Abstract
Beauveria bassiana is a biocontrol agent which shows entomopathogenecity on insect pests especially the Lepidopterons invading the agriculturally important crops. The mode of infection is through the cuticle by degrading the chitin present on it which is enabled by the exochitinase enzyme of Bbchit1 gene. A good quality genomic DNA was isolated from Beauveria bassiana NCIM 1216 and amplified with specific primers to isolate the gene corresponding to Bbchit1 which codes for the exochitinase enzyme that is responsible for pathogenesis. The Bbchit1 gene of B. bassiana was transformed with the binary plasmid pBANF-bar-pAN-Bbchit1, in which the Bbchit1 gene was placed downstream of the constitutive gpd promoter, which was mediated by A. tumefaciens, and transformants were selected on the basis of herbicide resistance. Fifty herbicide resistant colonies were obtained and analyzed. The exochitinase produced by these transformants was observed maximum on the 7th day of inoculation in both which was 0.09 μmol/ml/min for the purified fraction and 0.06 μmol/ml/min for the crude extract. The chitinolytic activity was observed maximum at pH 5 and at temperature of 40°C. The genetically modified pure form can be used in the production of transgenic plants and in bringing out commercial formulation for the control of Lepidopteran pests.
Keywords: Beauveria bassiana, Bbchit1, Exochitinase
Introduction
Biological control agents are being considered as alternatives to synthetic chemical insecticides that are known to have toxic effects on non-target organisms, including animals and humans [1, 2]. Entomopathogenic fungi are key regulatory factors of insect populations in nature [3] and are attracting attention as biocontrol agents for insect pests [4]. However, mycoinsecticides constitute a very small percentage of the total insecticide market [5], even though entomopathogenic fungi are the only practical microbial control for insects that feed by sucking plant and animal juice and for the many acridid, lepidopteron and coleopteran pests which have no known viral or bacterial pathogens [6, 7]. Improvements in the effectiveness of mycoinsecticides may be obtained by optimizing the preparation and application of the inoculum [8]. Virulence of mycoinsecticides can also be achieved through the genetic modification [9, 10].
The insect cuticle, the first barrier against fungal pathogens, consists of a thin outer epicuticle, containing lipid and proteins, and a thick procuticle, consisting of chitin and proteins. Entomopathogenic fungi produce proteases, chitinases, and lipases which can degrade insect cuticle [11]. The entomopathogenic fungi M. anisopliae and Beauveria bassiana produce several chitinases [2, 12–14], which may have a variety of different functions. Some of these chitinases are important cuticle-degrading enzymes and act synergistically with proteases to hydrolyze insect cuticle [15]. Overexpression of the Bbchit1 gene from the entomopathogenic fungus Beauveria bassiana in transgenic B. bassiana can significantly enhance the virulence of B. bassiana for aphids. Insect bioassay results show significantly lower 50% lethal concentrations and 50% lethal times of the transformants overexpressing Bbchit1 gene compared to the values for the wildtype strain, suggesting that overproduction of chitinase of Bbchit1 can promote the infection efficiency of B. bassiana in aphids and accelerate infection.
In light of the importance of the Beauveria bassiana, a biocontrol agent the present work was carried out to study the selection of a suitable expression vector to carry Bbchit1 gene that expresses exochitinase responsible for pathogenesis.
Materials and Methods
Cloning of Bbchit1 Gene
Isolation of DNA from Beauveria bassiana NCIM 1216
The genomic DNA of Beauveria bassiana NCIM 1216 was prepared as per the protocol [16] with a slight modification.
