Abstract
The Universal Method (UM) described here will allow the detection of any bacterial rDNA leading to the identification of that bacterium. The method should allow prompt and accurate identification of bacteria. The principle of the method is simple; when a pure PCR product of the 16S gene is obtained, sequenced, and aligned against bacterial DNA data base, then the bacterium can be identified. Confirmation of identity may follow. In this work, several general 16S primers were designed, mixed and applied successfully against 101 different bacterial isolates. One mixture, the Golden mixture7 (G7) detected all tested isolates (67/67). Other golden mixtures; G11, G10, G12, and G5 were useful as well. The overall sensitivity of the UM was 100% since all 101 isolates were detected yielding intended PCR amplicons. A selected PCR band from each of 40 isolates was sequenced and the bacterium identified to species or genus level using BLAST. The results of the UM were consistent with bacterial identities as validated with other identification methods; cultural, API 20E, API 20NE, or genera and species specific PCR primers. Bacteria identified in the study, covered 34 species distributed among 24 genera. The UM should allow the identification of species, genus, novel species or genera, variations within species, and detection of bacterial DNA in otherwise sterile samples such as blood, cerebrospinal fluid, manufactured products, medical supplies, cosmetics, and other samples. Applicability of the method to identifying members of bacterial communities is discussed. The approach itself can be applied to other taxa such as protists and nematodes.
Keywords: Universal method, Bacterial identification, Universal primers, Golden mixture, Multiplex
Introduction
Historically, identification and classification of eubacteria are based on phenotypic characteristics [1–4]. Early molecular techniques used in bacterial classification were based on GC content [5], plasmid profiling, and compatibility to genetic transformation [6]. Currently, two fundamental molecular applications are being extensively utilized in bacterial detection and identification; these are based on hybridization and nucleotide sequencing. Hybridization based applications such as Southern [7, 8], PCR [9], real-time PCR [10, 11], microarray [12, 13], universal tagging method [14], and loop-mediated isothermal amplification (LAMP) [7] are sensitive and specific techniques for the detection and identification of microbes. Specific PCR primers have been employed to confirm the presence or absence of target microorganisms or specific features associated with them such as antibiotic resistance and virulence factors [1, 15–18]. Specific primers proved useful in assessing clinical samples for the presence of slow growing bacteria such as Mycobacterium tuberculosis [19–22] and Helicobacter pylori [8, 23]. Although specific primers are powerful tools showing superior sensitivities and specificities, yet they can not predict the presence or absence of non-target bacterial species in the tested sample. Detection and identification of non-target bacteria require a different approach; degenerate primers are used to amplify gyr B for taxonomic analysis of Pseudomonas putida [22] while PCR coupled to sequencing of the 16S rDNA is used to identify pathogenic Pseudomonas spp. [24] and others [25, 26]. PCR-RFLP analysis of 16S genes is used in the analysis of Campylobacter, Helicobacter, and Arcobacter [27, 28]. Though useful, most methods suffer from major limitations that can not be overlooked especially in clinical laboratories. In clinical samples the absolute presence or absence of bacteria must be determined. In addition, accurate identification of pathogenic bacteria is essential for therapy, post-treatment follow up, and epidemiological purposes.
To solve this problem, investigators [4, 17, 20, 25, 27, 29–31] sought universal primers. Unfortunately, “Universal Primers” do not live up to their name since they do not cover all bacteria [30]. The term is misleading and should be abandoned since most claimed universal primers, if not all, are not universal as exemplified in Tables 1, 2. Multiplexing [13, 32, 33] employs specific primer pairs, it is a time saving process that allows the simultaneous detection of a limited number of target microbes. However, multiplexing with specific primers will miss non-target microbes. Multiplex has been applied to respiratory viruses [32], middle ear bacteria [33], and others. Group-specific primers are utilized in the detection of sulfate-reducing bacteria [34]. Templex is an assay based on type-specific primers; it was developed for the simultaneous detection of 25 different human papilloma genotypes [35].
Table 1.
