Abstract
The distribution of coliphages infecting different Escherichia coli virotypes (EHEC, EIEC, EPEC, ETEC) and an avirulent strain (K-12) in sewage system of a hospital and a sewage treatment plant (STP) was investigated by culture-based agar overlay methods. Coliphages were found in all the samples except stool dumping site in the sewage system of the hospital and lagoon of the STP. Bacteriophage count (pfu/ml) infecting E. coli strains showed the following ascending pattern (EHEC < EIEC < EPEC < ETEC < E coli K-12) in all the collected samples except one. Phages capable of infecting avirulent E. coli K-12 strains were present in the highest number among all the examined locations. Phages specific for E. coli K-12 presented high diversity in plaque size on the bacterial lawn. Virulent E. coli specific coliphages rarely produced plaques with diameter of 1–2 mm or over. Conventional agar overlay method was found to be not satisfactory for phage community analysis from primary stool dumping site of the hospital, probably due to the presence of high concentration of antimicrobial substances. The gradual decrease seen in the five groups of coliphage quantity with the ongoing treatment process and then the absolute absence of coliphages in the outlet of the examined treatment plant is indicative of the usefulness of the treatment processes practiced there.
Keywords: E. coli, Virotypes, Bacteriophages, Sewage treatment plant, Hospital sewage
Introduction
Escherichia coli is the predominant facultative anaerobe of the human colonic flora. The organism typically colonizes the infant gastrointestinal tract within hours of life, and, thereafter, E. coli and the host derive mutual benefit [1]. Unfortunately, some E. coli strains are diarrheagenic due to presence of conserved features such as, their ability to colonize the intestinal mucosal surface despite peristalsis and competition for nutrients by the indigenous flora of the gut (including other E. coli strains). Once colonization is established, the pathogenetic strategies of the diarrheagenic E. coli strains exhibit remarkable variety. Three general paradigms have been described by which E. coli may cause diarrhea including, (i) enterotoxin production (ETEC and EAEC), (ii) invasion (EIEC), and/or (iii) intimate adherence with membrane signaling (EPEC and EHEC). Two another group of diarrheagenic E. coli includes diffusedly adherent E. coli (DAEC) and cytolethal distending toxin (CDT) producing E. coli. However, the interaction of the organisms with the intestinal mucosa is specific for each category [2]. Among the E. coli strains, the major pathotypes are enteropathogenic (EPEC) and enterotoxigenic (ETEC) E. coli [3]. EIEC strains are biochemically and genetically closely related to Shigella species and possess a virulence plasmid encoding the ability to penetrate intestinal epithelial cells. The plasmid virulence genes and other aspects of pathogenesis are virtually identical to those of Shigella species [4]. E. coli strains that cause hemorrhagic colitis (HC) and hemolytic uremic syndrome (HUS), express Shiga toxin (Stx), cause A/E lesion on epithelial cells and possess a about 60-MDa plasmid are denoted as EHEC [5, 6]. EPEC strains harbor locus of enterocyte effacement (LEE), but do not produce Stx [7, 8]. ETEC strains elaborate at least one member of two defined groups of enterotoxin: heat-stable toxin (ST) and heat-labile toxin (LT) [5]. Treatment of E. coli diarrhea with oral rehydration solution has substantially reduced mortality from dehydration but has little or no effect on the pathogen itself. Recent development of antibiotic resistant bacterial flora has challenged the sustainable use of antibiotics and now we need to consider for alternatives [9].
At 1917, bacteriophages were discovered by Fedrix D’Herelle and the discoverer himself found potential of bacteriophages to use in therapeutic purposes (phage therapy). Due to rapid development of antibiotic therapy and some other practical causes phage therapy faced critical skepticism at its early period [9]. Although recent interest in phage therapy has caused reappraisal of the method, we are at extreme shortage with data regarding ecological distribution of phages infecting bacterial species and strains under same species [10]. This lack of data is surprising because E. coli phages lambda and T4 are among the best characterized biological systems [11]. In E. coli, majority of genes encoding virulence factors reside in mobile genetic elements which also include prophages [2, 4, 12–14]. Exclusion of some of those prophages may alter the pathogenic potential of a particular strain. In addition, prophage provides bacteria superinfection immunity against the same bacteriophage [15, 16]. Under these circumstances it is not unrealistic to assume that E. coli virotypes arise due to the presence of relevant prophages in the bacterial chromosome or plasmids.