Beauveria bassiana mycelia were ground to a powder in a mortar under liquid nitrogen. Ten milligrams of powder were transferred to a microfuge tube and then, 300 ml lysis buffer (100 mM Tris–HCl pH 8.0, 50 mM EDTA, 3% SDS) was added. The tube was incubated at 65°C for 1 h before 300 ml Tris–EDTA (TE)-saturated phenol/chloroform (1:1) was added. The mixture was then vigorously vortex and centrifuged at 12,000 rpm for 20 min. The supernatant was transferred to a new microfuge tube and then, 0.1 vol. 3 M Sodium acetate and 1 vol. ice-cold isopropanol were added. The solution was mixed before incubation in ice for 30 min. The DNA pellet was spun down at 12,000 rpm for 20 min before being washed with 70% ethanol. The DNA pellet was dissolved in 100 ml TE buffer and then, at 37°C for 1 h. The DNA was purified by phenol: chloroform: isoamyl alcohol (25:24:1) extraction, then vortexing and by centrifuging at 13,000 rpm for 5 min. The top aqueous layer was transferred to a microfuge tube and re-extracted. The DNA was precipitated on ice by adding 0.1 vol. 3 M Sodium acetate and 1 vol. ice-cold isopropanol. The pellet was spun down and dissolved in TE buffer as described earlier. 4 ml RNase (1 mg/ml) was added to remove RNA.
Amplification of Bbchit1 Gene of Beauveria bassiana by Polymerase Chain Reaction
Primers used for Polymerase Chain Reaction
| Primers | Sequence |
|---|---|
| Bbchit1-1 | 5′-CAACATACCAATCATGGCTCCTTTTCTTCAAA-3′ |
| Bbchit1-2 | 5′-TTATTTTCGACTTTACAAGTACAATCCAT-3′ |
The Bbchit1 gene of Beauveria bassiana NCIM 1216 was amplified in a Master cycler gradient (Eppendorf, Germany). In Polymerase Chain Reaction (PCR), the specific primers Bbchit1-1 and Bbchit1-2 (Helini Biomolecules, Chennai) were used to amplify the genomic sequence of the open reading frame (ORF) of the gene.
PCR reactions contained 2 μg B. bassiana DNA, 1 unit Taq DNA polymerase (Helini Biomolecules, Chennai), 2 pmol primers, 0.2 mM dNTP, 1× PCR buffer, and deionized water to make the final volume to 50 ml. Thirty PCR cycles consisting of denaturation (30 s at 95°C), annealing (30 s at 60°C), and extension (120 s at 72°C) were performed. The PCR products were then checked on 1% agarose gel, and an expected band (~1 kb) was excised, extracted and digested with restriction enzymes for subcloning.
Agarose Gel Electrophoresis
Required amount of agarose (w/v) was weighed and melted in 1× TBE buffer (0.9 M Tris–borate, 0.002 M EDTA, pH 8.2). Then, 1–2 μl ethidium bromide was added from the stock (10 mg/ml). After cooling, the mixture was poured into a casting tray with an appropriate comb. The comb was removed after solidification and the gel was placed in an electrophoresis chamber containing 1× TBE buffer. The products were mixed with 6× loading buffer (0.25% bromophenol blue, 0.25% xylene cyanol FF, 30% glycerol in water) at 5:1 ratios and loaded into the well. Electrophoresis was carried out at constant 60 V [17].
Eluting DNA from Agarose Gel Fragments
Ethidium bromide stained agarose gel was visualized under a transilluminator. The fragment of interest was excised with a clean razor blade. After removing the excess liquid, the agarose fragment was placed in the spin column. The tube was centrifuged at 5,500 rpm for not more than 45 s for the elution of DNA. The eluent was checked by running on an agarose gel and observed on a transilluminator for the presence of ethidium bromide stained DNA. The eluted DNA was used directly in manipulation reactions. This DNA fraction was sequenced (Helini Biomolecules, Chennai).
DNA Manipulation
The Bbchit1 gene was introduced into multiple cloning sites into the BamHI site and EcoRI of the pAN52-1 vector. Then it was digested with S2 nuclease to remove the ATG translation start codon in the gpd promoter, and religated. Then the vector was subjected for the sequence analysis to examine the orientation of the insert and to confirm that the ATG had been successfully removed. There were two orientations of the multiple cloning sites within pAN52-1. After confirming that there was no mutant by sequence analysis, the Bbchit1 open reading frame was excised from pGEM-T with EcoRI and SmaI, it was ligated into pBANF-bar-pAN-ES to form pBANF-bar-pAN-Bbchit1.
