Abstract
In this study, an incidence pattern of 1.7% for Yersiniaenterocolitica and 2.5% for Y. intermedia were observed in an analysis of 120 diversified food samples collected from the local market of Mysore, Southern India. Two native isolates characterized as Y. enterocolitica belonged to biotype 1B and revealed the presence of major virulence related traits such as regulator of virulence, mucoid Yersinia factor regulator, attachment invasion locus, heat stable enterotoxin, Yersinia type II secretory system and phospholipase A in PCR. Force type neighbor-joining phylograms generated for Y. enterocolitica based on PCR amplicons of rovA and ypl showed 100% homology with two to three strains of Y. enterocolitica and about 75% homology with several strains of Y. pestis.
Keywords: Yersinia enterocolitica, Yersinia intermedia, PCR detection, Virulence determinants
Introduction
Globally, food safety is a major concern of public health and one of the widely recognized and significant foodborne pathogenic bacterial species is that of Yersinia enterocolitica. The prime concern of significance arises due to the fact that strains of Y. enterocolitica harbouring virulent traits are able to survive in both vacuum packed and refrigerated foods and thereby gets implicated in foodborne outbreaks, worldwide [1]. In the Indian scenario, the earliest reports about the isolation of Y. enterocolitica have been those of clinical samples [2, 3]. Subsequently, few studies have revealed the prevalence of Y. enterocolitica and other Yersinia spp. in samples of different food products including traditional fast foods [4–7]. As most of the individuals with enteric Y. enterocolitica infection are asymptomatic or minimally symptomatic and do not seek medical attention, reliable population based estimates of incidences are lacking. Considering the importance of Y. enterocolitica in foods and the limited knowledge available with reference to Indian scenario, the objective of this study was to assess the prevalence of potential pathogenic Yersinia spp. in a diverse range of foods through the use of PCR detection method and also analyze the genetic relatedness with other species.
Materials and Methods
All dehydrated media, bacterial reagents, stains, octadiscs of antibiotics and PCR purification spin kit (HiPur A) used in this study were procured from HiMedia Laboratories, Mumbai, India. The water used in the experimental trials was Milli-Q water (A10 Elix 3, Millipore Corporation, Billerica, USA).
Isolation and Characterization of Yersinia spp.
A total of 120 traditional food samples were collected at various points of sales from the local market in sterile, disposable polythene bags and brought to the laboratory in insulated icebox within 30 min of collection and subjected to analysis. The collected samples included (i) processed rice and wheat-based foods added with vegetables, spices and seasonings with common names of pani puri, bhel puri, churmuri, masala puri and sev puri (n = 50), (ii) fried wheat-based and vegetable stuffed foods like bread sandwich and cutlets (n = 20), (iii) concentrated and sweetened milk sweet (peda) and ice cream (n = 20), (iv) raw meat of pork, chicken and lamb (n = 10), raw milk (n = 10) and vegetable salad (n = 10). Yersinia spp. were isolated from individual food samples following the alkaline post-enrichment protocol and surface plating of treated sample aliquots on pre-poured plates of Cefsulodin-Irgasan-Novobiocin (CIN) agar [8, 9]. Plates were incubated at 30°C for 24 h. Presumptive colonies of Yersinia were selected based on characteristic bull’s eye appearance with deep red center and translucent halo around the colony. The presumptive isolates were identified by morphological, cultural and biochemical characteristics according to the documented procedures and the results were assessed according to the standard schemes for Yersinia spp. [10, 11].
Biotyping, Antibiogram and Phenotypic Virulence Traits of Characterized Native Isolates of Y. enterocolitica
The food isolates identified as Y. enterocolitica CFR 2301 and 2302 were further categorized into their biotypes according to earlier described schemes [1, 12]. The isolates were subjected to individual tests such as Voges Proskauer reaction, esculin hydrolysis and production of indole, L ornithine decarboxylase, β-d-glucosidase, pyrazinamidase, lipase and acid from inositol, salicin, l-sorbose, trehalose, xylose, and sucrose. Further, the antibiogram pattern was obtained by subjecting these two isolates to standard disc-diffusion assay [13] using octadiscs of antibiotics in nutrient agar at 37°C. After 24 h of incubation, the diameter of zones formed around the individual antibiotic tested in the disc was measured and results analyzed according to the standard reference procedure of National Committee for Clinical Laboratory Standards. Similarly, the two isolates of Y. enterocolitica were subjected to temperature dependent auto-agglutination and crystal violet binding assay for assessing presence of virulence plasmid in determining phenotypic virulence trait [14, 15].
