Abstract
Urinary tract infection (UTI) with E. coli (UPEC) is one of the most common bacterial infections among human beings. In addition to the host predisposing factors, genes are also proposed to have an important role in the occurrence of UTIs. This study investigated the distribution of three pathogenic genes including aggR, aap and aatA among UPEC infected samples and their linkage with stbA, the essential gene for maintaining of pAA plasmid. A total of 244 samples were collected from patients with UTIs through clinical laboratories located in western side of Tehran (Iran) during years 2008–2009. E. coli isolation was performed according to standard laboratory methods. DNAs were extracted from samples using Boiling method, and the presence of aap, aggR, aatA and stbA genes were investigated by PCR. No pathogenic genes (aap, aggR, aatA) were found in 104 out of 244 UPEC samples, while 14 of them were carrying stbA gene. Out of 140 UPEC samples with pathogenic genes, 94 (46.6%) were carrying aap gene, 52 (23%) aggR gene, and 80 (35.4%) aatA gene. A total of 18 samples were also carrying all pathogenic genes together. Moreover, 44 out of 144 samples were carrying stbA gene. The results obtained by this study showed that the aggR, aap and aatA pathogenic genes have different existence patterns in different E. coli strains that infect different organs. Our study also showed that these three plasmid genes in EAEC strains are able to transpose in the genome and change their level of linkage with pAA plasmid essential gene stbA. Meanwhile, this study confirmed that aggR, aap and aatA genes are not specific to only EAEC strains.
Keywords: Urinary tract infections, Uropathogenic Escherichia coli, Plasmid pAA
Introduction
Urinary tract infections (UTIs) are among the most prevalent infections in human beings. Surveys conducted around the world have indicated that intestinal bacteria (family Enterobacteriaceae) are the main causes of UTIs, of which Escherichia coli is the most common one. To create an infection, fecal bacteria colonize at the entrance of the urinary tracts where they can enter into the internal organs such as bladder and kidney to make infections. Regarding the origin of intestinal bacteria and their moving out of the digestive system due to change in the conditions and horizontal transfer, they undergo some genetic differences [1, 10, 12]. Enteroaggregative E. coli (EAEC) is a pathogenic strain in the digestive tract that causes a severe and stable diarrhea [3].
The EAEC pathogenic genes, such as Fembria, enterotoxin, aggR, aap and aatA are mostly located on the 55,989 bp plasmid or pAA. Estimates show that 50–100% of EAEC bacteria carry pAA plasmid. The aap gene produces anti-aggregation protein that forms a bacterial capsule with the capacity to prevent bacterial accumulation and helps bacteria to disperse around.
The aatA gene produces a membrane protein that is part of aat-PABCE transmitter system and is necessary for translocation of pathogenic proteins such as aap. The AggR gene also produces a transcription activator for several pathogenic genes [2, 4–6, 13]. Since pAA plasmid carries many transposable elements, the presence of these genes is not in direct relationship with the presence of the plasmid.
In contrast, stbA (Stable plasmid inheritance protein A) gene, another gene located on pAA, has no role in the pathogenesis [2]. This gene produces a protein that is necessary for the low copy number plasmid to be passed to the daughter cells. Therefore, it is presumed that the transferring of plasmid to the daughter cells is directly dependent to the presence of stbA gene.
In this study, we examined the distribution of aap, aggR, aatA, and stbA genes and their linkage with pAA plasmid in the Iranian patients with UPEC infection.
Materials and Methods
A total of 244 Uropathogenic Escherichia coli were collected after culturing the samples obtained from UTI patients with (colony cunt > 100,000 CFU/ml) the highest level of infection on EMB and Blood agar medium. Sample collection was done through medical laboratories located in west of Tehran, Iran. The E. coli strains were determined using IMVIC biochemical tests and API kit (API Rapid 20E, Biomerieux, USA) according to manufacturer’s instructions.
