Abstract
A collection of 57 enterococcal isolates from different origin (including river, treatment plant, spring and garbage water, soil, animal, and vegetables from Aydın) was screened for the production of bacteriocins. Enterococci were identified at species levels as Enterococcus faecium (34), E. hirae (6), E. casseliflavus (4), E. durans (4), E. faecalis (4), E. mundtii (3) and E. avium (2). Of the 57 isolates 40 of them inhibited the growth of at least one indicator bacterium. Based on our PCR results 54 strains possesed enterocin genes. The genes of entA and entB were the most frequently detected structural genes among the PCR positive strains (54 and 53 strains, respectively) and the entB gene was always associated with entA gene. The highest combination of enterocin genes (24 of 54 strains) detected was entA, entB, entP and entL50A/B. The enterocins AS-48 and CylLLS genes were not found. Three enterococcal isolates, 2 E. faecium and 1 E. hirae were not harbour any of tested enterocin genes. No correlation between the presence of enterocin structural genes and the origin of the strain was detected, also no relationship seemed to exist between the tested enterocin genes and the activity spectra of isolates. Genes encoding bacteriocins are widely disseminated among enterocci from different origin and more studies should be done for evaluate industrial potential of bacteriocins.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-011-0143-0) contains supplementary material, which is available to authorized users.
Keywords: Bacteriocin, Enterococcus, Enterocin Structural Genes, PCR
Introduction
Enterococci represent lactic-acid producing bacteria that can be isolated from different ecosystems such as human, animals, water, soil, plants, food/feed, waste and poultry [1]. Enterococci can produce antimicrobial peptides, called bacteriocins or more specifically enterocins with inhibitory activity against strains closely related to the producer microorganism [2]. It is assumed that bacteriocin production is a bacterial defense mechanism, which gives the producer strain a competitive advantage towards non-producer and bacteriocin sensitive strains in the same niche. In recent years, Franz et al., proposed simplified classification scheme for enterocins; Class I enterocins (lantibiotic enterocins), Class II enterocins (small, non-lantibiotic peptides); Class III enterocins (cyclic enterocins); and Class IV enterocins (large proteins) [3]. Cytolysin belongs to Class I enterocins, it is a two-peptide bacteriocin and both structural subunits contain lanthionie residues [4]. Class II can be subdivided into three subclasses: II.1, enterocin of the pediocin family (enterocin A, enterocin P and bacteriocin 31); II.2, enterocins synthesized without a leader peptide (enterocin L50A/B, and enterocin Q); II.3, other linear, non-pediocin-type enterocins (enterocin B). Class III enterocins includes cyclic antibacterial peptides like enterocin AS-48 and Class IV enterocins are large proteins such as enterolysin A [5]. A major portion of the bacteriocin-producing enterococci have been isolated from foods (cheese, meat, fish and vegetables), animals, and humans [6]. Bacteriocin producing enterococci have also been isolated from municipial wastes [7], sewage sludge [8], rumen content of different ruminants and animal wastes [9].
Many bacteriocins from enterococci have been purified and genetically characterized over the years, and most of them have been obtained from Enterococcus faecalis and Enterococcus faecium. Although a screening for production of bacteriocins by E. faecalis and E. faecium was the subject of several previous studies [10–14], only little information is available about this feature in E. avium, E. hirae, E. mundtii, E. casseliflavus and E. durans isolated from different environments. The aim of this study was to screen the production of antimicrobial activities as well as the presence of bacteriocin structural genes in enterococcal isolates include diverse species isolated from different sources in Turkey.
Materials and Methods
Bacterial Strains
A collection of 57 Enterococcus strains were isolated from different sources including water, soil, animal and vegetable samples. The Enterococci isolated by using m-Enterococcus agar and species identification was based on 16S rDNA sequence analysis. The frozen stocks were stored at −80°C in skimed milk (20% v/v). The following cultures were tested for sensitivity to enterocins: Listeria sp. (food isolate), Listeria innocua DSM 20649, E.coli ATCC 35218, Enterococcus faecalis ATCC 51299, Bacillus cereus ATCC 11778, Staphylococcus aureus ATCC 25923, Micrococcus luteus ATCC 9341.