PCR Detection for Virulence Factors
In the present study, selected virulence determinant genes in Y. enterocolitica were included to assess the prevalence of these virulence traits in the native food isolates of Yersinia spp. by PCR. The target genes, their nucleotide sequences and amplification conditions for these PCR primers are presented in Table 1. The primers in this study were designed by subjecting selected sequences of target genes (documented in NCBI Gene Bank) to Primer 3 Software program, followed by subjecting to BLAST and selection of primer sets with lowest E values. The synthesized primers were obtained from a commercial company (Sigma-Aldrich, Bangalore, India). Isolation of total genomic DNA of individual isolates was carried out with a slight modification of the Triton-X method of Blais and Phillippe [16]. Amplifications for the individual target genes (includes a multiplex for ail and yst) were carried out in a 25-μl reaction mix containing 4 μl of template DNA and subjected to PCR in an automated DNA thermal Cycler (Eppendorf, Master Cycler, Cedex, France) according to the conditions detailed in Table 1. The PCR products in aliquots of 12 μl were run in 1.5% agarose gels in 1X Tris acetate buffer for 1 h at 120 V and stained in 0.5 μg/ml ethidium bromide solution [17] and documented in Gel Documentation System (Vilber Lourmat, France).
Table 1.
Nucleotide sequence of specific primers and PCR conditions used in the detection of Yersinia species
| Target gene and Accession No. | Primer designation | DNA sequences (5′–3′) (bp) | PCR product | References |
|---|---|---|---|---|
| Attachment invasion locus | ail | 5′ CTATTGGTTATGCGCAAAGC 3′ | 359 | Fenwick and Murray [27] |
| M29945 | 5′ TGCAAGTGGGTTGAATTGCA 3′ | |||
| Heat stable enterotoxin | yst | 5′ TCTTCATTTGGAGCATTCGG 3′ | 159 | Present studya |
| X65999.1 | 5′ ATTGCAACATACATCGCAGC 3′ | |||
| Yersinia type II secretion system | yts1 | 5′ GCAGTAAAAGGCAACATCAGCG 3′ | 224 | Iwobi et al. [25] |
| AJ344214 | 5′ AAACAACGCGCATGACGACTTC 3′ | |||
| Regulator of virulence | rovA | 5′ CCATTAATATTGGATGCCAGA 3′ | 154 | Present studya |
| AF171097.1 | 5′ AATGCACCCCTGTCAGACTT 3′ | |||
| Phospholipase | ypl | 5′ CACAGCAAGCGGACTATTCA 3′ | 328 | Present studya |
| AM286415 | 5′ ATGACCAGTGCATCACCAAA 3′ | |||
| Mucoid Yersinia factor regulator | myfF | 5′ AGCGGGCTAAAGTTTGAGGT 3′ | 170 | Present studya |
| U12766.1 | 5′ TTTAGCGATCCTTTCGGTTG 3′ | |||
| PCR conditions for all the primers | ||||
| 94°C 5′; 94°C 1′; 55°C 1′; 72°C 1′; 72°C 8′; (35 cycles) | ||||
aPrimers were designed through Primer 3 Software Program, followed by BLAST and selection of primer sets with lowest E values
Phylogenetic Analysis of the Sequences of Amplified PCR Products of rovA and ypl
The PCR amplified products of rovA and ypl in the native isolates of Y. enterocolitica CFR 2301 and Y. intermedia CFR 2303 were purified using a commercial PCR purification Spin Kit (HiPur A) and subjected to sequence analysis by a commercial company (Sigma-Aldrich, Bangalore, India). The resulting nucleotide sequences for rovA and ypl were subjected to BLAST programme of NCBI (online access) to assess the per cent homology with closely related strains/variants documented in Gene Bank databases and also generate a Force Type of Neighbor-Joining method based similarity network tree. A pair-wise alignment of nucleotide sequences obtained for Y. enterocolitica CFR 2301 and Y. intermedia CFR 2303 with those strains actually used in designing of primers earlier was performed using Lalign Software [18] and per cent homology was recorded.