The total DNAs were extracted from bacteria by boiling method. The Multiplex PCR method introduced by Cerna et al. [2] was used to detect aggR, aap and aatA pathogenic genes. The StbA gene amplified separately using a set of primers designed by GeneRunner software (GenBank Accession no. CU928159).
A full loop of bacterial colony resuspended into 200 μl of distilled water. The suspension was boiled for 5 min and then centrifuged at 14,000 rpm for 5 min to form cell lysate pellet.
A 5-μl aliquot of the supernatant was used as template for PCR amplification. Multiplex-PCR was carried out using thermal cycler (Eppendorf, Germany) in a total volume of 50 μl containing 10 pmol of each three pairs of primers (Table 1), 25 μmol of deoxynucleoside triphosphates, 5 μl of 10× Taq buffer [50 mM KCl, 10 mM Tris–HCl (pH 8.3)], 2 mM MgCl2 and 2.5 U of Taq polymerase (Fermentas, USA).
Table 1.
Primers used for PCR
| Primer concentration (pmol) | PCR product size (bp) | Primer sequence | Gene | Reference |
|---|---|---|---|---|
| 10 | 310 | F CTT GGG TAT CAG CCT GAA TG | aap | [2] |
| R AAC CCA TTC GGT TAG AGC AC | ||||
| 15 | 457 | F CTA ATT GTA CAA TCG ATG TA | aggR | [2] |
| R AGA GTC CAT CTC TTT GAT AAG | ||||
| 20 | 629 | F CTG GCG AAA GAC TGT ATC AT | aatA | [2] |
| R CAATGT ATA GAA ATC CGC TGT T | ||||
| 10 | 200 | F CAAACCTGGCTATTGCT | stbA | Current study |
| R ATCCACTATTACATCATCGAAC |
F forward, R reverse
The Multiplex-PCR condition was set up as follows: 5 min at 95°C as initial denaturation; 45 s at 95°C as denaturation phase, 35 s at 57°C as annealing phase, and 45 s at 72°C as extension phase, for 40 cycles; and 5 min at 72°C as final extension. The stbA gene was also amplified in a total volume of 25 μl containing 10 pmol of each primers (Table 1), 25 μmol of deoxynucleoside triphosphates, 3 μl of template, 2.5 μl of 10× Taq buffer [50 mM KCl, 10 mM Tris–HCl (pH 8.3)], 1 mM MgCl2 and 2 U of Taq polymerase (Fermentas, USA).
The stbA PCR condition was set up as follows: initial denaturation for 5 min at 95°C; 40 cycles of denaturation for 45 s at 95°C, annealing for 35 s at 57°C, and extension for 45 min at 72°C; followed by the final extension for 5 min at 72°C. PCR products were separated by gel electrophoresis on a 2% agarose gel. In order to confirm the accuracy of genes amplified in this study, a PCR product of each gene was sent for sequencing to the Macrogen Company (South Korea) and the result was confirmed by NCBI Blast Tool.
Results
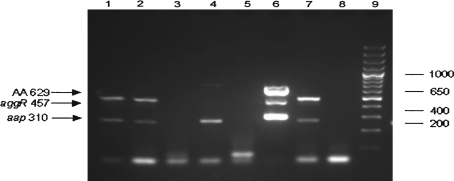
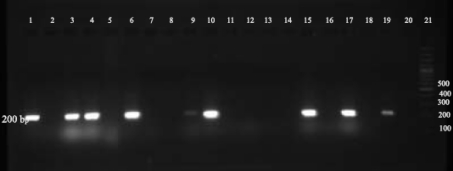
The results obtained by amplification of aggR, aap and aatA genes are shown in Fig. 1. The result obtained by amplification of stbA gene is also shown in Fig. 2.
Fig. 1.
Gene amplification result of pathogenic genes aggR, aap and aatA run on 2% agarose gel. Rows 1–7E. coli isolated from patients, row 8 negative control and row 9 size marker 100 bp
Fig. 2.
