Abstract
Soil and sediment samples obtained from Orange MR dye contaminated habitat were screened for heterotrophic bacterial population. The heterotrophic bacterial density of dye-contaminated soil was 2.14 × 106 CFU/g. The generic composition of heterotrophic bacterial population was primarily composed of 10% of Proteus sp., 15% Aeromonas sp., 20% Bacillus sp., 25% Pseudomonas sp. and 30% Micrococcus sp. The bacterial strain that decolorized the azo dye Orange MR up to 900 ppm was identified as Micrococcus sp. The optimum inoculum load, pH and temperature were found to be 5%, 6 and 35°C, respectively. The rate of decolorization was assessed using spectrophotometer at 530 nm and the percentage of decolorization was ascertained. The autochthonous bacterial isolate was able to utilize the dye as both nitrogen and carbon source.
Keywords: Azo dye, Orange MR, Autochthonous bacteria, Micrococcus, Decolorization
Introduction
Azo dyes accounts for the greatest part of all textile dye stuffs produced and have been the most universally used synthetic dyes in textile, food, leather, pharmaceutical, paper and cosmetic industries [1–3] and eventually more than 7 × 105 metric tons of these dyes are globally produced every year [4]. Azo dyes are synthetic organic colourants characterized by great structural variety. They have the same chromophore N=N and different auxochromes such as NH2, NR2 and OH groups [5]. Azo dyes at high concentrations and their intermediate amines pose environmental hazards due to their toxicity and potential carcinogenicity [6]. Microbes can degrade azo dyes both aerobically and anaerobically [7] and aerobic degradation is more efficient than anaerobic degradation. In the present study an effort was made to decolourise the Orange MR, azo dye aerobically by the autochthonous bacteria. Orange MR is an azo dye extensively used in textile dyeing and hence this dye has been selected for the present study. An attempt was made to isolate autochthonous bacteria capable of degrading Orange MR under laboratory conditions with a view of extending its utility in eco-restoration process.
Materials and Methods
Segregation of Dye Degrading Bacteria
Soil samples were collected from five different azo dye contaminated sites in Kanchipuram town, Tamil Nadu, India. The total heterotrophic bacterial population of the soil samples were analysed by the pour plate technique employing nutrient agar. Morphologically dissimilar and prominent colonies were picked and restreaked on nutrient agar to ensure purity and identified up to the generic level as per the scheme of previous work of Simudu and Aiso [8] and Bergey’s Manual [9] was also referred in the identification procedures.
Selection and Detection of Dye Degrading Bacteria
Nutrient agar plates incorporated with Orange MR azo dye at different concentrations (ranging from 100 to 1000 ppm) were prepared. To this 0.1 ml of the soil suspension was spread plated and incubated at 37°C for 24–36 h. Three bacterial strains such as Micrococcus sp., DBS1, DBS 2 and DBS 3 which were able to resist high azo dye concentration of 900 ppm were isolated from nutrient agar plates incorporated with dye. To evaluate their dye degradation efficiency these isolates were streaked on dye incorporated plates. Micrococcus sp. DBS 2 was able to produce clear and distinct zone of clearance of dye around the growth. This strain was selected for further analysis.
Scanning of Dye for its Absorbance Maxima, Decolorization and Optimization Studies
The maximum absorbance of the dye incorporated broth was determined. It was found to be 530 nm and hence the decolorization was assayed at 530 nm using a spectrophotometer. The 250 ml nutrient broths was incorporated with dye at ten different concentrations from 100 to 1000 ppm were inoculated with 24 h old bacterial culture of Micrococcus sp. DBS 2 at 5% (v/v) concentration and incubated at 37°C for 48 h. Aliquots were withdrawn from the culture broth at 12 h intervals and centrifuged at 10,000 rpm for 10 min in a tabletop centrifuge and the bacterial cells were removed as pellets. The supernatant was analysed spectrophotometrically at 530 nm and the decolorization was calculated using the following formula described in the earlier work of Sani and Banerjee [10]
 |
As a preamble to use Micrococcus sp., for decolorization experiments, optimization studies with regard to pH, temperature and inoculum load were also carried out. Five different sets of nutrient broth (250 ml), each incorporated with nine different concentrations (100–900 ppm) of Orange MR was prepared separately. Each set was inoculated with different amounts of inoculums ranging from 1 to 5% (i.e., 1 ml inoculum/100 ml of broth; for 5%: 5 ml of inoculum/100 ml of broth). The flasks were incubated at 37°C for 48 h. At 12 h interval, the decolorization was monitored spectrophotometerically, using uncoloured broth as control. The optimum amount of inoculum (i.e., 5% consisting of 9 × 104 counts) capable of producing maximum percentage of decolorization was taken as standard for further analysis. The pH of the seven different sets of nutrient broths (250 ml) each incorporated with three different concentrations of 300, 500 and 700 ppm of the dye separately were adjusted to pH 4, 5, 6, 7, 8, 9 and 10 (using 0.1 N, HCl and NaOH) and were inoculated with 5% (consisting of 9 × 104 counts) of 24 h old bacterial culture of DBS-2. All the seven sets were incubated at 37°C for 48 h. Aliquots were withdrawn from the culture broth at a regular interval of 12 h and analysed for decolorization. The optimal pH was assessed on the basis of maximum decolorization. Four different sets of sterile nutrient broth (250 ml) each incorporated with three different concentrations of 300, 500 and 700 ppm of Orange MR was prepared separately and inoculated with 5% (consisting of 9 × 104 counts) of 24 h old bacterial culture. Each set of inoculated flasks was incubated at different temperatures of 30, 35, 40 and 45°C. The optical density was measured at 12 h interval up to 48 h, using uninoculated broth as control. From the optical density, the percentage of decolorization was calculated. The temperature, which affected maximum efficiency of the bacterial strain, was used as another important environmental parameter.