Results
Prevalence of Potential Pathogenic Yersinia spp.
Cultural and biochemical characterization of 110 presumptive isolates of Yersinia obtained from 120 food samples resulted in the identification of 2 isolates as Y. enterocolitica and 3 as Y. intermedia. In relation to the number of samples analyzed, the incidence pattern was 1.7% for Y. enterocolitica and 2.5% for Y. intermedia (Table 2). One isolate each of Y. enterocolitica CFR 2301 and CFR 2302 was obtained from a sample of vegetable cutlet (5%) and pork intestine (10%), respectively, while two isolates of Y. intermedia CFR 2303 and 2304 were obtained from samples of pani puri and one isolate CFR 2305 from ice cream.
Table 2.
Distribution pattern of characterized isolates of Yersinia spp. among the analyzed food samples
| Sample type | Samples(s) positive for Yersinia spp. (n) | Identified species (%) | |
|---|---|---|---|
| Processed rice and wheat-based traditional fast foods added with, vegetables, spices and seasonings [pani puri, bhel puri, churmuri masala puri and sev puri] (n = 50) | 2 | 4 | Y. intermedia |
| (pani puri) | CFR 2303 and 2304 | ||
| Fried wheat-based and vegetable stuffed foods [Bread sandwiches and cutlets] (n = 20) | 1 | 5 | Y. enterocolitica |
| (vegetable cutlet) | CFR 2301 | ||
| Traditional concentrated and sweetened milk sweet [peda] and ice cream (n = 20) | 1 | 5 | Y. intermedia |
| (Ice cream) | CFR 2305 | ||
| Raw meat [pork, chicken, lamb] (n = 10) | 1 | 10 | Y. enterocolitica |
| (pork intestine) | CFR 2302 | ||
| Raw milk (n = 10) | Nil | Nil | |
| Vegetable salad (n = 10) | Nil | Nil | |
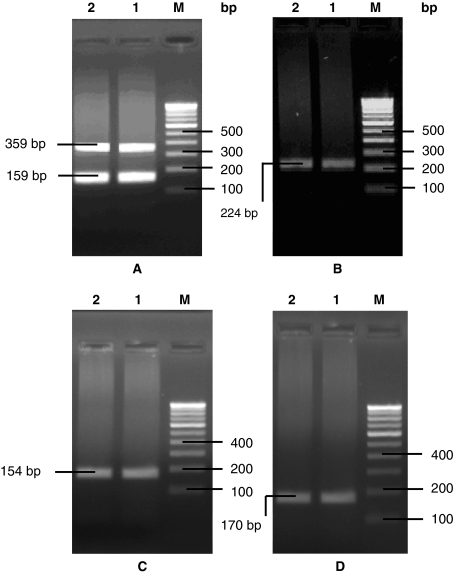
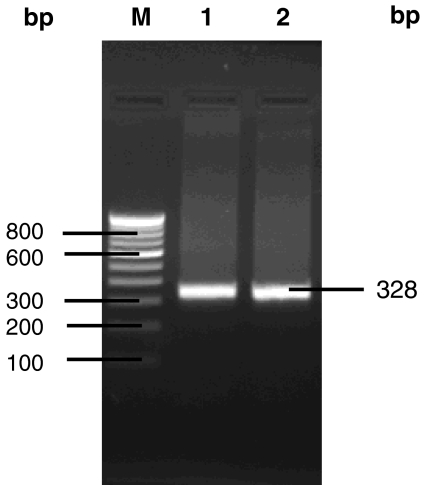
Both the native isolates of Y. enterocolitica CFR 2301 and 2302 were negative for esculin hydrolysis, pyrazinamidase activity and acid from salicin, which indicated these isolates as belonging to biotype 1B. Further, both these isolates revealed almost a similar antibiogram pattern (Table 3), except for differences in relation to ceftazidime, cephotaxime, nitrofurantoin, carbenicillin and streptomycin. These isolates were negative for the analyzed phenotypic virulence characteristics. However, in PCR, Y. enterocolitica CFR 2301 and 2302 exhibited positive amplification with the virulence linked primers of ail and yst (including multiplex), yts1, ypl, rovA, and myfF (Figs. 1, 2). The usually considered non-pathogenic isolates of Y. intermedia CFR 2303, 2304 and 2305 showed positive PCR amplification with only the primers of ypl.