The stbA gene amplification results of UPEC samples on 2% agarose gel. Rows 1–19 PCR products of some samples, row 20 negative control and row 21 100 bp size marker
The genotypes of all samples are presented in Table 2. Out of 244 UPEC samples, 104 had no pathogenic genes (aap, aggR, and aatA) while 14 of them were carrying stbA gene. Of 140 UPEC samples with pathogenic genes, 94 (46.6%) were carrying aap gene, 52 (23%) aggR gene, and 80 (35.4%) aatA gene, with a total of 18 samples carrying all three pathogenic genes together. 44 out of 144 samples were also carrying stbA gene (Table 2).
Table 2.
The results of presence or absence aggR, aap, aatA and stbA genes by PCR
| UPEC samples n = 244 |
aatA | aap | aggR | stbA |
|---|---|---|---|---|
| 104 | (−) | (−) | (−) | 14/104(+) |
| 26 | (−) | (+) | (−) | 6/26(+) |
| 18 | (−) | (−) | (+) | 8/18(+) |
| 28 | (+) | (−) | (−) | 4/28(+) |
| 16 | (−) | (+) | (+) | 2/16(+) |
| 34 | (+) | (+) | (−) | 12/34(+) |
| 18 | (+) | (+) | (+) | 12/18(+) |
Discussion
Pathogenic Escherichia coli strains including EAEC as the cause of diarrhea and UPEC as the cause of UTI, have the common pathogenic factors including adhesion and Fimbria [1]. In UPEC strains, kps, afa, sfa, auf, pap, usp, ireA, prs, aer, cnf and hly are the genes which participate in the pathogenicity; while aap, aatA, aggc, agg3c, aggR, eae, ehly, iha, irp2, lpfA, pet, pic, pils, shf, astA genes are those participating in EAEC pathogenicity [5–7, 9]. In the investigation carried out by Abe et al. [1], it was shown that UPEC strain might acquire same properties as EAEC strain, which means some genes can be shared between these two strains. In the study done by Cerna et al. [2] it was determined that 24 (86%) out of 28 EAEC strain were carrying pAA pathogenic genes. 23 samples had all three aatA, aggR, aap plasmid genes, while one sample carried only aap gene. Out of 70 genes amplified by PCR, 23 genes (32.8%) were aggR, 23 genes (32.8%) were aatA (AA), and 24 genes (34.4%) were aap [2]. The study done by Gadsden et al. [8] showed that unlike the EAEC strain, UPEC strain has no aatA, aggR, aap genes. Also, Monteiro et al. [11] demonstrated that aap gene is not specific for EAEC strain. However, the results obtained by our study showed that 140 (57.4%) out of 244 UPEC samples contained one, two or three aatA, aggR, aap genes. We identified 226 genes by PCR, of which 94 (46.6%), 52 (23%), and 80 (35.4%) were aap, aggR, and aatA, respectively. In our study, the stbA stabilizer gene was also amplified by PCR to determine the presence of pAA plasmid. To the best of our knowledge, this study is the first study done in Iranian population.
Interestingly, it seems that the gene existence patterns for aggR, aap and aatA plasmid genes tend to change when the bacterium moves from the intestine to the urinary tract from 86% in the intestine to 57.4% in the urinary tract. In our study, only 44 out of 140 samples with aggR, aap, and aatA genes had stbA gene; while one out of 14 samples with none of aggR, aap and aatA genes had stbA gene. These results show that the pathogenic genes can exit from the plasmid and re-locate in another place in the genome, and therefore, greatly decrease their linkage with pAA plasmid. Meanwhile, this study shows that the aggR, aap and aatA pathogenic genes are not specific to the EAEC strain. It seems that despite the numerous transposons on plasmids and bacterial genome and also a decreased level of evolutionary pressure, a logical change occurs in the existence patterns of these genes.
Acknowledgments
This study was part of a MSc thesis done in the Islamic Azad University, Tonekabon branch. The authors would like to gratitude all people who helped and participated in sample collection.