Data on the distribution of bacteriophages are not only interesting for their potential clinical use but would add to our understanding of the natural diversity of phages. Molecular studies focusing on specific viral genes elucidate the large genetic variation in viral communities in aquatic systems [17]. Current work was designed to examine the diversity of coliphages infecting four E. coli virotypes in comparison with coliphages capable of infecting E. coli K-12 strains from hospital sewage and sewage treatment plant (STP). Furthermore, we considered the analysis of the effectiveness of the sewage treatment processes practiced currently in Bangladesh.
Materials and Methods
Bacterial Strains and Media
A total of five E. coli strains (Table 1) were used for phage isolation. Escherichia coli O157:H7 reference strain NCTC 12079 was used as representative of EHEC. EIEC, EPEC, and ETEC strains were local isolates and have been collected from Environmental Microbiology and Molecular Ecology laboratory, Department of Microbiology, University of Dhaka and were further confirmed by PCR upon receiving [2]. Luria–Bertani (LB) medium was used for the propagation of bacterial species and subsequent bacteriophage isolation.
Table 1.
E. coli strains used in the study
| Bacterial strains |
|---|
| E. coli O157:H7 NCTC 12079 (EHEC) |
| Enteroinvasive E. coli (EIEC) |
| Enteropathogenic E. coli (EPEC) |
| Enterotoxigenic E. coli (ETEC) |
| Escherichia coli K-12 |
Sampling Site
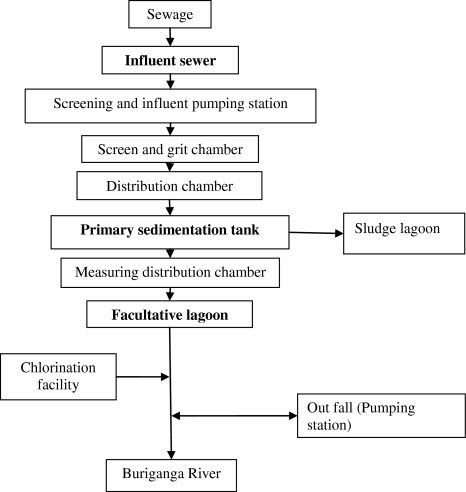
Raw hospital sewage (500 ml) was collected from the three different locations in the largest (1,400 bed) general hospital in Bangladesh, Dhaka Medical College Hospital (DMCH) sewage. It included (i) fresh patient excretion (collected at a place in sewer system where it is collected from all over the hospital and kept for 24 h), (ii) excreta dumping site (dumped for 15 days) and (iii) sewage outlet. Sewage sample was collected from three different points at STP, Pagla, Narayanganj that receives sewage from all over the Dhaka city. The points were (i) influent sewer, (ii) primary sedimentation tank, and (iii) facultative lagoon (Fig. 1).
Fig. 1.
Overview of the treatment process used at STP in Pagla, Narayanganj (Bold marks show our sampling points)
Sample Preparation
Each liquid sample (500 ml) was passed through the Whatman #1 filter paper two times to remove most of the suspended debris. 1.5 ml of filtrate was then collected into sterile Eppendorf tube and centrifuged at 10,000 rpm for 10 min. The supernatant was then filtered by 0.22 μm low protein binding syringe filter (Whatman U.S.A. catalogue No. 6780-2502) to remove the bacterial population present there.
For semisolid samples (deposited stool), one gram semisolid sample was added to 99 ml sterile 1× PBS in a 250 ml conical flask. Flasks were incubated at 37°C for 3–4 h to allow bacteriophages to come into solution. 50 ml suspension was then taken into 50 ml tubes and centrifuged at 10,000 rpm for 10 min and supernatant was collected. The supernatant was filtered with 0.22 μm low protein binding syringe filter and collected into sterile McCartney bottle.
Bacteriophage Isolation and Enumeration
The standard protocol for phage propagation by agar overlay technique was adapted in the current research [18]. One loopful of selected bacterial colony was inoculated into LB broth and incubated for 3 h in a shaking incubator with 150 rpm at 37°C. After incubation the culture was centrifuged at 10,000 rpm and the supernatant was discarded. The pellet was resuspended in 10 mM MgSO4 solution and kept at 4°C until used. 0.1 ml of bacterial suspension and 0.1 ml of processed sewage sample was mixed without vortexing and incubated for 20 min at 37°C in a water bath for attachment/adsorption of the present phages in the sewage sample to their host bacteria. Afterward, the mixture was added to 5 ml previously melted soft LB agar kept at 47°C. This preparation was mixed gently and poured onto LB Agar plates and kept for solidification. Finally, the plates were incubated at 37°C in an inverted position. Plaques were counted after 24 h and expressed as plaque forming unit (pfu) per ml.