Table 3.
Antibiogram of native food isolates of Y. enterocolitica
| Antibiotic | Food isolates | |
|---|---|---|
| Y. enterocolitica CFR 2301 | Y. enterocolitica CFR 2302 | |
| Ampicillin | R | R |
| Augmentin | R | R |
| Norfloxacin | S | S |
| Co-Trimoxazole | S | S |
| Gentamycin | S | S |
| Tobramycin | S | S |
| Cephoxitin | S | S |
| Ceftazidime | S | R |
| Cephotaxime | S | I |
| Nalidixic acid | S | S |
| Nitrofurantoin | I | R |
| Netillin | S | S |
| Ofloxacin | S | S |
| Carbenicillin | S | I |
| Kanamycin | S | S |
| Streptomycin | I | S |
| Tetracycline | S | S |
| Nystatin | R | R |
| Novobiocin | R | R |
S sensitive; R Resistant; I Intermediate
Fig. 1.
PCR amplicons with ail and yst (a), yts1 (b), rovA (c) and myfF (d) primers in isolates of Y. enterocolitica CFR 2301(lane 1) and Y. enterocolitca CFR 2302 (lane 2), Lane M 100 bp marker
Fig. 2.
PCR amplicons with ypl primers in isolates of Y. enterocolitica CFR 2301 (lane 1) and Y. intermedia CFR 2303 (lane 2), Lane M 100 bp marker
Relatedness of Native Isolates of Yersinia spp.
The resultant partial nucleotide sequences of respective PCR amplicons were 119 bases for rovA and 296 bases for ypl in Y. enterocolitica and 296 bases for ypl in Y. intermedia. The phylogram generated from partial nucleotide sequences of rovA of Y. enterocolitica CFR 2301 showed almost 98% sequence homology with the gene of putative inner membrane protein in Y. enterocolitica subsp. enterocolitica 8081 and rovA in another strain of Y. enterocolitica (Table 4). The same native isolate revealed a low sequence homology of 71% with the gene represented as protein of unknown function DUF 1656 in Serratiaproteamaculans 568 and 100% homology with Homosapiens chromosome 3 clone RP11-92124 with no details of the gene, but for only the clone (Fig. 3a). In a similar manner, the phylogram based on partial nucleotide sequences of ypl revealed a homology of 95% with strains of Y. enterocolitica and 79% with seven cultures of Y. pestis (Fig. 3b), wherein this homology was for the gene of phospholipase A (Table 5). In the case of Y. intermedia CFR 2303, the phylogram generated from the partial nucleotide sequences of ypl showed a homology of 82% with two cultures of Y. enterocolitica, which were present in the earlier generated phylogenetic tree for rovA specific primers. A homology of 77% each was observed with three cultures of Y. pseudotuberculosis and six cultures of Y. pestis. Besides, there was a homology of 70% with Serratia spp. and 80% with Pasteurellamultocida (phylogram not shown).
Table 4.