Bacteriocin Production Assay
For detection of antimicrobial activity, 50 μl of an overnight culture of the indicator strain was added to 5 ml of molten soft BHI broth (Merck, Darmstadt, GERMANY) supplemented with 0.7% agar, mixed, and poured onto a BHI agar plate (Merck, Darmstadt, GERMANY). A single colony of each enterococci to be tested for antimicrobial activity was trasferred with a sterile toothpick to the agar plate seeded with the indicator bacteria [15]. Plates were incubated for 24 h at 37°C in aerobic conditions. The antimicrobial activity was visually detected by observing clear inhibition zones around the tested strain.
PCR Detection of Enterocin Structural Genes
Structural genes (cylLLS, entA, entB, entP, entAS-48, entL50A/B and bac31) of different bacteriocins (Cytolysin L, enterocin A, enterocin B, enterocin P, enterocin AS-48, enterocin L50 A/B and bacteriocin 31 respectively) were studied by PCR in all isolated enterococci using primers included in Table 1. PCR was performed using Techne T-3000 thermocycler. All the PCR reactions were carried out in a final volume of 20 μl containing 1× PCR buffer, 1.5 mM MgCl2, 200 mM each of the four dNTPs, 0.5 mM of each primer and 1.25 units of Taq DNA polymerase. The cycles used were 94°C for 5 min, 94°C for 30 s, 55°C (Ent A, B and P), 57°C (Bac 31 and AS-48), 58°C (Cylls and Ent L50A/B) for 30 s and 72°C for 45 s for the next 35 cycles, 72°C for 5 min were used fort the last cycle. Positive and negative controls were included in all assays, except for entAS-48 and CylLLS.
Table 1.
Specific primers used for PCR detection of enterocin structural genes
| Gene | Sequence (5′-3′) | Fragment (bp) | References |
|---|---|---|---|
| entA | F: GGTACCACTCATAGTGGAAA | 138 | [10] |
| R: CCCTGGAATTGCTCCACCTAA | |||
| entB | F: CAAAATGTAAAAGAATTAAGTACG | 201 | [10] |
| R: AGAGTATACATTTGCTAACCC | |||
| entP | F: GCTACGCGTTCATATGGTAAT | 87 | [10] |
| R: TCCTGCAATATTCTCTTTAGC | |||
| entL50A/B | F: ATGGGAGCAATCGCAAAATTA | 274 | [10] |
| R: TAGCCATTTTTCAATTTGATC | |||
| AS-48 | F: GAGGAGTATCATGGTTAAAGA | 339 | [10] |
| R: ATATTGTTAAATTACCAA | |||
| bact31 | F: CCTACGTATTACGGAAATGGT | 130 | [10] |
| R: GCCATGTTGTACCCAACCATT | |||
| cylLLS | F: GGCGGTATTTTTACTGGAGT | 248 | [4] |
| R: CCTACTCCTAAGCCTATGGTA |
Results and Discussion
The Enterococci used in this study were isolated from different water, soil, animal and vegetable samples in Aydın (Turkey). Based on 16S rDNA sequence analysis strains were identified as E. faecium (34), E. hirae (6), E. faecalis (4), E. casseliflavus (4), E. durans (4), E. mundtii (3), E. avium (2). Strains were collected from the following sources river water (n = 9), soil (n = 4), treatment water plant (n = 12), sheep and cattle rectums and caecums (n = 11), spring water (n = 11), garbage water (n = 3), vegetable (n = 7) (Table 2).
Table 2.