Preservation of Bacteriophages
Isolated plaques were picked up by slight suction with micropipette tips. The picked plaque was resuspended in 1 ml SM-gelatin buffer containing 0.58% NaCl, 0.2% MgSO4·7H2O, and 5 ml 1 M Tris–Cl and 0.5 ml 2% gelatin in 100 ml preparations. Then, the tubes were kept on rocker for 1 h to allow phages get free from the agar medium. Finally, 50 μl of chloroform was added to each tube and kept at 4°C in dark.
Results and Discussion
Nine percent mortality of all age group of people in Bangladesh are due to diarrhea (Mortality country fact sheet 2006, WHO). Diarrheagenic E. coli is one of the main causes of diarrheal diseases and still remains a major public health problem in developing countries including Bangladesh. From July 1991 to May 1992, a survey on children of below 5 years of age (n = 1,053) in Bangladesh was conducted and had reported that ETEC (12%) and EPEC (15.5%) strains were significantly associated with diarrhea. EIEC was not isolated from any sample whereas EHEC (0.008%) was isolated only from the control children i.e. children without diarrhea. Enteroaggregative (9.5%) and diffuse adherent E. coli strains (8.2%) were also isolated from children with diarrhea [3]. From the early days of bacteriophage research, lytic activity of certain phages on bacterial strains has proved to be a reliable method for the identification of bacteria [19]. In 2004, a research was conducted at ICDDR,B (Bangladesh) for the isolation of E. coli specific bacteriophages from the stool of pediatric diarrheal patients in Bangladesh [11]. They had also surveyed coliphages in the environmental water from neighborhood of the infected patients. Despite a high fecal pollution of environmental water in Dhaka (because people often defecate in trenches along the streets), only a low level of coliphages was detected in the environmental water samples. Authors [11] correlated this finding with the observation that coliphages decay rapidly in the environment [10]. As phages reach environment from patients through sewage system, our purpose in this work was to find out distribution of bacteriophages capable of infecting different E. coli virotypes in sewage and comparison of this outcome with the circulation of the non virulent E. coli K-12 strain specific phages in the same environment.
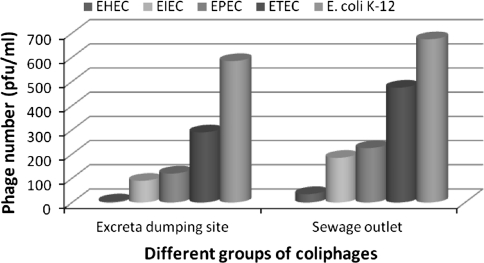
While investigating samples from DMCH i.e. fresh patient excretion (collected at a place in sewer system where it is collected from all over the hospital and kept for 24 h), and sewage outlet, coliphages infecting avirulent strain, K-12 was seen to have the highest prevalence (5.85 × 102 pfu/ml and 6.75 × 102 pfu/ml respectively) and EHEC specific coliphages showed the lowest prevalence (5.0 pfu/ml and 3.5 × 101 pfu/ml, respectively) among the five groups of coliphages studied. Interesting observation was drawn in distribution of coliphages infecting E. coli virotypes in these samples. In both the samples i.e. fresh patient excretion (collected at a place in sewer system where it is used to be kept for 24 h), and sewage outlet, coliphage count infecting E. coli strains showed the following ascending pattern EHEC < EIEC < EPEC < ETEC < E coli K-12 (Fig. 2). There is a dictum that phages are found where ever bacteria flourish. The virus-to-bacterium ratio (VBR) has been used to study the affiliation between bacteriophages and host bacteria in various aquatic ecosystems [10]. VBR values are found to be higher for more nutrient-rich productive environments, and even in oligotrophic environments. All the locations of sewage collection for this study were qualitatively and quantitatively rich in organic material, and hence had supported high bacterial growth of variety of species and which in turn produced greater numbers of viruses. Fast growth and high productivity of bacteriophages under these nutrient-replete conditions are also due, in part, to higher infection rates and larger burst sizes.
