Abstract
Nucleic acid tests that detect HIV infection at an early phase are available and have been applied on individual dried blood spot (DBS). The present study was undertaken with an aim to evaluate the feasibility of performing PCR for HIV-1 DNA on pools of DBS as an alternative to individual testing. Standardization of PCR by a modified Amplicor HIV-1 DNA assay version 1.5 (Roche molecular diagnostics, USA), on pooled DBS was performed using five confirmed HIV reactive samples with known low viral load of HIV-1 and HIV non-reactive samples in pools of 5, 10 and 20 DBS. After successful standardization of pooling procedure, a total of 183 pools (of 10 DBS each) were prepared from 1,823 DBS samples, collected from a population-based study that tested negative for HIV antibodies and p24 antigen. All these pools were screened for HIV-1 DNA by the Amplicor assay. Standardization of pooling procedure indicated that pooling of 10 DBS gave an optimum result. Out of 183 pools tested, one pool of 10 samples was positive and of these ten DBS that were tested individually to identify the positive DBS, one sample was detected to be positive for HIV-1 DNA. Our study demonstrates that PCR for HIV-1 DNA can be successfully performed on pools of DBS. However, this may be needed only on specialized studies of HIV and not for routine epidemiology studies as only a very small fraction of cases would be missed if only antibody/antigen testing were done.
Keywords: Dried blood spots (DBS), Polymerase chain reaction (PCR), Pooling strategy, HIV screening
Introduction
Whole blood dried onto filter paper constitutes a potentially useful material for molecular testing of viruses, including HIV [1–3]. Though HIV is a RNA virus, once inside the host CD4 cell following infection, HIV RNA is converted to DNA. This viral DNA, after combining with host DNA, exists as Proviral DNA. Dried blood spots (DBS) represent a valuable and comprehensive genetic repository because genomic DNA can be isolated from blood samples processed on these filter papers for use in polymerase chain reaction (PCR) [4]. Besides the extraction of DNA from DBS, amplification from punches of DBS directly taken into the PCR reaction mixture has been reported [5].
In spite of continuous improvements of the serological assays, a residual risk that is extremely small still exists attributed largely during the antibody-negative, pre seroconversion window period, the period between infection and detectable seroconversion [1, 6]. Direct detection of the virus can be achieved by nucleic acid test (NAT), which enables detection of the viral nucleic acid. Previous studies reported the detection of HIV DNA approximately 1 week earlier than detection of p24 antigen and about 2 weeks earlier than antibody detection [6, 7].
PCR technique was introduced in 1983 and the use of DBS for PCR testing was first shown to have utility in 1987 [2] using in-house technique. Later, DBS PCR was adapted to a standardized, commercially available microwell plate amplification and detection kit, Amplicor HIV-1 produced by Roche molecular systems [8]. Though several studies have extracted DNA and RNA from a single DBS and performed PCR for HIV DNA, there have been no reports of performing HIV PCR on DBS that have been pooled together. The principle of pooling is simply to mix a predetermined number of samples in a pool and test the pool as one sample. Since pooling is a cost effective and time saving way of performing nucleic acid tests, PCR on pooled DBS is especially suited for samples that require testing on large scale (e.g. samples from general population).
With the absence of availability of any publications on HIV PCR on pooled DBS, there has been a need of standardization and evaluation of the feasibility of performing PCR for HIV-1 DNA from pooled DBS. The present study was undertaken with an aim to evaluate the feasibility of performing PCR for HIV-1 DNA on pools of DBS as an alternative to individual testing.
Materials and Methods
Standardization of HIV-1 DNA PCR on Pooled DBS
DBS from five confirmed HIV reactives with known HIV-1 viral load (ranging between 500 and 600 copies/ml) in the corresponding plasma sample and 32 HIV ELISA non-reactive samples were used for standardization. All the five HIV reactive DBS were positive for HIV-1 DNA when tested individually to confirm that DNA could be detected from DBS. Earlier reports have shown a good correlation of HIV-1 viral load (quantitative test) in paired DBS and plasma samples [3, 9]. Initially, the HIV status was determined on DBS collected from all these individuals using a 4th generation HIV ELISA [10].
Three groups of pools with 5, 10 and 20 DBS in each were prepared as shown in Table 1. In each pool only one known HIV reactive sample was included whereas the rest of the DBS were from confirmed HIV non-reactive samples. Thus using five confirmed HIV reactive DBS a total of 15 pools were prepared that included five pools of 5 DBS in each pool, five pools of 10 DBS in each pool and five pools of 15 DBS in each pool. Viral load assay was performed with Roche Amplicor version 1.5 monitor assay, Roche molecular diagnostics, Branchburg, USA. For obtaining DNA from DBS, a 6 mm punch was taken by following the protocol as mentioned in several other studies [8, 11, 12].