Sources, harboured enterocin genes and inhibitory spectrum of enterococcal isolates
| Isolate | Source | Enterocin genes | Inhibition | ||||
|---|---|---|---|---|---|---|---|
| Listeria sp. | L. innocua | E. faecalis | B. cereus | S. aureus | |||
| E. faecium HBE-1 | River water | A, B, bac31, L50A/B | + | + | + | – | – |
| E. faecium HBE-2 | River water | A, B, L50A/B | + | + | + | – | – |
| E. faecium HBE-3 | River water | A, B, L50A/B | +++ | +++ | ++ | – | – |
| E. casseliflavus HBE-4 | River water | A, B, L50A/B | – | + | – | – | + |
| E. faecium HBE-5 | River water | A, B | – | – | – | – | – |
| E. casseliflavus HBE-6 | River water | A, B | + | + | – | + | + |
| E. faecium HBE-7 | River water | A, B, P | +++ | +++ | ++ | – | + |
| E. faecium HBE-8 | River water | A, B, P, L50A/B | +++ | +++ | ++ | – | + |
| E. faecium HBE-9 | River water | A, B, P | +++ | +++ | ++ | – | + |
| E. hirae HBE-10 | Agricultural soil | – | – | – | – | – | – |
| E. hirae HBE-11 | Agricultural soil | A, B | – | – | – | – | – |
| E. durans HBE-12 | Agricultural soil | A, B | +++ | +++ | ++ | – | + |
| E. faecium HBE-13 | Agricultural soil | A, B | – | – | – | – | + |
| E. faecium HBE-14 | Treatment plant water | A, B, P | – | – | – | – | – |
| E. faecalis HBE-15 | Treatment plant water | A, B, P | – | – | – | – | – |
| E. faecalis HBE-16 | Treatment plant water | A, B, P, L50A/B | – | – | – | – | – |
| E. faecium HBE-17 | Treatment plant water | A, B, P, L50A/B | +++ | +++ | ++ | – | + |
| E. faecium HBE-18 | Treatment plant water | A, B, P | – | – | – | – | + |
| E. faecium HBE-19 | Treatment plant water | – | – | – | – | – | + |
| E. faecium HBE-20 | Treatment plant water | A, B, P | – | – | – | – | + |
| E. faecium HBE-21 | Treatment plant water | A, B, P | – | – | – | – | + |
| E. faecalis HBE-22 | Treatment plant water | A, B, P | – | – | – | – | – |
| E. faecium HBE-23 | Treatment plant water | A, B, P | ++ | ++ | ++ | – | + |
| E. faecium HBE-24 | Treatment plant water | A, B, P | ++ | ++ | ++ | – | + |
| E. faecium HBE-25 | Treatment plant water | A, B, P | – | – | – | – | – |
| E. faecium HBE-26 | Sheep rectum | A, B, P | + | – | – | + | + |
| E. faecium HBE-27 | Sheep rectum | A, B, P | – | – | – | + | + |
| E. hirae HBE-28 | Sheep caecum | A, B, P | – | – | – | – | – |
| E. hirae HBE-29 | Sheep caecum | A, B, P, L50A/B | – | – | – | – | – |
| E. faecium HBE-30 | Sheep caecum | A, B,P, L50A/B | – | – | – | – | + |
| E. faecium HBE-31 | Cattle rectum | A, B, L50A/B | ++ | ++ | + | – | + |
| E. faecium HBE-32 | Cattle rectum | A, B, L50A/B | ++ | ++ | + | + | |
| E. faecium HBE-33 | Cattle rectum | A, B | – | – | – | – | + |
| E. faecium HBE-34 | Cattle caecum | A, B, L50 A/B | – | – | – | – | + |
| E. faecium HBE-35 | Cattle caecum | A, B, P, L50A/B | +++ | +++ | ++ | – | + |
| E. faecium HBE-36 | Cattle caecum | – | – | – | – | – | + |