Fig. 2.
Relative numbers of coliphages (pfu/ml) infecting different E. coli virotypes in sewage samples collected from DMCH
With the sample from primary stool dumping site of DMCH (used to be dumped here for 15 days), proper bacterial lawn formation failed. Liquid waste along with stool arrives here from the first collection point and stays for 15 days. We have collected the surface fluid from this site. Large empty area appeared on bacteria inoculated LB plates instead of equally distributed lawn grown as controls (without any sewage sample or phage suspension). Those empty locations were further checked for the presence of phages and the result was negative. Failure of proper lawn formation may indicate the presence of elevated amount of antibacterial substances, such as antibiotics, disinfectants, antiseptics in the sewage from this particular location. Therefore, isolation of phages from such sources may demand different strategies to protect the bacterial growth from antibacterial substances.
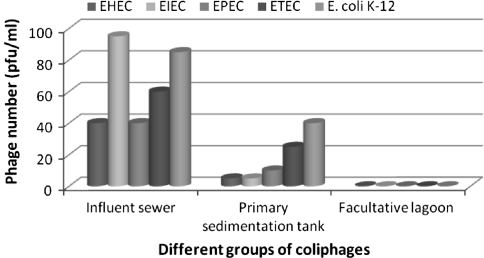
The count (pfu/ml) for each group of coliphage was the highest in the influent sewer (all the incoming sewage from all over the Dhaka city gathers here first; Fig. 1) among the three collected samples from the three different points at STP, Pagla, Narayanganj. In primary sedimentation tank, a relatively lower pfu/ml was documented for all the five groups of coliphages and the final outlet in the facultative lagoon contained absolutely no coliphages at all. The gradual decrease found in all the examined groups of coliphage quantity with the ongoing treatment process and then the absolute absence of coliphages in the outlet of the examined treatment plant is indicative of the usefulness of the treatment processes practiced there [20]. Over ten Lagoons, each spreading over huge surface area is there exposed to intensive sunlight due to the tropical location of the country. Sunlight UV was previously identified as the major destructive factor, causing up to 5% phage infectivity loss per hour for surface water due to thymine dimer formation [11]. Furthermore, exposure to treatment of sunlight has been found to result in doubling the virus inactivation rates [10]. These could be significant reasons behind the inclusive absence of coliphages in the lagoon sample (Fig. 3). Relative distribution of different virotypes (pathotypes) of E. coli has been investigated in clinical specimens before but their frequency and survival in nature has not been thoroughly explored yet. As far as we are concerned, there is no work so far reporting pattern of phage distribution specific for these different E. coli virotypes in natural ecosystem. Interestingly, similar distribution pattern among the five groups of coliphages (EHEC < EIEC < EPEC < ETEC < E coli K-12) was observed in the two samples from this STP (Fig. 3) and in the two samples from DMCH (Fig. 2). However, in influent sewer of this STP, coliphages infecting EIEC strain were the most abundant in quantity. The signaling of a potential infectious disease outbreak in advance may be a valuable tool for the implementation of various preventive measures in the community. The possible role of vibriophages for monitoring cholera epidemic has been described before [21]. Bacteriophages specific for different diarrheagenic E. coli should be further investigated to assess their potency to be used in signaling of a probable diarrheal outbreak in advance.
Fig. 3.
Gradual decrease in the numbers (pfu/ml) for all the groups of coliphages in the sewage sample while treated at the STP, Pagla, Narayanganj
Phages against different E. coli virotypes examined in this work had exhibited profound diversity in plaque size which ranged from less than 1 to 5 mm (Table 2). Surprisingly, coliphages infecting non virulent E. coli K-12 were always the most abundant in number among all the collected samples and showed the most diversity in plaque size distribution. For the other coliphages infecting virulent E. coli strains (EHEC, EIEC, EPEC, and ETEC), plaque size was rarely recorded exceeding 1–2 mm diameter.
Table 2.
