sequences can represent distinct eco and genotypes. They isolated 11 strains of Brevundimonas alba from a freshwater community by employing conventional plating and MPN dilution series. All the 11 strains had identical 16S rRNA gene sequences and each strain utilized a specific combination of 59 C-substrates and the NOI were low which suggested that each strain occupied different ecological niche.
The genomes of certain phylogenetically identical strains exhibit profound differences. Escherichia coli K-12 and 0157:H7 differ not only in genome size (by 0.89 Mb) but also in a considerable number of chromosomal genes. Twenty-five percent of the genes present in the enterohemorrhagic organism E.coli 0157:H7 are not found in the nonpathogenic organism E. coli K-12, whereas 12% of the genes in the latter organism are absent in the former organism [27]. Nevertheless, some of the 16S rRNA gene sequences (e.g., the two rrsE genes) are identical in the two organisms. Similarly, genomic fingerprinting [28–30] and analysis of fosmid libraries of DNA fragments from marine samples [31] have indicated that nonpathogenic bacteria with identical 16S rRNA gene sequences but distinctly different genomes coexist in natural ecosystems [24]. The term micro diversity has been used to describe the phenomenon of phylogenetically closely related but physiologically distinct bacterial populations [9]. In order to assess the extent of microdiversity present in a natural habitat, the niche separation between the different genotypes with identical 16S-rRNA genes, and finally the potential limitations of 16S-rRNA-based methods, more information about the genetic and ecophysiological differences of such bacteria is required.
Acknowledgments
This work was performed at Department of Biotechnology, BU, Bhopal with kind cooperation of Dr. Anil Prakash, Head, Department of Biotechnology & Bioinformatics Centre, Bhopal.
Footnotes
The Editor-in-Chief has retracted this article because of significant overlap with a previously published article by Jaspers and Overmann. All authors agree to this retraction.
Change history
3/21/2019
The Editor-in-Chief has retracted this article [1] because of significant overlap with a previously published article by Jaspers and Overmann [2]. All authors agree to this retraction.
Change history
3/21/2019
The Editor-in-Chief has retracted this article [1] because of significant overlap with a previously published article by Jaspers and Overmann [2]. All authors agree to this retraction.
References
- 1.Brock T. The study of microorganisms in situ: progress and problems. In: Gray TRG, Jones JG, editors. Ecology of microbial communities (Fletcher M. Cambridge: Cambridge University Press; 1987. [Google Scholar]
- 2.Wilson EO (1994) The diversity of life. Penguin, London
- 3.Finlay BJ. Global dispersal of free living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 4.Curtis TP, Sloan WT, Scanneu J. Estimating prokaryotic diversity its limits. Proc Natl Acad Sci USA. 2002;99:10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtis TP, Sloan WT. Prokaryotic diversity its limits: microbial community structure in nature implications for microbial ecology. Curr Opin Microbiol. 2004;7:221–226. doi: 10.1016/j.mib.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Amann R, Ludurg W, Schleifer KH. Phylogentic indentification insitu detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pernthaler J, Glockner FO, Unterholzner S, Altreider A, Psenner R, Amann R. Seasonal community population dynamics of pelagic bacteria and archaea in a high mountain lake. Appl Environ Microbiol. 1998;60:4299–4306. doi: 10.1128/aem.64.11.4299-4306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward DM, Ferris MJ, Nold SC, Bateson MM. A natural view of microbial biodiversity within not spring cyanobacterial mat communities. Microbial Mol Biol Rev. 1998;62:1353–1370. doi: 10.1128/mmbr.62.4.1353-1370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore LR, Rocap G, Chisholm SW. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 10.Postius C, Ernst A. Mechanism of dominance: coexistence of picocyanobacterial genotypes in a freshwater ecosystem. Arch Microbiol. 1999;172:69–75. doi: 10.1007/s002030050742. [DOI] [PubMed] [Google Scholar]
- 11.Gray ND, Head IN. Linking genetic identities function in communities of uncultured bacteria. Environ Microbiol. 2001;3:481–492. doi: 10.1046/j.1462-2920.2001.00214.x. [DOI] [PubMed] [Google Scholar]
- 12.Böddinghaus B, Wolters J, Heikens W, Bottger EC. Phyhogenetic analysis and identification of deferent serovars of Mycobacterium intracellulare at the molecular level. FEMS Microbiol Lett. 1990;70:197–204. doi: 10.1016/S0378-1097(05)80039-4. [DOI] [PubMed] [Google Scholar]