| E. faecalis HBE-37 | Spring water | A, B, P, L50A/B | – | – | – | – | – |
| E. avium HBE-38 | Spring water | A, B, P, L50A/B | – | – | – | – | + |
| E. avium HBE-39 | Spring water | A, B, P, L50A/B | – | – | – | – | – |
| E. casseliflavus HBE-40 | Spring water | A, B, P, L50A/B | – | – | – | + | + |
| E. casseliflavus HBE-41 | Spring water | A, B, P, L50A/B | – | + | – | – | + |
| E. mundtii HBE-42 | Spring water | A, B, P, L50A/B | – | + | – | – | – |
| E. faecium HBE-43 | Spring water | A, P, L50A/B | – | – | – | – | + |
| E. faecium HBE-44 | Spring water | A, B, P, L50A/B | – | – | – | – | + |
| E. hirae HBE-45 | Spring water | A, B, P, L50A/B | – | – | – | – | – |
| E. durans HBE-46 | Spring water | A, B, P, L50A/B | + | + | – | – | – |
| E. durans HBE-47 | Spring water | A, B, L50A/B | – | – | – | – | – |
| E. faecium HBE-48 | Garbage water | A, B, P, L50A/B | +++ | +++ | ++ | – | – |
| E. faecium HBE-49 | Garbage water | A, B, P, L50A/B | +++ | +++ | ++ | – | + |
| E. durans HBE-50 | Garbage water | A, B, P, L50A/B | + | + | + | – | + |
| E. mundtii HBE-51 | Vegetable | A, B, P, L50A/B | ++ | ++ | + | + | – |
| E. hirae HBE-52 | Vegetable | A, B, L50A/B | – | – | – | – | – |
| E. mundtii HBE-53 | Vegetable | A, B, P, L50A/B | – | – | – | – | – |
| E. faecium HBE-54 | Vegetable | A,B, P, L50A/B | +++ | +++ | ++ | – | + |
| E. faecium HBE-55 | Vegetable | A, B, P, L50A/B | +++ | +++ | ++ | – | + |
| E. faecium HBE-56 | Vegetable | A, B, P, L50A/B | +++ | +++ | ++ | – | – |
| E. faecium HBE-57 | Vegetable | A, B, P, L50A/B | – | – | – | – | – |
+ weak inhibition, ++ moderate inhibition, +++ strong inhibition
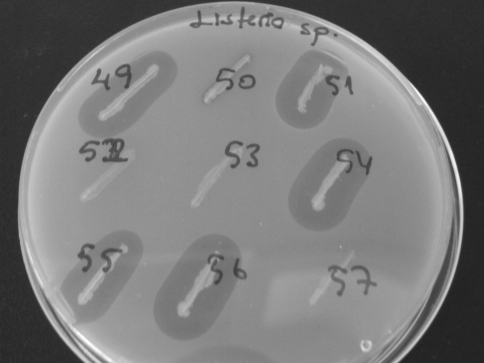
The 57 enterococcal isolates were screened for antibacterial activity since they may be produce enterocins to control the growth of the tested indicator bacteria (Fig. 1). Table 2 shows the profile of the inhibitory effects exhibited by the enterococcal strains, as well as the specific bacteriocin genes harboured by them.
Fig. 1.
Examples of enterococcal isolates and their inhibition zones against Listeria sp
Of the 57 isolates screened, 40 of them (70.2%) were found to be effective against at least one of the indicator bacteria. The isolates having antilisterial activity included 18 E. faecium, 3 E. casseliflavus, 3 E. durans and 2 E. mundtii. Of the 40 isolates with antibacterial activity, 12 of them were found to be inhibitory only against to S. aureus (11 E. faecium and 1 E. avium). The similar results were reported in previous studies that most enterocin producing enterococci displayed antilisterial activity, while a fewer portion of them also showed antibacterial spectrum activity against Staphylococcus spp., Bacillus spp. and Clostridium spp [3, 10, 16–18]. No correlation was found between the origin of the strains and the inhibitory spectrum.