Table 1.
Pooling procedure for qualitative DNA PCR standardisation
| Number of DBS pooled together in each pool | Number of pools prepared with each positive DBS | DBS extracted in 200 µl chelex using 6 mm punch | Amount of DNA taken in each test pool (µl) |
|---|---|---|---|
| 5 | 5 | 1 | 10 |
| 10 | 5 | 2 | 10 |
| 20 | 5 | 2 | 5 |
Preparation of DNA Pools from 5 DBS
For the 5 DBS pooling process, only one confirmed HIV-1 positive DBS sample with known viral load copies (S1) was taken along with rest four confirmed HIV negative samples (S2, S3, S4, S5). A 6 mm punch from each DBS was extracted separately with 200 µl chelex reagent provided in the Amplicor HIV-1 DNA test, version 1.5 by following all the steps for extraction as mentioned in kit insert [13]. After extracting all the 5 DBS separately, 10 µl from each extracted DNA sample was pooled together into one PCR amplification tube, thus making the final volume as 50 µl. This pooled extract was further processed by a qualitative PCR.
Preparation of DNA Pools from 10 DBS
For the 10 DBS pooling process, only one confirmed HIV positive DBS sample with known viral load copies (S1) was taken along with rest nine confirmed HIV negative samples (S2, S3, S4, S5, S6, S7, S8, S9, S10). Instead of a single DBS, two 6 mm punches, each taken from 2 separate samples were extracted with 200 µl chelex reagent provided in the Amplicor HIV-1 DNA test, version 1.5 by following all the steps for extraction as mentioned in kit insert [13]. Thus five smaller pools of 2 DBS each were prepared for extraction. After extracting all the five pools of 2 DBS each, 10 µl from each was pooled together into one PCR amplification tube, thus making the final volume as 50 µl. This pooled extract was further processed by a qualitative PCR.
Preparation of DNA Pool from 20 DBS
For the 20 DBS pooling process, only one confirmed positive DBS sample with known viral load copies (S1) was taken long with rest 19 confirmed HIV negative samples (S2–S20). Instead of a single punch, two 6 mm punches, each taken from two separate DBS were extracted with 200 µl chelex reagent provided in the Amplicor HIV-1 DNA test, version 1.5 by following all the steps for extraction as mentioned in kit insert [13]. Thus ten smaller pools of 2 DBS each were prepared for extraction. After extracting all the ten pools of 2 DBS each, 5 µl from each was pooled together into one PCR amplification tube, thus making the final volume as 50 µl. This pooled extract was further processed by a qualitative PCR. Table 1 presents the detailed pooling procedures.
HIV-1 DNA PCR on Pooled DBS from Known HIV-1 Negative Individuals
Dried blood spots (DBS) were collected as part of population-based HIV study of 12,617 respondents (aged 15–49 years) from south India [14] where a blood sample by the finger-prick method was obtained from these respondents on filter paper (Whatman No. 3; Whatman International Ltd, Maidstone, Kent, UK), preferably six drops of blood, without any anticoagulant on it. These blood drops were allowed to dry and were stored in sealed polythene bags with desiccant in the field office at room temperature and within 1 week were transported to the testing laboratory. In the laboratory, DBS were stored under refrigeration at 2–8°C until testing for HIV-1 DNA was performed, maximum being within 1 year of collection. Of these 12,617 respondents tested for HIV infection, 240 were found to be HIV positive. Of the samples negative for both antibody and antigen, a subset of 585 samples belonging to people who were considered at relatively high risk of HIV and another set of 10% of all samples (1238) negative for HIV antibody or antigen underwent qualitative PCR (Amplicor 1.5; Roche molecular diagnostics, Branchburg, USA) testing for HIV viral nucleic acid to detect very recent infections. After the successful standardisation of pooled PCR, DBS from these 1,823 individuals who were negative for both HIV antigen and antibodies were tested for HIV-1 DNA by PCR by the optimized pooling procedure (i.e. using 10 DBS pooling strategy).
DNA from 10 DBS each was pooled together, except for a single pool that had only 3 DBS pooled together as described above. To identify the positive DBS from the positive pool, the DNA extracted from all the ten individual DBS that were included in the positive pool were processed again by HIV-1 PCR.