Plaque size distribution and differential count (pfu/ml) of five different groups of isolated coliphages
| Site | Point of sample collection | Coliphages isolated against | Count (pfu/ml) of five different groups of coliphages according to their plaque size | |||
|---|---|---|---|---|---|---|
| E. coli strain | <1 mm | 1–2 mm | 2–3 mm | 3–5 mm | ||
| DMCH Sewage | Excreta kept here for 24 h | EHEC | 5 | 0 | 0 | 0 |
| EIEC | 45 | 20 | 25 | 0 | ||
| EPEC | 15 | 75 | 30 | 0 | ||
| ETEC | 105 | 145 | 40 | 0 | ||
| E. coli K-12 | 240 | 210 | 100 | 35 | ||
| Excreta dumping site (excreta dumped here for 15 days) | Proper bacterial lawn formation failed so that enumeration of coliphages was not possible | |||||
| Sewage outlet | EHEC | 35 | 0 | 0 | 0 | |
| EIEC | 55 | 130 | 0 | 0 | ||
| EPEC | 75 | 116 | 34 | 0 | ||
| ETEC | 195 | 131 | 149 | 0 | ||
| E. coli K-12 | 90 | 335 | 175 | 75 | ||
| Treatment plant, Pagla | Influent sewer | EHEC | 52 | 33 | 0 | 0 |
| EIEC | 40 | 0 | 0 | 0 | ||
| EPEC | 41 | 54 | 0 | 0 | ||
| ETEC | 40 | 0 | 0 | 0 | ||
| E. coli K-12 | 60 | 0 | 0 | 0 | ||
| Primary sedimentation tank | EHEC | 0 | 30 | 10 | 0 | |
| EIEC | 5 | 0 | 0 | 0 | ||
| EPEC | 5 | 0 | 0 | 0 | ||
| ETEC | 5 | 0 | 5 | 0 | ||
| E. coli K-12 | 26 | 0 | 0 | 0 | ||
| Sewage | Facultative lagoon | EHEC | 0 | 0 | 0 | 0 |
| EIEC | 0 | 0 | 0 | 0 | ||
| EPEC | 0 | 0 | 0 | 0 | ||
| ETEC | 0 | 0 | 0 | 0 | ||
| E. coli K-12 | 0 | 0 | 0 | 0 | ||
From the overall study, it becomes obvious that coliphages against several E. coli strains are abundant in sewage in Bangladesh. There are hypothesis that bacteriophages are intimately associated with bacterial virulence genes [22]. The occurrence of virotype specific coliphages may be implicated with acquiring virulence by host bacterial species and should be further investigated. In a country like Bangladesh from tropical territory, bacteria of Enterobacteriaceae family got ample opportunity to survive and multiply in the environment. Therefore, this environment is very amenable for evolution of bacteriophages against emerging bacterial strains of either drug resistance or increased virulence capacity. Phage therapy could help us initially to encounter those emerging bacterial strains than any other conventionally practiced therapeutic approach. Our research reports the distribution pattern of different types of phages capable of infecting different diarrheagenic E. coli strains in nature. Further study should be done to analyze in vitro and in vivo potentiality of these diarrheagenic E. coli specific coliphages for their role in biocontrol.
Contributor Information
Muntasir Alam, Email: muntasir@icddrb.org.
Tasmia Farzana, Email: tasmia.farzana@gmail.com.
Chowdhury Rafiqul Ahsan, Phone: +02-9661920-73, Email: crahsan@yahoo.com.
Mahmuda Yasmin, Phone: +02-9661920-73, Email: yasmin962001@yahoo.com.
Jamalun Nessa, Phone: +02-9661900-73, FAX: +880-02-8615583, Email: jamalun_nessa@hotmail.com.