Utilization of Dye as Carbon and Nitrogen Source
The bacterial strain Micrococcus sp. DBS 2 was tested for its dye degrading capacity. Two sets of mineral salt medium was made each by adding per litre of distilled water; K2HPO4 (0.8 g), KH2PO4 (0.2 g), CaSO4·2H2O (0.05 g), MgSO4·7H2O (0.5 g), FeSO4·7H2O (0.09 g), (NH4)2 SO4 (1.0 g) [11]. To one set dye was added at three different concentrations of 300, 500 and 700 ppm as nitrogen source and 1% glucose was added as carbon source. To another source the azo dye was added as carbon source and ammonium nitrate (0.03 g/100 ml) was added as nitrogen source. Both the sets were inoculated with 5% (consisting of 9 × 104 counts) of saline washed 24 h old bacterial culture and incubated at 35°C for 10–15 days. The culture broths were assayed for decolorization after the incubation period.
Results and Discussion
The total heterotrophic bacterial population (THBP) of the dye contaminated soil was determined by adopting pour plate technique and it was found to be 2.14 × 106 CFU/g. The occurrence of azo dye resistant bacteria in significant population at higher concentration emphasized the metabolic potential of autochthonous microorganisms. Knap and Newby [7] have reported the occurrence of dye resistant microorganisms in the contaminated soil. Mariappan et al. [12] have recorded an appreciable amount of THBP in the azo dye contaminated soil ranging from 13.2 × 107 (site 4) to 32 × 107 (site 1). When compared to the present study comparatively higher number of total viable bacterial count (8 × 106 CFU/ml) was reported for the azo dye-contaminated water of river Bhadar by Doctor et al. [13]. The occurrence of high bacterial load in the dye-contaminated soil may be either due to the enrichment of the dye degradable population or the dilution of the dye which might have lowered the toxicity of the dye.
The THBP bacterial strains were isolated and identified up to the generic level. The dye contaminated soil was dominated by 30% Micrococcus sp. followed by 25% Pseudomonas sp., 20% Bacillus sp., 15% Aeromonas sp. and 10% Proteus sp. as furnished in Table 1. This observation was found to be in accordance with the previous studies of Grigbsy et al. [14] on dye degradation using Sreptomyces chromofuscus and Mariappan et al. [12] on Orange G by Pseudomonas sp, Bacillus sp. and Escherichia sp. Sharma et al. [15] collected various soil and sludge samples from the vicinity of textile dyeing industries and waste disposal sites and identified five bacterial isolates belonging to the genera Bacillus, Alkaligenes and Aeromonas. From the dye contaminated water of river Bhadar different bacterial species such as 80.0% E. coli, 15.0% Pseudomonas aeruginosa and 5.0% Klebsiella pneumoniae were identified by Doctor et al. [13].
Table 1.