References
- 1.Abe CM, Salvador FA, Falsetti IN, Vieira MA, Blanco J, Blanco JE, Blanco M, Machado AM, Elias WP, Hernandes RT, Gomes TA. Uropathogenic Escherichia coli (UPEC) strains may carry virulence properties of diarrhoeagenic E. coli. FEMS Immunol Med Microbiol. 2008;52:397–406. doi: 10.1111/j.1574-695X.2008.00388.x. [DOI] [PubMed] [Google Scholar]
- 2.Cerna JF, Nataro JP, Garica TE. Multiplex PCR for detection of three plasmid-borne genes of enteroaggregative Escherichia Coli strains. J Clin Microbiol. 2003;41:2138–2140. doi: 10.1128/JCM.41.5.2138-2140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weintraub A. Enteroaggregative Escherichia coil: epidemiology, virulence and detection. J Med Microbiol. 2007;56:4–5. doi: 10.1099/jmm.0.46930-0. [DOI] [PubMed] [Google Scholar]
- 4.Huang DB, Mohanty A, DuPont HL, Okhuysen PC, Chiang T. A review of an emerging enteric pathogen: enteroaggreative Escherichia coil. J Med Microbiol. 2006;55:1303–1311. doi: 10.1099/jmm.0.46674-0. [DOI] [PubMed] [Google Scholar]
- 5.Park HK, Jung YJ, Chae HC, Shin YJ, Woo SY, Park HS, Lee SJ. Comparison of Escherichia coli uropathogenic genes (kps, usp and ireA) and enteroaggregative genes (aggR and aap) via multiplex polymerase chain reaction from suprapubic urine specimens of young children with fever. Scand J Urol Nephrol. 2009;43:51–57. doi: 10.1080/00365590802299338. [DOI] [PubMed] [Google Scholar]
- 6.Buckles EL, Mougeot B, Molina A, Lockatell CV, Johnson DE, Drachenberg CB, Burland V, Blattner FR, Donnenberg MS. Identification and characterization of a novel Uropathogenic Escherichia coli-associated fimbrial gene cluster. Infect Immun. 2004;72:3890–3901. doi: 10.1128/IAI.72.7.3890-3901.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boisen N, Struve C, Scheutz F, Krogfelt KA, Nataro JP. New adhesin of enteroaggregative Escherichia coli related to the Afa/Dr/AAF family. Infect Immun. 2008;76:3281–3292. doi: 10.1128/IAI.01646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gadsden FW, Johnson JR, Wain J, Okeke IN. Enteroaggregative Escherichia coli related to Uropathogenic Clonal Group A. Emerg Infect Dis. 2007;13:757–760. doi: 10.3201/eid1305.061057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lloyd AL, Rasko DA, Mobley HT. Defining genomic islands and uropathogen-specific genes in Uropathogenic Escherichia coli. J Bacteriol. 2007;189:3532–3546. doi: 10.1128/JB.01744-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson JR, Russo TA. Molecular epidemiology of extraintestinal pathogenic (uropathogenic) Escherichia coli. Int J Med Microbiol. 2005;295:383–404. doi: 10.1016/j.ijmm.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Monteiroa BT, Camposb LC, Sircilia MP, Franzolina MR, Bevilacquaa LF, Nataroc JP, Eliasa WP. The dispersin-encoding gene (aap) is not restricted to enteroaggregative Escherichia coli. Diagn Microbiol Infect Dis. 2009;65:81–84. doi: 10.1016/j.diagmicrobio.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 12.Wiles TJ, Kulesus RR, Mulvey MA. Origins and virulence mechanisms of Uropathogenic Escherichia coli. Exp Mol Pathol. 2008;85:11–19. doi: 10.1016/j.yexmp.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imuta N, Nishi J, Tokuda K, Fujiyama R, Manago K, Iwashita M, Sarantuya J, Kawano Y. The Escherichia coli efflux pump TolC promotes aggregation of enteroaggregative Ecoli 042. Infect Immun. 2008;76:1247–1256. doi: 10.1128/IAI.00758-07. [DOI] [PMC free article] [PubMed] [Google Scholar]