Transformation and Screening
Transformants were selected for resistance to 60 g of the herbicide phosphinothricin per ml. Wet mycelia (2.5 g) of herbicide resistant transformants were transferred from Potato Dextrose broth to basal salt medium supplemented with 2% glucose and 0.5% sodium nitrate was grown at 25 ± 2°C for 24 h at 100 rpm. Higher chitinase activity should be detected in liquid cultures of the transformants which overproduced Bbchit1 than in broth cultures of the wild-type strain. SDS-polyacrylamide gel electrophoresis was employed to confirm the expression of Bbchit1 and overexpression by assaying chitinase activity.
Gene Expression Analysis
To analyze the regulatory effect of glucose on the production of chitinase in the media containing colloidal chitin, the screened culture was transferred into potato dextrose broth suspended with 1% [wt/vol.] colloidal chitin (HI-MEDIA). After 24 h, the samples were collected at 48-h intervals by filtration through Whatman filter paper number 5. The filtrates were lyophilized, dissolved in distilled water and desalted by using 75% Ammonium sulphate precipitation. The protein so obtained was centrifuged and the pellet obtained was dialysed. Twenty micrograms of protein was then subjected to SDS-PAGE.
Chitinase Purification
Culture fluid of cloned vector containing culture grown in basal salt medium containing 1% (wt/vol.) colloidal chitin for 1 week was filtered through Whatman filter paper number 5 and the filtrate was subjected to precipitation with ammonium sulphate (75%, wt/vol.). The pellet was harvested by centrifugation and dissolved in 100 mM Tris–HCl buffer (pH 8.0). After dialysis against the same buffer, the crude extract was subjected to anion-exchange chromatography with a DEAE cellulose column (Helini Biomolecules, Chennai). After the column was washed with three column volumes of 20 mM Tris–HCl buffer (pH 7.8), it was eluted with a 100-ml linear gradient of NaCl (0–1 M) in the washing buffer at a flow rate of 1 ml/min. Fractions with chitinase activity were collected and concentrated against PEG 20,000 for further analysis.
Analysis of Chitinase Enzyme of Beauveria bassiana by SDS-PAGE
Protein samples (50 μg) were diluted fivefold in electrophoresis sample buffer and subjected to 12.5% polyacrylamide gel containing 0.1% SDS. The medium ranged molecular weight marker (HELINI, Chennai) mixed with the sample buffer was also loaded in one of the wells. Electrophoresis was carried out at constant voltage of 75 V. Gels were stained with Coomassie Brilliant Blue solution overnight and then destained. Protein bands with chitinolytic activity were detected using SDS-polyacrylamide gel containing 0.01% glycol chitin by the method previously described [18].
Assay of Chitinase Activity
Chitinase activity assay was performed [19]. One unit of chitinase activity was defined as the amount of enzyme that released sugars equivalent to 1 μmol of N-acetylglucosamine per/h at 37°C.
Factors Affecting the Activity of Chitinases
The activity of the exochitinase enzyme was determined at various pH starting from 4 to 9 and at various temperatures starting from 25 to 60°C to assess its maximum activity.
Virulence of Transformants
To test the virulence of individual transformants, third-instar Leaf web formers feeding on Oil palm were used for bioassay. The larvae were sprayed with 107 fungal spores/ml and also with the crude extract. The excessive liquid on the insect body was removed with dry filter paper. The control larvae were treated with water plus 0.05% (v/v) Tween 80. All the experimental and control larvae were fed on Oil palm leaves. The number of insects with normal body colour and the number of dead insects were recorded every 8 h.
Results and Discussion
Cloning of Bbchit1 Gene
DNA Extraction, Purification and Quantification
The DNA pellet was white, thick thread like mass. This DNA obtained was further quantified by spectrophotometry and agarose gel electrophoresis. It was observed that Beauveria bassiana DNA fragments were observed to emit orange fluorescence under UV lamp. The A260/A280 ratio for Beauveria bassiana DNA was found to be 1.9 spectrophotometrically. This study revealed the method adopted for extraction; purification and quantification of DNA were found to be suitable for molecular studies of Beauveria bassiana NCIM 1216.