Percentage prevalence of different bacterial genera in the dye contaminated soil
| Bacterial genera | Total no. of isolates | No. of strains identified | Percentage prevalence |
|---|---|---|---|
| Aeromonas sp. | 40 | 6 | 15 |
| Bacillus sp. | 40 | 8 | 20 |
| Micrococcus sp. | 40 | 12 | 30 |
| Proteus sp. | 40 | 4 | 10 |
| Pseudomonas sp. | 40 | 10 | 25 |
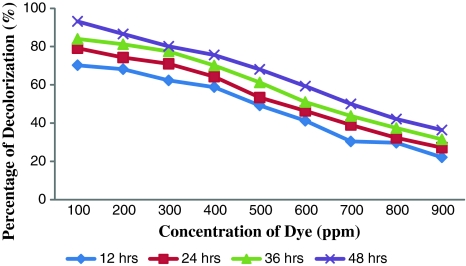
The dye resistant potential of the autochthonous bacteria of the dye contaminated soil was determined and presented in Table 2. Only six colonies were observed at the concentration of 900 ppm and this clearly indicated the deleterious effect of the dye on the native bacteria. The dye resistant bacterial isolates were characterized as Micrococcus sp. and labeled as DBS 1, DBS 2 and DBS 3. Among these three strains DBS 2 was able to produce prominent and distinct zone of clearance of dye around their growth up to 900 ppm and beyond this concentration this strain was unable to decolourise Orange MR. Based on the efficiency of decolorization, DBS 2 was employed for further investigations. Micrococcus sp. DBS 2 was able to decolorize the dye up to 900 ppm and the decolorization after 12, 24, 36 and 48 h was shown in Fig. 1. The decolorization of Orange MR was 36.34% after 48 h. At low concentration of 100 ppm the percentage of decolorization was 93.18% within 48 h but at high concentrations of 900 ppm the decolorization percentage was less i.e., 36.34% even after the prolonged incubation period of 48 h. The percentage of dye decolorization was inversely proportional to the concentration of dye (Fig. 1). Zimmermann et al. [16] reported the degradative potential of Pseudomonas KF 46 on the azo dye Orange II. Similar result was obtained by Bumpus and Brock [17] in their experiment on the degradation of crystal violet by P. chyrosporium.
Table 2.
Total dye resistant bacterial population in the dye contaminated sediment samples
| Concentration of dye (ppm) | Bacterial load (CFU/g) |
|---|---|
| 100 | TNTC |
| 200 | 250 × 104 |
| 300 | 185 × 104 |
| 400 | 120 × 104 |
| 500 | 84 × 104 |
| 600 | 40 × 104 |
| 700 | 18 × 104 |
| 800 | 11 × 104 |
| 900 | 6 × 104 |
TNTC too numerous to count
Fig. 1.
Decolorization of Orange MR by DBS 2 after 12, 24, 36 and 48 h
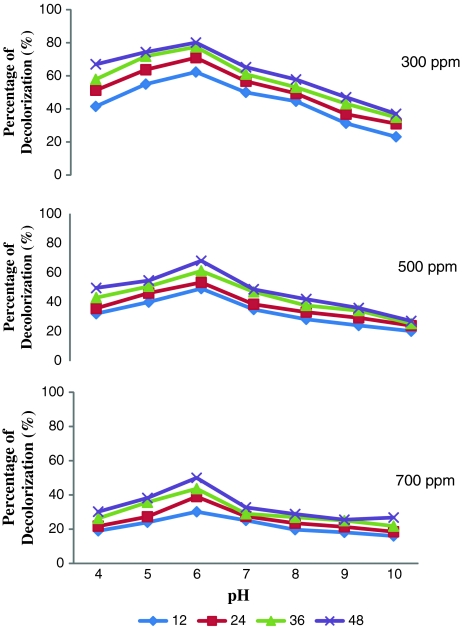
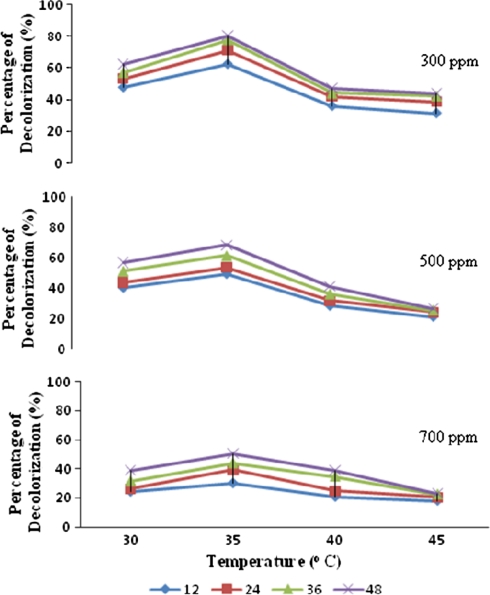
As a preamble to use this strain for decolorization experiments optimization studies were carried out. The inoculum load of 5% was observed to bring about efficient decolorization of Orange MR within 48 h. The optimum pH was 6 and the temperature optimum of DBS 2 was found to be at 35°C (Figs. 2, 3, respectively). From the data presented in Table 3 it could be inferred that the chosen bacterial strain had the ability to utilize Orange MR both as a source of carbon and nitrogen. Grigsby et al. [14] reported an enhanced degradation of azo dyes at 37°C by native micro flora. Mariappan et al. [12] recorded maximum degradation of the azo dye orange G by Pseudomonas sp SACO1 at 37°C and Escherichia sp. SACO 2 at 44°C. It was well documented that Knapp and Newby [7] employed neutral pH for the