Examples of claimed 16S universal and general primers
| Primer | Primer sequence; This study (underlined sequences) | Reference |
|---|---|---|
| U1 | 5′-CCAGCAGCCGCGGTAATACG-3′ | [48], This study: QUGP-F6a |
| U2 | 5′-ATCGG(C/T)TACCTTGTTACGACTTC-3′ | [48], This study: QUGP-R1a |
| 909 bp | 5′-AAACTCAAAGGAATTGAC-3′ | [17] |
| 907R | 5′-CCGTCAATTCMTTTGAGTTT-3′ | [28] |
| 909 bpR | 5′-GACGGGCGGTGTGTACCAA-3′ | [17], This study: QUGP-R2a |
| 8F | 5′-GGATCCAGACTTTGATYMTGGCTCAG-3′ | [20] |
| UnF | 5′-GAGTTTGATCCTGGCTCAG-3′ | [28], This study: QUGP-F1a |
| TPU1 | 5′-AGAGTTTGATCMTGGCTCAG-3′ | [4] |
| EubB | [20], This study: QUGP-F1a | |
| RTU8 or EubA | 5′-AAGGAGGTGATCCANCCRCA-3′ | [20] |
| 6R | 5′-AGAAAGGAGGTGATCCAGCC-3′ | [4] |
| [22] | ||
| UnR | 5′-GGACTACCAGGGTATCTAAT-3′ | [27], This study: QUGP-R3a |
| 11F | 5′-TGGCGAAGGCGGCCCCCTGGA-3′ | [35] |
aUnderlined sequence are shared by QUGPs
Table 2.
Can a 16S primer be universal when its ≥16 b?
| Bacteriumb | Published primers/universal primers | Al-Quds University general primers (QUGP) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RTU8a | 909 bpa | 909 bp Ra | 8F | F1 | F3 R3 | F4 R4 | F5 R5 | F6 R6 | R1b | R2 | R7 | |
| Acidobacteria bacterium | Y | Y | Y | Y | Y | R | Y | Y | Y | Y | Y | Y |
| Acinetobacter sp. | Y | N | Y | Y | M | Y | Y | Y | Y | Y | Y | Y |
| Aeromonas hydrophila | Y | N | Y | Y | M | Y | Y | Y | Y | Y | Y | Y |
| Agrobacterium tumefaciens | N | Y | Y | N | N | Y | Y | Y | Y | Y | Y | Y |
| Anabaena variabilis | Y | Y | Y | Y | Y | N | Y | Y | Y | Y | Y | N |
| Aquifex aeolicus VF5 | Y | N | Y | Y | Y | R | Y | M | Y | Y | N | N |
| Arthrobacter aurescens | N | Y | Y | N | N | Y | Y | Y | Y | Y | Y | N |
| Arthrobacter sp. FB24 | Y | Y | Y | Y | Y | Y | Y | Y | Y | T | Y | N |
| Bacillus cereus | N | Y | Y | N | N | Y | Y | Y | Y | Y | Y | Y |
| Bacillus subtilis | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Bifidobacterium adolescentis | Y | N | N | N | N | M | Y | Y | Y | T | Y | N |
| Bordetella pertussis | Y | Y | Y | Y | Y | R | Y | Y | Y | M | Y | Y |
| Brucella abortus | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Burkholderia cenocepacia | Y | Y | Y | N | N | Y | Y | Y | Y | M | Y | Y |
| Campylobacter jejuni | N | N | Y | Y | Y | R | Y | Y | Y | M | Y | Y |
| Chlamydophila pneumoniae 1 | Y | N | Y | N | N | R | N | N | M | M | Y | N |
| Chlamydophila pneumoniae | N | N | Y | N | N | M | N | N | Y | Y | Y | N |
| Corynebacterium jeikeium | Y | Y | N | Y | Y | Y | Y | Y | Y | Y | Y | N |
| Dehalococcoides ethenogenes | N | Y | Y | N | N | M | Y | Y | Y | Y | Y | N |
| Deinococcus geothermalis | Y | Y | Y | N | N | M | M | Y | Y | M | Y | Y |
| Desulfovibrio vulgaris | Y | N | Y | Y | M | R | Y | Y | Y | M | Y | N |
| Escherichia coli O157:H7 | Y | N | Y | Y | M | Y | Y | Y | Y | Y | Y | Y |
| Fusobacterium nucleatum | N | Y | Y | Y | Y | Y | Y | Y | Y | N | Y | Y |
| Haemophilus influenzae | Y | N | Y | Y | M | Y | Y | Y | Y | Y | Y | Y |
| H. pylori | N | N | N | Y | Y | Y | Y | Y | Y | Y | T | Y |
| Lactobacillus acidophilus | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | N | Y |
| Legionella pneumophila | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Magnetococcus sp. | Y | Y | Y | N | N | Y | Y | Y | Y | Y | Y | M |
| M. tuberculosis | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | N |
| Mycoplasma pneumoniae | N | N | Y | Y | Y | N | N | Y | N | T | Y | M |