PCR Procedure
PCR for detecting the presence of HIV-1 proviral DNA in all the above cases was performed by using Amplicor HIV-1 DNA version 1.5 (Roche molecular diagnostics, Branchburg, USA). The Amlicor HIV-1 DNA test, version 1.5 is based on four major processes: sample preparation; PCR amplification of target DNA using HIV-1 specific complementary primers; hybridization of the amplified products to oligonucleotide probes specific to the target; and detection of the probe-bound amplified products by colorimetric determination [13].
Primers
Primers SK145 and SKCC1B supplied in the commercial kit of Amlicor HIV-1 DNA test, version 1.5 are used to amplify a 155-nucleotide sequence of the HIV-1 gag gene [13].
RT and downstream PCR primer SKCC1B complementary to nucleotides 1485–1512 of HIV-1HXB2 is (5′-TACTAGTAGTTCCTGCTATGTCACTTCC-3′) and upstream primer SK145 (5′-AGTGGGGGGACATCAAGCAGCCATGCAAAT-3′).
Preparation of Samples from DBS [2, 13]
A 6 mm punch of DBS was taken into a 1.5 ml screw-cap tube containing 1.0 ml of blood wash solution (BLD WS) and was incubated for 30 min at room temperature. After micro-centrifugation for 3 min at maximum speed, supernatant was aspirated; 1.0 ml of BLD WS was added to each tube, recapped and vortexed. Tubes were microcentrifuged for 3 min at maximum speed. All this was repeated again. Supernatant was aspirated, being careful to avoid disturbing the DBS. The dry DBS was extracted immediately (alternatively it can be stored at −70°C until extraction). 200 µl working Extraction Reagent was added to each DBS and vortexed. Tubes were incubated for 30 min at 60°C ± 2°C in a dry heat block containing sand and then for 30 min at 100°C ± 2°C in a dry heat block containing sand. Samples were vortexed briefly and microcentrifuged for 3 s. 50 µl of samples was added to appropriate PCR reaction tubes containing the working master mix. Rest of the steps for PCR test was performed as mentioned in the kit insert [13].
Results
Standardization of HIV-1 DNA PCR on Pooled DBS
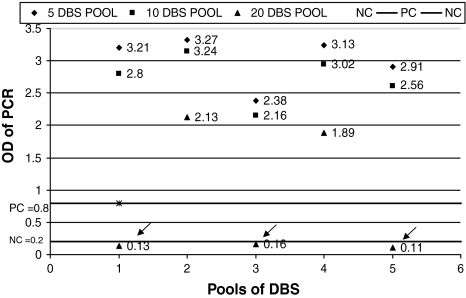
All the negative and positive controls in the HIV-1 DNA PCR runs were valid. Results of all the 15 DNA pools from the DBS are presented in Fig. 1 and Table 2. Internal control (IC) that serves as an extraction and amplification control was observed to have sufficiently high OD values for all 15 pools, indicating that the control DNA in all the pools was properly amplified.
Fig. 1.
Individual results of DBS pooled together for standardization [Arrowheads indicate the pools (20 DBS pooled together) that were reported as negative in spite of a positive sample among each of them]
Table 2.
Standardization results of HIV-1 DNA PCR on pooled DBS
| No. of DBS pooled together | Range of PCR OD for HIV-1 DNA | Range of OD for Internal Control | No. of pools tested | No. of positive pools | No. of negative pools |
|---|---|---|---|---|---|
| 5 DBS | 2.38–3.0 | 2.35–3.0 | 5 | 5 | 0 |
| 10 DBS | 2.16–3.0 | 2.45–3.0 | 5 | 5 | 0 |
| 20 DBS | 0.16–2.13 | 2.48–3.0 | 5 | 2 | 3 |
Specificity, sensitivity, positive and negative predictive values of the assay were all observed to be 100% when tested on pools of 5 and 10 DBS as all the pools of 5 and 10 DBS pooled together were positive for HIV-1 PCR. It was noted that DNA could be detected from only 2/5 pools from the 20 DBS pooled together thus indicating a sensitivity of only 60% and a negative predictive value of 98% though both specificity and positive predictive values remained to be 100%. Thus pooling of 20 DBS that indicated 98.06% overall accuracy of the assay was ruled out. Standardization of pooling procedure accordingly indicated that pooling of 10 DBS gave an optimum result due to its cost effectiveness compared to 5 DBS pooled together and hence was then adopted for pooling the study samples.