degradation of azo dyes using bacterial consortia. Zimmermann et al. [16] also employed neutral pH for using Pseudomonas KF 46 to degrade azo dyes. Similar pH dependent dye degradation efficiency of Bacillus sp and Pseudomonas sp was reported by Mariappan et al. [12]. They reported that the maximum decolorization was brought about by Bacillus sp., at a pH of 10 and by Pseudomonas sp., at pH 8. The chosen bacterial strain, DBS 2 had the ability to utilize Orange MR both as a source of carbon and nitrogen. Student’s t test analysis was carried out on the ability of DBS 2 to utilize the dye as carbon and nitrogen source. The mean values of different concentrations of orange MR i.e., 100, 200 and 300 ppms showed extremely high significant difference (P < 0.0001). It becomes explicit from Student’s t test that the efficacy of the isolated strain DBS 2 to utilize the dye as nitrogen source was higher than its ability to utilize the dye as carbon source (Table 3). Similar utilization of azo dyes as carbon source and nitrogen source by the native bacterial isolates was reported by Mariappan et al. [12]. More often azo dye degrading organisms metabolize and utilize azo dye as nitrogen source despite of the strength of azo bond. Razoflores et al. [18] have remarked such potentials of utilizing azo dyes as energy, nitrogen and carbon sources by anaerobic bacteria. Hu [19] has reported the ability of Pseudomonas luteola to grow and bring about 95% decolorization of azo dye when exposed to media with azo dye RP2B without nitrogen supplementation. Similar observation was made by O’Neill et al. [20] also.
Fig. 2.
Decolorization of different concentrations of Orange MR by DBS 2 at different time intervals and at different pH
Fig. 3.
Decolorization of different concentrations of Orange MR by DBS 2 at different time intervals and at different temperatures
Table 3.
Utilization of Orange MR as carbon and nitrogen source by DBS 2
| Concentration of dye (ppm) | As carbon source | Percentage of decolorization | |||||
|---|---|---|---|---|---|---|---|
| Mean ± SD | As nitrogen source | Mean ± SD | t test value | df | Level of significance | ||
| 100 | 15.42 | 15.42 ± 3.6 | 78 | 78 ± 1.4 | 28.0616 | 4 | P < 0.0001 |
| 19.02 | 79.4 | ||||||
| 11.82 | 76.6 | ||||||
| 200 | 11.21 | 11.21 ± 2.5 | 85.3 | 85.3 ± 2.0 | 40.0828 | 4 | P < 0.0001 |
| 13.71 | 83.3 | ||||||
| 8.71 | 87.3 | ||||||
| 300 | 6.57 | 6.57 ± 1.5 | 90 | 90 ± 1.5 | 80.1570 | 4 | P < 0.0001 |
| 7.57 | 91.5 | ||||||
| 5.57 | 88.5 | ||||||
The preliminary investigation on occurrence of azo dye resistant bacterial population and their decolorization potential provide vital basic data on the use of native micro flora in azo dye degradation. The significant role of autochthonous bacteria in bioremediation has been emphasized in the present study. Thus, Micrococcus DBS 2 isolated in the present study affirmed as a promising candidate for the biodegradation of dye industry effluent. Further studies on analytical aspects of the biotransformation and fate of metabolites would make it possible to utilize those bacteria for bioremediation of Orange MR polluted habitats.