Sequence homology of Y. enterocolitica CFR 2301 for rovA with closely related strains of Y. enterocolitica appearing in the phylogram
| Y. enterocolitica subsp. enterocolitica 8081/Putative inner membrane protein | ||
| 8 | 65 | |
| 2301 | ATTCGCGCATATGATGGATCAGTAGCCATAACACTAAACCTAGCATAACCGCCTTAAA | |
| 8081 | ATTCCGCAGCATATGATGGATCAGTAGCCATAACACCAAACCTAGCATAACCGCCTTAAA | |
| 66 | 118 | |
| 2301 | AATTGGCGGGAAATAGATAGAGGCACCTAAAACCAAGTCTGACAGGGGTGCAT | |
| 8081 | AATTGGCGGGAAATAGATAGAGGCACCTAAAACCAAGTCTGACAGGGGTGCAT | |
| Y. enterocolitica/Transcriptional regulator rovA | ||
| 8 | 65 | |
| 2301 | ATTCGCGCATATGATGGATCAGTAGCCATAACACTAAACCTAGCATAACCGCCTTAAA | |
| YE | ATTCCGCAGCATATGATGGATCAGTAGCCATAACACCAAACCTAGCATAACCGCCTTAAA | |
| 66 | 118 | |
| 2301 | AATTGGCGGGAAATAGATAGAGGCACCTAAAACCAAGTCTGACAGGGGTGCAT | |
| YE | AATTGGCGGGAAATAGATAGAGGCACCTAAAACCAAGTCTGACAGGGGTGCAT | |
2301—Y. enterocolitica CFR 2301; 8081—Y. enterocolitica subsp. enterocolitica 8081; YE—Y. enterocolitica
Fig. 3.
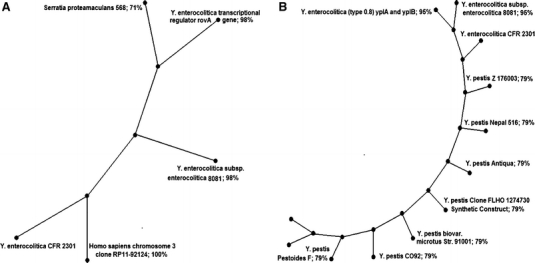
Phylogram of Y. enterocolitica CFR 2301 based on sequence analysis of PCR amplicons with rovA (a) and ypl (b) specific primers
Table 5.
Sequence homology of Y. enterocolitica CFR 2301 for ypl with closely related strains of Y. enterocolitica and Y. pestis appearing in the phylogram
| Y. enterocolitica subsp. enterocolitica 8081/Phospholipase A | ||
| 8 | 67 | |
| 2301 | TTACGCTCCTGCTGCGAGAAGCATCGGCGGATTTACACGGTTGGGTGATGCCGCGCTTGC | |
| 8081 | TTACGCTCCTGCTGCGAGAAGCATCGGCGGATTTACACGGTTGGGTGATGCCGCGTTGC | |
| 68 | 127 | |
| 2301 | TTTCGGCGGGGATAGATCCGGCGAGCCTATCTGATACAGCTTCAGGGTTTCAGGCTGGGA | |
| 8081 | TTTCGGCGGGGATAGATCCGGCGAGCCTATCTGATACAGCTTCAGGGTTTCAGGCTGGGA | |
| 128 | 187 | |
| 2301 | TTTACAGTGATAATCAACAGTATGTCCTCTCTTTCGCGGGTACCAATGATATTCANGATT | |
| 8081 | TTTACAGTGATAATCAACAGTATGTCCTCTCTTTCGCGGGTACCAATGATATTCAGGATT | |
| 188 | 247 | |
| 2301 | GGTTAAGTAATATCCGGCAAGCAACANGTTATGAGGATGTTCAATANAATCAGGNAGTNG | |
| 8081 | GGTTAAGTAATATCCGGCAAGCAACAGGTTATGAGGATGTTCAATATAATCAGGCTGTTG | |
| 248 | 288 | |
| 2301 | CNCTGGGGAAAACCNCTAAAATGGCNTTTGGTGATGCACTG | |
| 8081 | CGCTGGGGAAAACCGCTAAAATGGCATTTGGTGATGCACTG | |
| Y. pestis KIM/Phospholipase A | ||
| 97 | 155 | |
| 2301 | TCTGATACAGCTTCAGGGTTTCAGGCTGGGATTTACAGTGATAATCAACAGTATGTCCT | |
| Z 176003 | TCTGATAGC-GCTTCAGGCTTTCTCGCGGGGATTTACAGTGATAATCAACAGTATGTCTT | |
| 156 | 215 | |
| 2301 | CTCTTTCGCGGGTACCAATGATATTCANGATTGGTTAAGTAATATCCGGCAAGCAACANG | |
| Z 176003 | ATCTTTTGCAGGCACTAATGATCGGCACGATTGGTTGAGTAATATCCGACAGGCGGTGGG | |
| 216 | 275 | |
| 2301 | TTATGAGGATGTTCAATANAATCAGGNAGTNGCNCTGGGGAAAACCNCTAAAATGGCNTT | |
| Z 176003 | CTATGAGGATGTGCAATACAATGAAGCGGTGGCTCTGGGAAAAACAGCAAAAATGGCTTT | |
| 276 | 288 | |
| 2301 | TGGTGATGCACTG | |
| Z 176003 | TGGTGATGCGCTG | |
2301—Y. enterocolitica CFR 2301; 8081—Y. enterocolitica subsp. enterocolitica 8081; Z 176003, Y. pestis Z 176003
Discussion