| Neisseria meningitidis | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Nocardioides sp. | Y | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | N |
| Prochlorococcus marinus | N | Y | Y | N | N | R | Y | Y | Y | Y | Y | Y |
| P. putida | N | N | Y | N | N | Y | Y | Y | Y | Y | Y | Y |
| Ralstonia eutropha | N | Y | Y | Y | Y | Y | Y | Y | Y | T | Y | Y |
| Rhodopirellula baltica | N | Y | N | Y | N | M | R | R | R | T | Y | Y |
| Rickettsia akari | N | Y | N | Y | Y | M | Y | Y | T | M | N | Y |
| Rickettsia rickettsii | N | Y | N | Y | Y | M | Y | Y | T | M | N | Y |
| Salmonella enterica | Y | N | Y | Y | M | Y | Y | Y | Y | Y | Y | Y |
| Staphylococcus aureus | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Streptococcus pyogenes | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Streptomyces avermitilis | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | N |
| Streptomyces coelicolor | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | N |
| Syntrophobacter fumaroxidans | Y | Y | Y | Y | Y | Y | N | Y | Y | Y | Y | Y |
| Thermotoga lettingae | Y | N | N | N | N | R | Y | Y | Y | T | N | M |
| Thermus thermophilus | N | Y | Y | Y | Y | M | N | Y | Y | Y | Y | Y |
| Treponema pallidum | Y | Y | Y | N | N | M | N | Y | R | Y | Y | Y |
| Vibrio cholerae O395 | Y | N | Y | Y | M | Y | Y | Y | Y | N | Y | Y |
| V. parahaemolyticus | N | N | Y | Y | M | Y | Y | Y | Y | Y | Y | Y |
| Wolbachia endosymbiont | N | Y | N | Y | M | Y | N | Y | Y | N | M | Y |
Y primer sequence is present, M 5′ or central mismatch was detected, T 3′-end mismatch was found. R Only the reverse primer may be functional. N the sequence is completely absent or partially present
Published a RTU8, 909 bp, 909R, and 8F are as in Table 1
bDistribution of primers was analyzed in other bacteria: Bacteroides fragilis, Bdellovibrio bacteriovorous, Clostridium tetani, Enterococcus faecalis, Listeria monocytogenes, and Ochrobactrum anthropi
A robust method, required for bacterial identification, has been perused by several investigators [4, 14, 26, 36–40]; studies on universal and multiplex primers]. With the current number of eubacterial species surpassing 7166 species [41], the UM described here [42] should fulfil this requirement.
The UM had integrated several general primers, PCR amplification, DNA sequencing, and sequence alignment (BLAST) [43, 44] in one system designed for the detection and identification of bacteria. First, the capacity of the UM and primer mixture to detect bacteria is described. Second, the applicability of the UM in bacterial identification was repeatedly demonstrated. Third, the UM should allow the detection of bacterial DNA in any given sample including clinical samples, water samples, medical supplies, drugs, and others. Fourth, the detection and subsequent identification of novel bacterial species should become a simple process especially when all known bacteria have been sequenced and deposited in accessible data bases. The ability of the UM to identify bacteria is dependent on the availability of sequenced bacterial genomes (specifically rDNA sequences) of identified bacteria in the nucleotide data bases available for alignment analyses. This threat to the UM and other alignment dependent methods will fade away as more bacteria are sequenced. However, it is unlikely to dissipate completely and will continue to challenge the UM and other similar methods.