Amplification of Bbchit1 Gene of Beauveria bassiana NCIM 1216 by PCR and Sequencing
The amplified fragment of DNA when analyzed by agarose gel electrophoresis indicates that it was of good quality. Then the sample was eluted and sequenced, the sequence of the amplified product was as follows.
CAACATACCAATCATGGCTCCTTTTCTTCAAACCAGCCTCGCGCTCCTTCCATTGTTGGCTTCCACCATGGTCAGCGCCTCGCCCTTGGCGCCGCGAGCCGGCACCTGCGCCACCAAAGGCCGGCCGGCCGGCAAAGTGCTCCAGGGCTACTGGGAGAACTGGGACGGTGCCAAGAACGGGGTGCACCCTCCGTTTGGCTGGACGCCCATCCAAAACCCCGACATTCGCAAGCACGGCTACAACGTCATCAATGCTGCCTTTCCCATCATCCAGCCTGACGGCACCGCGCTCTGGGAGGACGGCATGGACACGGGCGTCAAGGTGGCGAGCCCGGCCGACATGTGCGAGGCCAAGGCAGCAGGTGCCACCATCTTGATGTCGATTGGCGGTGCTACTGCGGCCATTGACCTGAGCTCGTCGGCTGTGGCTGACAAGTTTGTCTCGACCATTGTGCCGATTCTGAAAAAGTACAACTTTGACGGCATTGATATCGACATTGAATCCGGCCTCACAGGCAGCGGAAACATAAACACCCTGTCCACCTCGCAGACCAACCTGATTAGAATCATTGACGGCGTTCTCGCGCAGATGCCCGCCAACTTTGGCTTGACCATGGCGCCAGAGACTGCCTACGTTACCGGTGGGACTATTACGTACGGATCAATCTGGGGCTCTTACCTCCCCATTATCAAAAAGTACCTGGACAATGGTCGTCTCTGGTGGCTCAACATGCAGTACTACAATGGCGAAATGTACGGCTGCTCCGGCGACTCGCACAAGGCCGGTACTGTCGAAGGATTCATTGCTCAGACCGACTGCCTGAACAAGGGACTTAGTATTCAGGGCGTGACAATCACGATTCCCTATGACAAGCAAGTGCCTGGCCTTCCTGCCCAGCCTGGGGCTGGCGGCGGCCACATGTCCCCGTCCAACGTGGCGCAAGTTCTCTCCCACTACAAGGGCGCTTTGAAGGGATTGATGACTTGGTCTCTGAACTGGGACGGCTCCAAGAATTGGACATTTGGCGACAATGTCAAGGGGACTTTGGGGACTGCGTAATAAAAGCTGAAATGTTCATGTTAGGTA
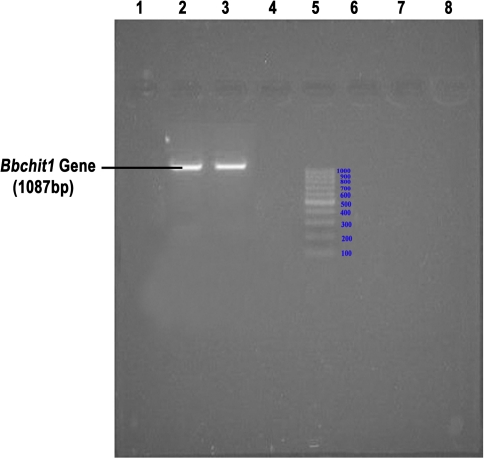
The sequence of the amplified product contains 1087 bases (Fig. 1). The sequence when made a BLAST hit, has evidenced that it was having an open reading frame coding for the enzyme exochitinase encoded by 1051 bases. The sequence of the exochitinase enzyme was deposited in NCBI Database and its GI: 194025184. Hence, this gene was further used for cloning in a suitable vector for the gene expression studies.