In the background of heat sensitive nature of Y. enterocolitica, the isolation of this species from a heat processed wheat-based and vegetable stuffed (vegetable cutlet) product in our study is of interest. Considering the protocol involved for isolation of Y. enterocolitica from foods and related samples, their low recovery is usually attributed to presence of low viable populations of this organism in a given environment and also a poor competitor amongst co-existing microflora. The pattern of foods consumed and prevailing environmental parameters in most of the temperate countries have revealed higher incidences of Y. enterocolitica in clinical and food samples. Most of the studies with foods have focused on meat-based food products and the incidence pattern has been highly variable [19]. On the other hand, in the prevailing tropical climatic conditions of India, a lower percent incidence has been recorded in food samples evaluated a different time periods [4, 7, 20]. As an exception, the study with raw milk samples revealed an incidence of 55% with almost all the isolates obtained being identified as Y. enterocolitica [6].
It has been identified, that in the United States, most of the foodborne outbreaks caused due to Y. enterocolitica has been attributed to biotype 1B, which were generally referred to as ‘American strains’. However, in recent years, biotype 1B isolates have been reported from other countries of Europe (Germany), Asia, Africa, Australasia and Middle East [21, 22]. In the Indian scenario, isolates of Y. enterocolitica obtained from clinical samples and food products including meat-based products have been those of non-pathogenic biotype 1A [3–5, 23]. As Y. enterocolitica is known to be of an infective type foodborne pathogen, it becomes significant to evaluate the antibiogram pattern for its usefulness in public health and/or as markers in genetic related research investigations. The antibiogram pattern of the two isolates of Y. enterocolitica CFR 2301 and 2302 showed an almost similar pattern with those reported in few of the earlier investigations, except for the sensitivity to carbenicillin [24]. Both the isolates were sensitive to ofloxacin, while it was shown to be resistant in the case of isolates obtained from clinical samples [2].
The native isolates of Y. enterocolitica did exhibit the prevalence of yst and ail genes, which are unique to pathogenic isolates of this species. The native isolates obtained in our study showed the presence of the yts1 gene, which is invariably present only in highly pathogenic group of Y. enterocolitica [25]. Besides, both the native isolates of Y. enterocolitica were also positive for two other important virulence factors such as rovA and myfF (Fig. 1), which are known to regulate a number of genes essential for pathogenesis.
The occurrence of Y. intermedia in this study is of quite interest, as not much has been reported about isolation of this species in Indian scenario [3]. The absence of classical virulence markers like yst and ail in Y. intermedia isolates may give an indication that they are non-pathogenic. In the present study, Y. intermedia isolates were found to give amplification product for phospholipase gene (ypl), which codes for phospholipase A and known to be a virulent factor [26]. It was of interest to observe that the phylogram generated for Y. intermedia CFR 2303 based on partial nucleotide sequence of ypl had good homology with cultures of Y. enterocolitica, Y. pestis and Y. pseudotuberculosis, an indication of potential pathogenicity. Additional investigations may help to understand homology among enzyme and target gene as well as the role of this factor in possible pathogenicity among cultures of Y. intermedia.