HIV-1 DNA PCR on Pooled DBS from Known HIV-1 Negative Individuals
Only one pool from the 183 pools was positive by PCR. From the single PCR positive study DBS pool, after performing PCR on ten individual samples, 1 DBS was found to be positive for HIV-1 DNA PCR. The results of PCR on pooled DBS are presented in Fig. 2. By including this one HIV-1 positive sample that was detected by PCR, total HIV positive samples increased from 240 to 241. This single positive sample was 0.4% of the total 241 HIV positive cases from this population-based study. Pooling strategy also resulted in 88% reduction in overall cost compared to PCR testing on individual sample. It also resulted in a considerable reduction in testing time; ten times less than individual testing.
Fig. 2.
Results of HIV-1 DNA PCR on pooled DBS detected as negative for HIV antibody and antigen
Discussion
In spite of a person screened as negative by a 4th generation HIV assay, only direct monitoring by sensitive NAT would provide accurate data of HIV infection in the population being tested. However, NAT is too expensive and labor-intensive for routine screening of single specimens. Because of the high cost of PCR on individual samples, costs saving strategies have been devised for large numbers of samples. One such strategy is to mix a predetermined number of the samples to be assayed to make a testing pool and process this pool as one sample. A positive result will indicate the presence of the viral DNA in one or more samples within the pool and will necessitate retesting of each of the samples that constituted the pool, so as to identify the truly positive sample(s). On the other hand, a negative result would indicate that none of the samples in the pool contain the viral DNA.
The issue of sensitivity of PCR on pooled samples primarily depends on dilution of the viral DNA. Hence, the number of samples in the pool (pool size) has to be optimized. In our study, the optimum pool size was determined by using varying pool sizes, each containing one known HIV-1 positive sample. The viral DNA could be detected in all the pools with 5 and 10 DBS. However, the 3/5 pools with 20 DBS were recorded false negative, due to the dilution of the DNA below undetectable levels. Since pooling of 10 DBS was more cost effective compared to pools of 5 DBS, it was considered as the optimum pool size and hence was used in our study.
Adopting the pooling strategy on 1,823 DBS from our study samples resulted in detection of an additional one positive sample by HIV-1 DNA PCR, making the total number of HIV positive DBS to 241 out of 12,617 total study samples. This PCR positive sample was actually from the subset of 585 samples belonging to people who were considered at relatively high risk of HIV and probably was from an individual who was either in the early sero-conversion phase of HIV infection or was on anti retroviral therapy, the details of which were not available. Due to insufficient sample, a quantitative PCR could not be performed that would have indicated the viral load of the positive sample and the probable time since sero-conversion. Another set of 10% of all samples (1238) negative for HIV antibody or antigen that underwent qualitative PCR in pools of ten samples remained negative for HIV-1 DNA.
Pooling strategy also resulted in a considerable reduction in overall cost and testing time compared to PCR testing on individual testing. In spite of several advantages of PCR, one limitation of the PCR test is that though theoretically 100% sensitivity and specificity can be achieved, the observed sensitivity of HIV-1 DNA PCR assays in clinical practice is reported to be 96–99% [15–17] due to the undetectable levels of viral DNA in the eclipse phase of infection i.e. 3–7 days from exposure and infection [6].
Pooling strategy can be more cost effective for testing HIV prevalence in research studies conducted on specialized population where the prevalence rates are low. However, such pooling strategy for PCR if applied to screen high risk populations, the number of HIV case detections can be increased cost effectively, only if all samples that are negative for HIV Ag or Ab are pooled together, similar to the strategy adapted in our study. But if all samples from high risk population studies are pooled together, as an initial step, then the pooling strategy may lose its cost effectiveness as due to high prevalence of HIV infection, quite a lot of of the pools will be positive and all samples in the positive pool will need individual testing for detection of HIV-1 DNA in the sample that may lead to higher cost and also will be more time consuming.
Our study has demonstrated that PCR for HIV-1 DNA can be successfully performed on pools of DBS with time and cost savings without compromising the sensitivity of the PCR.
Footnotes
An erratum to this article can be found at http://dx.doi.org/10.1007/s12088-011-0192-4
Contributor Information
Vemu Lakshmi, Email: vemulakshmigorthi@gmail.com.
Talasila Sudha, Email: s_talasila@hotmail.com.
Dandona Rakhi, Email: rakhi.dandona@phfi.org.
Lalit Dandona, Email: lalit.dandona@phfi.org.