Fig. 1.
Agarose gel showing the amplified Bbchit1 gene. Well 2,3: Amplified PCR Product of Bbchit1; Well 5: DNA Ladder of 1 kb. c. Construction and characterization of B. bassiana transformants overproducing exochitinase of Bbchit1
Construction and Characterization of B. bassiana Transformants Overproducing Exochitinase of Bbchit1
Beauveria bassiana NCIM 1216 was transformed with the binary plasmid pBANF-bar-pAN-Bbchit1 (Fig. 2). The Bbchit1 gene was placed downstream of the constitutive gpd promoter, which was mediated by A. tumefaciens. The transformants were selected on the basis of herbicide resistance. Fifty herbicide resistant colonies were obtained and analyzed in basal salt medium supplemented with glucose that repressed native Bbchit1 production. The chitinase produced by these transformants was analysed further by isolating the protein in pure form and assaying its enzyme activity.
Fig. 2.
Vector with the cloned insert of Bbchit1 gene. PgpdA. nidulans gpd promoter, bar the sequence encoding the phosphinothricin acetyltransferase gene, TtrpC terminator sequence of the A. nidulans TrpC gene, Nos-proA.tumefaciens Nos gene promoter, Nos-terA. tumefaciens Nos gene terminator, NPTII kanamycin resistance gene, LB left border, RB right border
Gene Expression Analysis
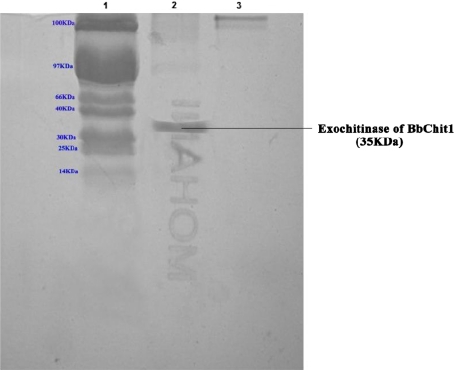
The enzyme exochitinase was precipitated by saturating with 70% Ammonium sulphate. This enzyme fraction upon subjecting to dialysis, the pure enzyme was obtained. This pure enzyme was further purified by Anion exchange chromatography and the fractions containing enzyme were collected. The enzyme containing fractions were checked by SDS-PAGE and the band corresponding to 35 KDa denotes that the eluent was exochitinase (Fig. 3). This was in accordance with the findings of Trudel and Asselin [20].
Fig. 3.
SDS-PAGE showing the purified chitinase. Well 1: 100 KDa Protein Marker; Well 2: Exochitinase of Bbchit1
Chitinase Activity Shown by Culture Filtrates
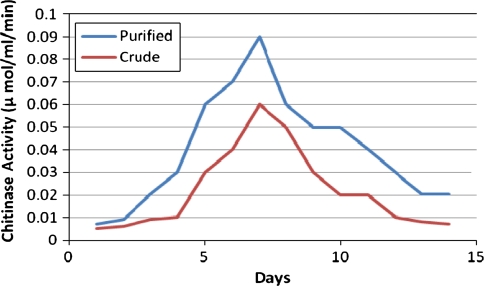
The chitinase activity of culture filtrate in purified form and the crude form at all the days of inoculation to 15 days was carried out to assess its chitinolytic activity. It was observed that maximum chitinolytic activity was observed on the 7th day of inoculation in both which was 0.09 μmol/ml/min for the purified fraction and 0.06 μmol/ml/min for the crude extract. It was noticed that with the increase in time of incubation, the activity increased rapidly and with the further increase in the time of incubation, the activity fell down (Fig. 4).
Fig. 4.
Chitinase enzyme activity. Peaks showing Chitinase activity of Purified and Crude forms
Chitinase Activity at Different pH
The chitinolytic activity of culture filtrates of Beauveria bassiana NCIM 1216 at varied pH conditions were carried out to assess its chitinolytic activity. It was observed that maximum chitinolytic activity was observed at pH 5 which was 0.06 μmol/ml/min. Its maximum chitinolytic activity was observed at the mild acidic range and its activity decreased as the basicity was increased. This could be concluded from the observations of pH levels 4–9. The organism was capable to show chitinolytic activity maximum at pH 5 (Fig. 5).