It could be inferred from the generated phylogram that the prevalence of a predominant virulence factor such as rovA may not be commonly present among all the cultures of Y. enterocolitica. This may be due to few of the pathogenic traits being present in the isolates, as could be seen in the phylogenetic tree generated for pathogenic native isolate CFR 2301 and Y. intermedia CFR 2303 based on a less pathogenic determinant namely phospholipase A (ypl).
The present study established the prevalence of isolates of Y. enterocolitica in food samples, which belonged to pathogenic biotype 1B and harbored certain potent virulent traits. These isolates being obtained from heat processed product do indicate the questionable hygiene and sanitation practices in food chain operation. At the same time, excessive use of cold chain needs to be re-looked in view of the psychotropic character of Y. enterocolitica. This study also revealed the prevalence of isolates of Y. intermedia, a finding that needs more focus and attention from public health point of view.
Acknowledgments
The authors are thankful to Dr. V. Prakash, Director, CFTRI, Mysore for providing the facilities and interest in present work. The first author is grateful to Council of Scientific and Industrial Research, New Delhi for awarding the Research Fellowship.
References
- 1.Ramesh A, Padmapriya BP, Bharathi S, Varadaraj MC. Yersinia enterocolitica detection and treatment. In: Cabellero BL, Trugo C, Finglas PM, editors. Encyclopedia of food sciences and nutrition. 2. New York: Academic Press/Elsevier; 2003. pp. 6245–6252. [Google Scholar]
- 2.Lal M, Kaur H, Gupta LK. Yersinia enterocolitica gastroenteritis—a prospective study. Indian J Med Microbiol. 2003;21:186–188. [PubMed] [Google Scholar]
- 3.Singh I, Bhatnagar S, Virdi JS. Isolation and characterization of Yersinia enterocolitica from diarrhoeic human subjects and other sources. Curr Sci. 2003;84:1353–1355. [Google Scholar]
- 4.Singh I, Virdi JS. Isolation, biochemical characterization and in vitro tests of pathogenicity of Yersinia enterocolitica isolated from pork. Curr Sci. 1999;77:1019–1021. [Google Scholar]
- 5.Warke R, Kamat A, Kamat M, Thomas P. Incidence of pathogenic psychrotrophs in ice creams sold in some retail outlets in Mumbai, India. Food Control. 2000;11:77–83. doi: 10.1016/S0956-7135(99)00027-4. [DOI] [Google Scholar]
- 6.Kushal R, Anand SK. Isolation, biochemical characterization and antibiotic susceptibility of Yersinia enterocolitica isolates from milk. J Food Sci Technol. 2001;38:129–134. [Google Scholar]
- 7.Satishbabu HN, Rati ER. Prevalence of Yersinia enterocolitica in pani puri—a popular street food of India. J Food Sci Technol. 2003;40:303–305. [Google Scholar]
- 8.Schiemann DA. Synthesis of a selective agar medium for Yersinia enterocolitica. Canadian J Microbiol. 1979;25:1298–1304. doi: 10.1139/m79-205. [DOI] [PubMed] [Google Scholar]
- 9.Aulisio CCG, Mehlman IJ, Sanders AC. Alkali method for rapid recovery of Yersinia enterocolitica and Yersinia pseudotuberculosis from foods. Appl Environ Microbiol. 1980;39:135–140. doi: 10.1128/aem.39.1.135-140.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiemann DA, Wauters G. Yersinia. In: Vanderzant C, Spittstoesser DF, editors. Compendium for the microbiological examination of foods. Washington: American Public Health Association; 1992. pp. 433–450. [Google Scholar]
- 11.Cappucino JG, Sherman N. Microbiology: a laboratory manual. 6. Singapore: Pearson Education; 2004. [Google Scholar]