References
- 1.Allain J, Laurian Y, Paul DA, Senn D, and members of the AIDS-haemophilia (1986) French Study Group. Serological markers in early stages of human immunodeficiency virus infection in haemophiliacs. Lancet 2:1233–1236 [DOI] [PubMed]
- 2.Cassol S, Salas T, Arella M, Neumann P, Schechter MT, O’Shaughnessy M. Use of dried blood spot specimens in the detection of human immunodeficiency virus type I by the polymerase chain reaction. J Clin Microbiol. 1991;29:667–671. doi: 10.1128/jcm.29.4.667-671.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uttayamakul S, Likanonsakul S, Sunthornkachit R, et al. Usage of dried blood spots for molecular diagnosis and monitoring HIV-1 infection. J Virol Methods. 2005;128(1–2):128–134. doi: 10.1016/j.jviromet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Gupta BP, Jayasuryan N, Jameel S. Direct detection of hepatitis B virus from dried blood spots by polymerase chain reaction amplification. J Clin Microbiol. 1992;30:1913–1916. doi: 10.1128/jcm.30.8.1913-1916.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huges D, Hurd C, Williamson LM. Genotyping for human platelet antigen-1 directly from dried blood spots on cards. Blood. 1996;88:3242. [PubMed] [Google Scholar]
- 6.Busch MP, Satten GA. Time course of viremia and antibody seroconversion following human immunodeficiency virus exposure. Am J Med. 1997;102:117–124. doi: 10.1016/S0002-9343(97)00077-6. [DOI] [PubMed] [Google Scholar]
- 7.Petersen LR, Satten GA, Dodd R, Busch M, Kleinman S, Grindon A, Lenes B, the HIV Seroconversion Study Group Duration of time from onset of human immunodeficiency virus type 1 infectiousness to development of detectable antibody. Transfusion. 1994;34:283–289. doi: 10.1046/j.1537-2995.1994.34494233574.x. [DOI] [PubMed] [Google Scholar]
- 8.Cassol S, Butcher A, Kinard S, et al. Rapid screening for early detection of mother-to-child transmission of human immunodeficiency virus type 1. J Clin Microbiol. 1994;32:2641–2645. doi: 10.1128/jcm.32.11.2641-2645.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alvarez-Munoz MT, Zaragoza-Rodriguez S, Rojas-Montes O, et al. High correlation of human immunodeficiency virus type-1 viral load measured in dried-blood spot samples and in plasma under different storage conditions. Arch Med Res. 2005;36(4):382–386. doi: 10.1016/j.arcmed.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Lakshmi V, Sudha T, Bhanurekha M, Dandona L. Evaluation of the murex HIV Ag/Ab combination assay when used with dried blood spots. Clin Microbiol Infect. 2007;13:1134–1136. doi: 10.1111/j.1469-0691.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- 11.Driver GA, Patton JC, Moloi J, et al. Low risk of contamination with automated and manual excision of dried blood spots for HIV DNA PCR testing in the routine laboratory. J Virol Methods. 2007;146:397–400. doi: 10.1016/j.jviromet.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 12.Luo W, Yang H, Rathbun K, Pau CP, Ou CY. Detection of human immunodeficiency virus type 1 DNA in dried blood spots by a duplex real-time PCR assay. J Clin Microbiol. 2005;43(4):1851–1857. doi: 10.1128/JCM.43.4.1851-1857.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Instruction manual, Amplicor HIV-1 DNA test, version 1.5. Roche diagnostics, Singapore
- 14.Dandona L, Lakshmi V, Sudha T, Kumar GA, Dandona R. A population-based study of human immunodeficiency virus in south India reveals major differences from sentinel surveillance based estimates. BMC Med. 2006;4:31. doi: 10.1186/1741-7015-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mallet F, Hebrard C, Brand D, et al. Enzyme-linked oligosorbent assays for detection of polymerase chain reaction-amplified human immunodeficiency virus type 1. J Clin Microbiol. 1993;31:144–1449. doi: 10.1128/jcm.31.6.1444-1449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauviago S, Barlet V, Guettari N, et al. Standardized nested polymerase chain reaction-based assay for detection of human immunodeficiency virus type 1 DNA in whole blood lysates. J Clin Microbiol. 1993;31:1066–1074. doi: 10.1128/jcm.31.5.1066-1074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whetsell AJ, Drew JB, Milman G, et al. Comparison of three nonradioisotopic polymerase chain reaction-based methods for detection of human immunodeficiency virus type 1. J Clin Microbiol. 1993;30:845–853. doi: 10.1128/jcm.30.4.845-853.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]