According to our PCR results occurence of enterocin structural genes seems to be widespread among enterococcal strains isolated from different sources and high frequency of the occurence of enterocin structural genes (94.7%) was observed (Fig. 2). It was reported that, in a collection of 61 enterococci isolated from environmental sources like surface and waste waters, sheep manure, enterocin structural gene positive strains were found to be 57.4% [22]. In another research, 64 of 122 (52.5%), inhibitory Enterococcus strains isolated from food and feed, animal, clinical and non-clinical samples, were found to harbour at least one enterocin structural gene [10]. None of the bacteriocin genes tested in this study were demonstrated in two strains of E. faecium (HBE-19, HBE-36) and E. hirae HBE-10, which showed antibacterial activity against S. aureus. Genes encoding new bacteriocins or other non-tested known bacteriocins could be included in those isolates. Although the detection of enterocin structural genes must not necessiraly correspond with the production of the bacteriocin, it can be speculated that various environments require various level of bacterial defense mechanism thus also the presence of bacteriocin genes as the basis for bacteriocin production [14].
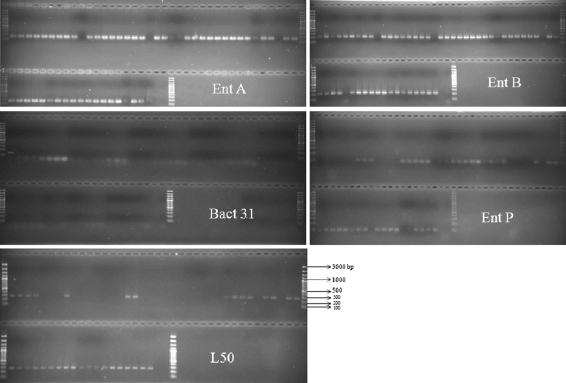
Fig. 2.
Amplification gel pictures of the enterocin structural genes of enteroccal isolates. Lanes 1,40, 41 and 63 are 100 bp-DNA ladder. Lanes after ladder are isolate numbers in order of HBE1-HBE57, last lane before ladder is negative control
Bacteriocin production seemed to be correlated to the species involved in this study; all E. hirae (6) and E. faecalis (4) were non-producers. The other strains had no activity against none of the indicator bacteria were E. faecium (4), E. mundtii (1), E. avium (1) and E. durans (1). All of these non-producer isolates except for E. hirae HBE-10, had various combination of enterocin genes. The detection of enterocin structural gene does not means the production of the corresponding enterocin [14] and existence of silent bacteriocin genes hes been previously reported in other studies [2, 19, 20]. It emphasized by Nes et al., [21] identification of putative bacteriocin genes does not necessarily mean that the relevant bacterium produces antimicrobial activity and a lack of detectable antimicrobial activity does not necessarily mean that genes involved in bacteriocin production are defective. First, because of some peptide bacteriocins act only a narrow range of target bacteria, it is of key importance to use a susceptible indicator. Secondly, the production of peptide bacteriocins is often regulated. Deficiency in production of antimicrobial activity is often due to a dysfunctional genetic system.
The structural genes of Enterocins A and B were shown to be most frequent genes (100 and 98.1%) among PCR positive strains, respectively in this study. Enterocin B gene was always found to be together with enterocin A gene. Similar to our results the enterocin B structural gene was always associated with the presence of enterocin B gene in previous studies due to the fact that no transport genes have been found for enterocin B producers [10, 23].
Enterocin P and Enterocin L50 A/B were found to be 72.2 and 62.9%, respectively. Enterocins A and P are belong to pediocin family and they are grouped in Class II.1 bacteriocins, which are very effective for growth of listeriae [24]. Enterocin B classified in the subgroup II.3, non-pediocin type enterocins. The individual peptides enterocin L50A and enterocin L50B possesed antimicrobial activity, with the L50A peptide being most active and enterocin L50 is grouped in Class II.c, leaderless type.
The combination of two different genes was observed in 6 strains (11.1% of PCR+ strains). Three different genes were detected in 21 strains (38.9%) and four different genes were present in 25 strains (46.3%). The wide enterocin genes distribution may be due to remarkable ability of enterococci to disseminate and receive genetic material between strains but also between genera [14].