- 12.Kandolo K, Wauters G. Pyrazinamidase activity in Yersinia enterocolitica and related organisms. J Clin Microbiol. 1985;21:980–982. doi: 10.1128/jcm.21.6.980-982.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bauer AW, Kirby WMM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standard single disc method. American J Clin Pathol. 1996;45:493–496. [PubMed] [Google Scholar]
- 14.Skurnik M, Boli I, Meikkinen H, Piha S, Wolf-Watz H. Virulence plasmid associated autoagglutination in Yersinia spp. J Bacteriol. 1984;158:1033–1036. doi: 10.1128/jb.158.3.1033-1036.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhaduri S, Conway LK, Lachica RV. Assay of crystal violet binding for rapid identification of virulent plasmid bearing clones of Yersinia enterocolitica. J Clin Microbiol. 1987;25:1039–1041. doi: 10.1128/jcm.25.6.1039-1042.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blais BW, Phillippe LM. A simple RNA probe system for analysis of Listeria monocytogenes polymerase chain reaction products. Appl Environ Microbiol. 1993;59:2795–2800. doi: 10.1128/aem.59.9.2795-2800.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Russel DW. Molecular cloning: a laboratory manual. 3. New York: Cold Spring Harbour Laboratory Press; 2001. [Google Scholar]
- 18.Huang X, Miller W. A time-efficient linear-space local similarity algorithm. Adv Appl Math. 1991;12:337–357. doi: 10.1016/0196-8858(91)90017-D. [DOI] [Google Scholar]
- 19.Capita R, Alonso-Calleja C, Prieto M, Garcia-Fernandez MDC, Moreno B. Incidence and pathogenicity of Yersinia spp. isolates from poultry in Spain. Food Microbiol. 2002;19:295–301. doi: 10.1006/fmic.2002.0492. [DOI] [Google Scholar]
- 20.Ram S, Khurana S, Khurana SB, Vadehra DV, Sharma S, Chhina RS. Microbiological quality and incidence of organisms of public health importance in food and water in Ludhiana. Indian J Med Res. 1996;103:253–258. [PubMed] [Google Scholar]
- 21.Schubert S, Bockemuhl J, Brendler U, Heesemann J. First isolation of virulent Yersinia enterocolitica O:8 biotype 1B in Germany. European J Clin Microbiol Infect Dis. 2003;22:66–68. doi: 10.1007/s10096-002-0859-1. [DOI] [PubMed] [Google Scholar]
- 22.Soltan-Dallal M, Tabarraie A, MoezArdalan K. Comparison of four methods for isolation of Yersinia enterocolitica from raw and pasteurized milk from northern Iran. International J Food Microbiol. 2004;94:87–91. doi: 10.1016/j.ijfoodmicro.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 23.Sachdeva P, Virdi S. Repetitive elements sequence (REP/ERIC)-PCR based genotyping of clinical and environmental strains of Yersinia enterocolitica biotype 1A reveal existence of limited number of clonal groups. FEMS Microbiol Lett. 2006;240:193–201. doi: 10.1016/j.femsle.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 24.Lee T, Lee S, Seok W, Yoo M, Yoon J, Park B, Moon K, Oh DH. Prevalence, antibiotic susceptibility and virulence factors of Yersinia enterocolitica and related species from ready-to-eat vegetables available in Korea. J Food Protect. 2004;67:1123–1127. doi: 10.4315/0362-028x-67.6.1123. [DOI] [PubMed] [Google Scholar]
- 25.Iwobi A, Heesemann J, Garcia E, Igwe E, Noelting C, Rakin A. Novel virulence associated type II secretion system unique to high pathogenicity Yersinia enterocolitica. Infect Imm. 2003;71:1872–1879. doi: 10.1128/IAI.71.4.1872-1879.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmiel DH, Wagar E, Karamanou L, Weeks D, Miller VL. Phospholipase A of Yersinia enterocolitica contributes to virulence in a mouse model. Infect Imm. 1998;66:3941–3951. doi: 10.1128/iai.66.8.3941-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fenwick SG, Murray A. Detection of pathogenic Yersinia enterocolitica by polymerase chain reaction. Lancet. 1991;337:496–497. doi: 10.1016/0140-6736(91)93436-D. [DOI] [PubMed] [Google Scholar]