References
- 1.Drasar BS, Hill MJ. Human intestinal flora. London, UK: Academic Press, Ltd.; 1974. pp. 36–43. [Google Scholar]
- 2.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albert MJ, Faruque SM, Faruque ASG, Neogi PKB, Ansaruzzaman M, Bhuiyan NA, Alam K, Akbar MS. Controlled study of Escherichia coli diarrheal infections in Bangladeshi children. J Clin Microbiol. 1995;33:973–977. doi: 10.1128/jcm.33.4.973-977.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaper JB, Mellies JL, Nataro JP. Pathogenicity islands and other mobile genetic elements of diarrheagenic Escherichia coli, chap 3. In: Kaper JB, Hacker JH, editors. Pathogenicity islands and other mobile virulence elements. Washington, DC: ASM Press; 1999. pp. 33–58. [Google Scholar]
- 5.Levine MM. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic and enteroadherent. J Infect Dis. 1987;155:377–389. doi: 10.1093/infdis/155.3.377. [DOI] [PubMed] [Google Scholar]
- 6.Levine MM, Edelman R. Enteropathogenic Escherichia coli of classic serotypes associated with infant diarrhea: epidemiology and pathogenesis. Epidemiol Rev. 1984;6:31–51. doi: 10.1093/oxfordjournals.epirev.a036274. [DOI] [PubMed] [Google Scholar]
- 7.Trabulsi LR, Keller R, Tardelli Gomes TA. Typical and atypical enteropathogenic Escherichia coli. Emerg Infect Dis. 2002;8:508–513. doi: 10.3201/eid0805.010385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 9.William CS. Bacteriophage therapy. Annu Rev Microbiol. 2003;55:437–451. doi: 10.1146/annurev.micro.55.1.437. [DOI] [PubMed] [Google Scholar]
- 10.Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 2000;64:69–114. doi: 10.1128/MMBR.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chibani-Chennoufi S, Sidoti J, Bruttin A, Dillmann ML, Kutter E, Qadri F, Sarker SA, Brūssow H. Isolation of Escherichia coli bacteriophages from the stool of pediatric diarrhea patients in Bangladesh. J Bacteriol. 2004;186:8287–8294. doi: 10.1128/JB.186.24.8287-8294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rietra PJ, Willshaw GA, Smith HR, Field AM, Scotland SM, Rowe B. Comparison of Vero-cytotoxin-encoding phages from Escherichia coli of human and bovine origin. J Gen Microbiol. 1989;135:2307–2318. doi: 10.1099/00221287-135-8-2307. [DOI] [PubMed] [Google Scholar]
- 13.Iguchi A, Thomson NR, Ogura Y, Saunders D, Ooka T, Henderson IR, Harris D, Asadulghani M, Kurokawa K, Dean P, Kenny B, Quail MA, Thurston S, Dougan G, Hayashi T, Parkhill J, Frankel G. Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 Strain E2348/69. J Bacteriol. 2009;191:347–354. doi: 10.1128/JB.01238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tauschek M, Gorrell RJ, Strugnell RA, Robins-Browne RM. Identification of a protein secretory pathway for the secretion of heat-labile enterotoxin by an enterotoxigenic strain of Escherichia coli. Proc Natl Acad Sci USA. 2002;99:7066–7071. doi: 10.1073/pnas.092152899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyman P, Abedon ST. Phage ecology of bacterial pathogenesis, chap 14. In: Abedon ST, editor. Bacteriophage ecology, population growth evolution and impact of bacterial viruses. Cambridge, UK: Cambridge University Press; 2008. pp. 353–385. [Google Scholar]
- 16.Boyd EF. Bacteriophages and bacterial virulence, chap 8. In: Kutter E, Sulakvelidze A, editors. Bacteriophages biology and application. USA: CRC Press; 2005. pp. 223–265. [Google Scholar]
- 17.Zhong Y, Chen F, Wilhelm SW, Poorvin L, Hodson RE. Phylogenetic diversity of marine cyanophage isolates and natural virus communities as revealed by sequences of viral capsid assembly protein gene g20. Appl Environ Microbiol. 2002;68:1576–1584. doi: 10.1128/AEM.68.4.1576-1584.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Russell DW. Molecular cloning, a laboratory manual, vol I. 3. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 2001. pp. 2.25–2.31. [Google Scholar]
- 19.Kasatiya S, Caprioli T, Champoux S. Bacteriophage typing scheme for Salmonellainfantis. J Clin Microbiol. 1979;10:637–640. doi: 10.1128/jcm.10.5.637-640.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leclerc H, Edberg S, Pierzo V, Delattre JM. Bacteriophages as indicators of enteric viruses and public health risk in groundwater. J Appl Microbiol. 2000;88:5–21. doi: 10.1046/j.1365-2672.2000.00949.x. [DOI] [PubMed] [Google Scholar]
- 21.Faruque SM, Naser I, Islam MJ, Faruque ASG, Ghosh AN, Nair GB, Sack DA, Mekalanos JJ. Seasonal epidemics of cholera inversely correlate with the prevalence of environmental cholera phages. Proc Natl Acad Sci USA. 2004;102:1702–1707. doi: 10.1073/pnas.0408992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patrick LW, Matthew KW. Bacteriophage control of bacterial virulence. Infect Immun. 2002;70:3985–3993. doi: 10.1128/IAI.70.8.3985-3993.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]