References
- 1.Michaels GB, Lewis DL. Microbial transformation rates of azo and triphenylmethane dyes. Environ Toxicol Chem. 1986;5:161–166. doi: 10.1002/etc.5620050206. [DOI] [Google Scholar]
- 2.Chung KT, Stevens SE., Jr Degradation of azo dyes by environmental microorganisms and helminthes. Environ Toxicol Chem. 1993;12:2121–2132. [Google Scholar]
- 3.Carliell CM, Barclay SJ, Naidoo N, Buckley CA, Mulholland DA, Senior E. Microbial decolorization of a reactive azo dye under anaerobic conditions. Water SA. 1995;21(1):61–69. [Google Scholar]
- 4.Keck A, Klein J, Kudlich M, Stolz A, Knackmass HJ, Mattes R. Reduction of azo dyes by Redox mediators originating in the Naphthalenesulfonic acid degradation pathway of Sphingomonas sp strain BN-6. Appl Environ Microbiol. 1997;63(9):3684–3690. doi: 10.1128/aem.63.9.3684-3690.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer U. Biodegradation of synthetic organic colourants. FEMS Symp. 1981;12:271–385. [Google Scholar]
- 6.Vyas BRM, Molitoris HP. Involvement of an extracellular H2O2-dependent liginolytic activity of the white rot fungus Pleurotus ostreatus in the decolorization of Remazol Brilliant Blue–R. Appl Environ Microbiol. 1995;61(11):3919–3927. doi: 10.1128/aem.61.11.3919-3927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knapp JS, Newby PS. The microbiological decolorization of an industrial effluent containing a diazo linked chromophore. Water Res. 1995;29(7):1807–1809. doi: 10.1016/0043-1354(94)00341-4. [DOI] [Google Scholar]
- 8.Simudu V, Aiso K. Occurrence and distribution of heterotrophic bacteria in seawater from the Kanagawa Bay. Bull Jap Sci Fish. 1962;28:1137. [Google Scholar]
- 9.Peter A, Sneath A, Elisabeth Sparbe M, editors. Bergey’s Manual of systematic bacteriology. Baltimore: Wlliams and Wilkins; 1986. [Google Scholar]
- 10.Sani RK, Banerjee UC. Decolorization of triphenylmethane dyes and textile and dyestuff effluent by Kurthia sp. Enzyme Microbiol Technol. 1999;24:433–437. doi: 10.1016/S0141-0229(98)00159-8. [DOI] [Google Scholar]
- 11.Steffan S, Bardi L, Marzona M. Azo dye biodegradation by microbial cultures immobilized in alginate beads. Environ Int. 2005;31:201–205. doi: 10.1016/j.envint.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 12.Mariappan C, Gayathri Devi TV, Yamuna RL, Palaniappan R, Selvamohan T. Orange-G Tolerance, utilization and degradation potentials of native bacterial isolates. Biosci Biotechnol Res Asia. 2003;01(2):87–91. [Google Scholar]
- 13.Doctor PB, Raiyani CV, Verma Y, Desai NM, Kulkarni PK, Ruparelia SG, Ghosh SK. Physico-chemical and microbiological analysis of dye contaminated river water. Indian J Environ Health. 1998;40(1):7–14. [Google Scholar]
- 14.Grigsby PMB, Gaszcfynskis NS, Crawford DL. Transformation of azo dye isomers by Streptomyces chromofuscus. Appl Environ Microbiol. 1996;62(5):1814–1817. doi: 10.1128/aem.62.5.1814-1817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma DK, Saini HS, Singh M, Chimni SS, Chandha BS. Isolation and characterization of microorganisms capable of decolorizing various triphenylmethane dyes. J Basic Microbiol. 2004;44(1):59–65. doi: 10.1002/jobm.200310334. [DOI] [PubMed] [Google Scholar]
- 16.Zimmermann H, Kulia HG, Leisinger T. Properties of purified orange II azo reductase enzyme initiating azo-dye degradation by Pseudomonas KF 46. Int J Biochem. 1982;129:197–203. doi: 10.1111/j.1432-1033.1982.tb07040.x. [DOI] [PubMed] [Google Scholar]
- 17.Bumpus JA, Brock BJ. Biodegradation of crystal violet by the white rot fungus Phanerochate chrysosporium. Appl Environ Microbiol. 1998;54:1143–1150. doi: 10.1128/aem.54.5.1143-1150.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Razoflores E, Luijnten M, Dondon BA, Letting G, Field JA. Complete biodegradation of the azo dye azodisalicyclate under anaerobic condition. Environ Sci Tech. 1997;31:2098–2103. doi: 10.1021/es960933o. [DOI] [Google Scholar]
- 19.Hu TL. Degradation of azo dye RP2B by Pseudomonas luteola. Water Sci Technol. 1998;38(4–5):299–306. [Google Scholar]
- 20.O’Neill C, Lopez A, Esteves S, Hawkes RF, Hawkes DL, Wilcox S. Azo dye degradation in an anaerobic-aerobic treatment system operating on stimulated textile effluent. Appl Microbiol Biotechnol. 2000;53(2):249–254. doi: 10.1007/s002530050016. [DOI] [PubMed] [Google Scholar]