Cytolysin and enterocin AS-48 genes were not found any of examined enterococci. Cytolysin is the only two-peptide lantibiotic isolated from enterococci with cytolytic (hemolytic) activity. Cytolysin is a virulence factor and consequently it is not considered useful as an antimicrobial agent [22]. The structural gene of bacteriocin AS-48 has been examined in enterococcal isolates and bacteriocins that very closely related or identical to peptide AS-48 were detected in E. faecalis and E. faecium isolates [25].
In conclusion, 70.2% of the enterococcal isolates included in this study showed antimicrobial activity against at least one of indicator bacteria and 94.7% of isolates harboured various enterocin structural genes. Enterocin structural genes are widely distributed and they can be found in possible combinations together. The presence of four enterocin genes in 25 strains in this study indicates the high genetic potential of many strains to produce various enterocins. Because of these properties screening of enterocin genes may offer great possibilities for isolation of bacteriocin producing strains and their use in the food industry.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
References
- 1.Devriese LA, Hommez J, Wijfels R, Haesebrouck F. Composition of the Enterococcal and Streptococcal intestinal flora of poultry. J Appl Bacteriol. 1991;71:46–50. [PubMed] [Google Scholar]
- 2.Poeta P, Igrejas G, Costa D, Sargo R, Rodrigues J, Torres C. Virulence factors and bacteriocins in faecal enterococci of wild boars. J Basic Microbiol. 2008;48:385–392. doi: 10.1002/jobm.200700327. [DOI] [PubMed] [Google Scholar]
- 3.Franz CM, Belkum MJ, Holzapfel WH, Abriouel H, Galvez A. Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol Rev. 2007;31:293–310. doi: 10.1111/j.1574-6976.2007.00064.x. [DOI] [PubMed] [Google Scholar]
- 4.Booth MC, Bogie CP, Sahl HG, Siezen RL, Hatter KL, Gilmore MS. Structural analysis and proteolytic activation of Enterococcus faecalis cytolysin, a novel lantibiotic. Mol Microbiol. 1996;21:1175–1184. doi: 10.1046/j.1365-2958.1996.831449.x. [DOI] [PubMed] [Google Scholar]
- 5.Nilsen T, Nes IF, Holo H. Enterolysin A, a cell wall degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl Environ Microbiol. 2003;69:2975–2984. doi: 10.1128/AEM.69.5.2975-2984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foulquie Moreno MR, Sarantinopoulos P, Tsakalidou E, DeVuyst L. The role and application of enterococci in food and health. Int J Food Microbiol. 2006;106:1–24. doi: 10.1016/j.ijfoodmicro.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 7.Lauková A, Jurin P. Distribution and characterization of Enterococcus species in municipal sewages. Microbios. 1997;97:73–80. [PubMed] [Google Scholar]
- 8.Marekova M, Laukova A, Skaugen M, Nes I. Isolation and characterization of a new bacteriocin, termed enterocin M, produced by environmental isolate Enterococcus faecium AL41. J Ind Microbiol Biotechnol. 2007;34:533–537. doi: 10.1007/s10295-007-0226-4. [DOI] [PubMed] [Google Scholar]
- 9.Laukova A, Marekova M. Production of bacteriocins by different Enterococcal isolates. Folia Microbiol. 2001;46:49–52. doi: 10.1007/BF02825884. [DOI] [PubMed] [Google Scholar]
- 10.Vuyst L, Foulquie Moreno MR, Revets H. Screening for enterocins and detection of hemolysin and vancomycin resistance in enterococci of different origins. Int J Food Microbiol. 2003;84:299–318. doi: 10.1016/S0168-1605(02)00425-7. [DOI] [PubMed] [Google Scholar]