Fig. 5.
Effect of pH on the Chitinase activity
Chitinase Activity at Different Temperatures
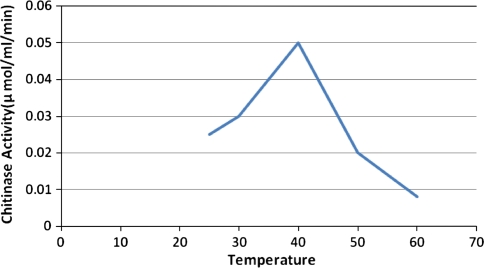
The chitinolytic activity of culture filtrate of Beauveria bassiana NCIM 1216 was carried out at varied temperature conditions to assess its chitinolytic activity. It was observed that maximum chitinolytic activity was observed at temperature 40°C which was 0.05 μmol/ml/min. Its chitinolytic activity was observed to increase from 25 to 40°C and its activity again tends to decrease as the temperature was increased. This could be concluded from the observations of temperature levels 25–60°C. The organism was capable to show maximum chitinolytic activity maximum at temperature of 40°C (Fig. 6).
Fig. 6.
Effect of temperature on the Chitinase activity
Virulence of Transformants
To determine the virulence of the transformants, we used third-instar Leaf web formers feeding on Oil palm as the experimental insect. All transformants displayed the same degree of virulence as Beauveria bassiana NCIM 1216. However, the cuticles of the larvae treated with purified enzyme extract began to whiten 24 h after the inoculation while the ones treated with Beauveria bassiana NCIM 1216 started to whitening 48 h after the inoculation. The data demonstrated that purified enzyme extract had greater virulence for the Leaf web formers feeding on Oil palm than the wild-type parental control (Fig. 7).
Fig. 7.
a Healthy Leaf web former; b Infected Leaf web former
Conclusion
A chitinase gene from B. bassiana NCIM 1216 was isolated from its genomic DNA by PCR. This gene was successfully cloned and expressed. The study revealed that Beauveria bassiana NCIM 1216 can be used as an effective biocontrol agent. The genetically modified form is more effective in expressing pathogenicity as the exochitinase activity was maximum compared to that of crude extract and it can be used for the production of transgenic plants and also in bringing a commercial formulation in a purified form of this pathogenic fungus that can be effectively used for the control of lepidopterans. Thus, the described transformation method is of great significance for manipulating the genetic make-up of the organism for improvement of its entomopathogenicity and also could provide a powerful tool for studying the functions of B. bassiana NCIM 1216 genes in general.
However, we realize that additional experiments are needed to demonstrate that the changes in morphology, growth characteristics, and virulence observed with purified enzyme extract are indeed due to the insertion of the foreign DNA.