- 13.Lebuhn M, Achourak W, Schloter M, Berge O, Meier H, Hartmann A, Heulin T. Taxonomic characterization of Ochrobactrum sp. isolated from soil samples and wheat roots, and description of Ochrobactrum tritici sp. nov. and Ochrobactrum grignunense sp. nov. Int J Syst Evol Microbiol. 2000;50:2207–2223. doi: 10.1099/00207713-50-6-2207. [DOI] [PubMed] [Google Scholar]
- 14.Trebesius K, Harmsen D, Rakin A, Schmelz J, Heesemann J. Development of RNA targeted PCR in situ hybridization with fluorescently labeled oligonucleotides for detection of Yersinia species. J Clin Microbiol. 1998;36:2557–2564. doi: 10.1128/jcm.36.9.2557-2564.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ash C, Farrow JAE, Dorsch M, Stackebrandt E, Collins MD. Comparative analysis of B. anthracis, B. cereus and related species on the basis of reverse transcriptase sequencing 16S rRNA. Int J Syst Bacteriol. 1991;41:343–346. doi: 10.1099/00207713-41-3-343. [DOI] [PubMed] [Google Scholar]
- 16.Read TD, Peterson SN, Tourasse N, Baillie LW, Paulsen IT, Nelson KE, Tettelin H, Fouts DE, Eisen JIA, Gill SR, Holtzapple EK, Okstad OA, Helgason E, Rilstone J, Wu M, Kolenay JF, Beanan MJ, Dedcon RJ, Brinkac LM, Gurinn M, Deboy RT, Madpu R, Daugherty SC, Durkin AS, Haft DH, Nelson WC, Peterson JD, Pep M, Khouri HM, Radune D, Benton JL, Mahamoud Y, Jiang L, Hance IR, Weidman JF, Berry KJ, Plant RD, Wolf AM, Watkins KL, Nierman WC, Hazen A, Cline R, Redmond C, Thwaite JE, White O, Salzberg SL, Thomason B, Friedlander AM, Kuchler TM, Hanna PC, Kslsto AB, Fraser CM. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature. 2003;423:81–86. doi: 10.1038/nature01586. [DOI] [PubMed] [Google Scholar]
- 17.King EO, Ward MK, Raney DE. Two simple media for demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 18.Bazzicalupo M, Fani R. The use of RAPD for generating specific DNA probes for microorganisms. In: Clapp J, editor. Methods in molecular biology. Totowa: Humana Press Inc; 1994. pp. 155–175. [DOI] [PubMed] [Google Scholar]
- 19.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson M, Lindow SE. Coexistence among epiphytic bacterial populations mediated through nutritional resource partitioning. Appl Environ Microbiol. 1994;60:4468–4477. doi: 10.1128/aem.60.12.4468-4477.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohlf FJ. NTSYSpc numerical taxonomy and multivariate analysis system for the IBM PC microcomputer (and compatibles) Setauket, NY: Applied Biostatistics; 1993. [Google Scholar]
- 22.Bachner BR, Savagean MA. Generalized indicator plate for genetic, metabolic and taxonomic studies with microorganisms. Appl Environ Microbiol. 1977;33:434–444. doi: 10.1128/aem.33.2.434-444.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirk JL, Beaudette LA, Hart M, Moutoglis P, Klironomos JN, Lee H, Trevors JT. Methods of studying soil microbial diversity. J Microbiol Methods. 2004;58:169–188. doi: 10.1016/j.mimet.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Rossellö-Mora R, Amann R. The species concept for prokaryates. FEMS Microbiol Rev. 2001;25:39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. [DOI] [PubMed] [Google Scholar]
- 25.Dykhuizen DE. Santa Rosalia revisited: why are there so many species of bacteria? Antonie Van Leeuwen. 1998;73:25–33. doi: 10.1023/A:1000665216662. [DOI] [PubMed] [Google Scholar]
- 26.Jaspers E, Overmann J. Ecological significance of microdiversity: identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Appl Environ Microbiol. 2004;70:4831–4839. doi: 10.1128/AEM.70.8.4831-4839.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perna NT, Plunkett G, Burland V, Man B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Danis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. Genome sequence of enterohaemorrhagic. Nature. 2001;0157:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 28.Sass H, Wieringa E, Cypionka H, Babenzien HD, Overmann J. High genetic physiological diversity of sulfate reducing bacteria isolated from oligotrophic lake sediment. Arch Microbiol. 1998;170:243–251. doi: 10.1007/s002030050639. [DOI] [PubMed] [Google Scholar]
- 29.Sikorski J, Nohle M, Wackernagel W. Identification of complex composition, strong strain diversity directional selection in local pseudomonas stutzeri populations from marine sediment and sials. Environ Microbiol. 2002;4:465–476. doi: 10.1046/j.1462-2920.2002.00325.x. [DOI] [PubMed] [Google Scholar]
- 30.Wieringa EJ, Overmann M, Cypionka H. Detection of abundant sulphate reducing bacteria in marine oxic sediment layers by a combined cultivation and molecular approach. Environ Microbiol. 2000;2:417–427. doi: 10.1046/j.1462-2920.2000.00123.x. [DOI] [PubMed] [Google Scholar]
- 31.Beja O, Koonin EV, Aravind L, Taylor LT, Seitz H, Stein JL, Bensen DC, Feldman RA, Swanson RV, Dehong EF. Comparative genomic analysis of archeal genotypic variants in a single population in two different oceanic provinces. Appl Environ Microbial. 2002;68:335–345. doi: 10.1128/AEM.68.1.335-345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]