- 11.Klibi N, Jouini A, Rojo-Bezares B, Masmoudi A, Ruiz-Larrea F, Boudabous A, Torres C. Phenotypic and genotypic characterization of bacteriocins in clinical enterococcal isolates of Tunisia. World J Microbiol Biotechnol. 2008;24:653–657. doi: 10.1007/s11274-007-9519-z. [DOI] [Google Scholar]
- 12.Sabia C, Niederhausern S, Guerrieri E, Messi P, Anacarso I, Monicardi G, Bondi M. Detection of bacteriocin production and virulence traits in vancomycin-resistant enterococci of different sources. J Appl Microbiol. 2008;104:970–979. doi: 10.1111/j.1365-2672.2007.03612.x. [DOI] [PubMed] [Google Scholar]
- 13.Strompfova V, Laukova A. In vitro study on bacteriocin production of Enterococci associated with chickens. Anaerobe. 2007;13:228–237. doi: 10.1016/j.anaerobe.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Strompfova V, Laukova A, Simonova M, Marcinakova M. Occurence of the structural enterocin A, P, B, L50B genes in enterococci of different origin. Vet Microbiol. 2008;132:293–301. doi: 10.1016/j.vetmic.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Del Campo R, Tenorio C, Jimenez-Diaz R, Rubio C, Gomez-Lus R, Baquero F, Torres C. Bacteriocin production in vancomycin-resistant and vancomycin-susceptible Enterococcus isolates of different origins. Antimicrob Agents Chemother. 2001;45:905–912. doi: 10.1128/AAC.45.3.905-912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cleveland J, Montville TJ, Nes IF, Chikindas ML. Bacteriocins: safe, natural antimicrobials for food preservation. Int J Food Microbiol. 2001;71:1–20. doi: 10.1016/S0168-1605(01)00560-8. [DOI] [PubMed] [Google Scholar]
- 17.Deegan LH, Cotter PD, Hill C, Ross P. Bacteriocins: biological tools for bio-preservation and shelf-life etension. Int Dairy J. 2006;16:1058–1071. doi: 10.1016/j.idairyj.2005.10.026. [DOI] [Google Scholar]
- 18.Giraffa G. Enterococcal bacteriocins: their potential as antilisteria factors in dairy technology. Food Microbiol. 1995;12:291–299. doi: 10.1016/S0740-0020(95)80109-X. [DOI] [Google Scholar]
- 19.Eaton TJ, Gasson MJ. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol. 2001;67:1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Semedo T, Santos MA, Lopes MF, Figueiredo Marques JJ, Barreto Crespo MT, Tenreiro R. Virulence factors in food, clinical and reference enterococci: a common trait in the genus? Syst Appl Microbiol. 2003;26:13–22. doi: 10.1078/072320203322337263. [DOI] [PubMed] [Google Scholar]
- 21.Nes IF, Diep DB, Holo H. Bacteriocin diversity in Streptococcus and Enterococcus. J Bacteriol. 2007;189:1189–1198. doi: 10.1128/JB.01254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pangallo D, Harichova J, Karelova E, Drahovska H, Chovanova K, Feriane P, Turna J, Timko J. Molecular investigation of enterococci isolated from different environmental sources. Biologia. 2004;59:829–837. [Google Scholar]
- 23.Franz CM, Worobo RW, Quadri LEN, Schillinger U, Holzapfel WH, Vederas JC, Stiles ME. Atypical genetic locus associated with constitutive production of enterocin B by Enterococcus faecium BFE 900. Appl Environ Microbiol. 1999;65:2170–2178. doi: 10.1128/aem.65.5.2170-2178.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eijsink VG, Skeie M, Middelhoven PH, Brurberg MB, Nes IF. Comparative studies of class IIa bacteriocins of lactic acid bacteria. Appl Environ Microbiol. 1998;64:3275–3281. doi: 10.1128/aem.64.9.3275-3281.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joosten HMLJ, Rodriguez E, Nunez M. PCR detection of sequences similar to the AS-48 structural gene in bacteriocin producing enterococci. Lett Appl Microbiol. 1997;24:40–42. doi: 10.1046/j.1472-765X.1997.00349.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.