References
- 1.Bidochka MJ. Monitoring the fate of biocontrol fungi. In: Butt TM, Jackson C, Magan N, editors. Fungi as biocontrol agents. Wallingford: CABI; 2001. pp. 193–218. [Google Scholar]
- 2.St. Leger RJ, Joshi L, Bidochka MJ, Rizzo NW, Roberts DW. Characterization and ultrastructural localization of chitinases from Metarhizium anisopliae, M. flavoviride, and Beauveria bassiana during fungal invasion of host (Manduca sexta) cuticle. Appl Environ Microbiol. 1996;62:907–912. doi: 10.1128/aem.62.3.907-912.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charnley AK. Entomopathogenic fungi and their role in pest control. In: Wicklow DT, Soderstrom BE, editors. The Mycota IV environmental and microbial relationships. Berlin: Springer; 1997. pp. 185–201. [Google Scholar]
- 4.Clarkson JM, Charnley AK. New insights into the mechanisms of fungal pathogenesis in insects. Trends Microbiol. 1996;4:197–203. doi: 10.1016/0966-842X(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 5.St. Leger RJ, Screen SE. Prospects for strain improvement of fungal pathogens of insects and weeds. In: Butt TM, Jackson C, Magan N, editors. Fungi as biocontrol agents. Wallingford: CABI; 2001. pp. 219–237. [Google Scholar]
- 6.Hajek AE, St. Leger RJ. Interactions between fungal pathogenesis and insect hosts. Annu Rev Entomol. 1994;39:293–322. doi: 10.1146/annurev.en.39.010194.001453. [DOI] [Google Scholar]
- 7.St. Leger RJ, Joshi L, Bidochka MJ, Roberts DW. Construction of an improved mycoinsecticide overexpressing a toxic protease. Proc Natl Acad Sci USA. 1996;93:6349–6354. doi: 10.1073/pnas.93.13.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prior C. Discovery and characterization of fungal pathogens for locust and grasshopper control. In: Lomer CJ, Prior C, editors. Biological control of locust and grasshopper. Wallingford: CABI; 1992. pp. 159–180. [Google Scholar]
- 9.St. Leger RJ, Roberts DW. Engineering improved mycoinsecticides. Trends Biotechnol. 1997;15:83–85. doi: 10.1016/S0167-7799(96)10071-8. [DOI] [Google Scholar]
- 10.Wood HA. Development and testing of genetically improved baculovirus insecticides. In: Shuler ML, Wood HA, Granados RR, Hammer DA, editors. Baculovirus expression systems and biopesticides. New York: Wiley; 1995. pp. 91–102. [Google Scholar]
- 11.Charnley AK, St. Leger RJ. The role of cuticle-degrading enzymes in fungal pathogenesis in insects. In: Cole GT, Hoch HC, editors. The fungal spore in disease initiation in plants and animals. New York & London: Plenum Press; 1991. pp. 267–287. [Google Scholar]
- 12.Bogo MR, Rota CA, Pinto H, Ocampos M, Correa CT, Ainstein HM, Schrank A. A chitinase encoding gene (chit1 gene) from the entomopathogen Metarhizium anisopliae: isolation and characterization of genomic and full-length cDNA. Curr Microbiol. 1998;37:221–225. doi: 10.1007/s002849900368. [DOI] [PubMed] [Google Scholar]
- 13.Kang SC, Park S, Lee DG. Isolation and characterization of a chitinase cDNA from the entomopathogenic fungus, Metarhizium anisopliae. FEMS Microbiol Lett. 1998;165:267–271. doi: 10.1111/j.1574-6968.1998.tb13156.x. [DOI] [PubMed] [Google Scholar]
- 14.Screen SE, Hu G, St. Leger RJ. Transformants of Metarhizium anisopliae sf. anisopliae overexpressing chitinase from Metarhizium anisop and N. Magan (ed.), Fungi as biocontrol agents. Wallingford: CABI; 2001. [DOI] [PubMed] [Google Scholar]
- 15.St. Leger RJ, Cooper RM, Charnley AK. Cuticle degrading enzymes of entomopathogenic fungi: regulation of production of chitinolytic enzymes. J Gen Microbiol. 1986;132:1509–1517. [Google Scholar]
- 16.Reader U, Broda P. Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol. 1985;1:17–20. doi: 10.1111/j.1472-765X.1985.tb01479.x. [DOI] [Google Scholar]
- 17.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 18.St. Leger RJ, Staples RC, Roberts DW. Entomopathogenic isolates of Metarhizium anisopliae, Beauveria bassiana, and Aspergillus flavus produce multiple extracellular chitinase isozymes. J Invertebr Pathol. 1993;61:81–84. doi: 10.1006/jipa.1993.1014. [DOI] [Google Scholar]
- 19.Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugars. Anal Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 20.Trudel J, Asselin A. Detection of chitinase activity after polyacrylamide gel electrophoresis. Anal Biochem. 1989;178:362–366. doi: 10.1016/0003-2697(89)90653-2. [DOI] [PubMed] [Google Scholar]