Rational and Hypothesis
DNA sequencing and sequence alignment have been widely applied and accepted as methods of bacterial detection and identification [2, 8, 18, 21, 24–26]. Phylogenetic analysis of bacteria can be based on their 16S DNA sequences [2]. The UM took advantage of this fact and the fact that rDNA sequences of different bacteria will eventually be accumulated by the different nucleotide data banks. The integration of these facts together with general primer mixtures capable of amplifying any bacterial rDNA in one system called “The UM for detection and identification of bacteria” was accomplished. The assembly of general primer mixtures capable of amplifying 16S genes of bacteria represent the key to successful application of the UM. The assembled primer mixtures should contain a minimum of two forward and two reverse general primers. More than one mixture may be required to detect all bacteria. A single mixture may be assembled that potentially can substitute for all mixtures. Successful detection and amplification can then be utilized in bacterial identification and other applications.
Materials and Methods
DNA Preparation
Bacterial isolates were labeled as Al-Quds University Bacterial Collection (QUBC). Fresh pure bacterial cells were treated with 100 μl of freshly prepared lysozyme (Sigma Chemical Co.; 5 mg per ml of sterile J-buffer; 100 mM Tris–HCl, 100 mM EDTA, 150 mM NaCl; pH 8; autoclaved) [23], incubated in a microfuge tube for 30 min at 35°C, cells or incompletely lysed cells were collected by centrifugation, the pellet was lysed with 20 μl of 0.5 N NaOH and 20 μl of 1% SDS. The cell lysate were boiled for 10 min, then diluted to 200 μl with sterile water.
Primer Design
The 16S rDNA sequence of H. pylori HPAG1, selected for its clinical importance [42], was used to BLAST all 940 microbial genomes. The BLAST results showing minimal similarity to H. pylori, were then scanned for conserved regions ≥16 bases long. In the second stage, >50 representative 16S rDNA sequences were searched for the presence or absence of each putative primer sequence. The presence of primer sequence in published rDNA was determined as shown in Table 2. Primer sequences were refined to arrive at the final primers listed in Tables 3, 4. Some primer sites were used in both directions; forward and reverse. New Al-Quds University General Primers (QUGP) Forward (F) or Reverse (R) were longer primers and were based on the short general primers, their sequences are shown in Tables 3, 4 They were designated QUGP-Fn3, QUGP-Fn5, QUGP-Fn6, QUGP-Rn3, and QUGP-Rn1, additional primers (QUGP-R2 and its long form QUGP-Rn2) were introduced. QUGP-F4 and QUGP-R4 were not modified since they showed high melting temperature (Tm). Primers were commercially synthesized (Hy Labs, Jerusalem or Danyel, Rehohot). Multiplex mixtures were prepared and tested (Table 5) to define a Golden (G) mixture that will react positively with the vast majority, if not all, bacteria. Each working primer mixture contained 10 pmol of each respective primer/μl.
Table 3.
Al-Quds University 16S General PCR and Sequencing Primers (QUGP-16S; this studya) based on 16S sequence of H. pylori gi|108562424:c1140006-1138506
| Primer | PCR and sequencing primers | Oligomer: location | Tm (ºC) |
|---|---|---|---|
| QUGP-F1 | 5′-AGTTTGATCCTGGCTCAG-3′ | 18: 10–27a | 53.7 |
| QUGP-F1b | 5′-AGTTTGATCATGGCTCAG-3′ | 18: 10–27 | 51.40 |
| QUGP-F3 | 5′-GATACCCTGGTAGTCCA-3′ | 17: 753–769 | 52.80 |
| QUGP-R3 | 5′-TGGACTACCAGGGTATC-3′ | 17: 769–752 | 52.80 |
| QUGP-F4 | 5′-CCGCCTGGGGAGTACG-3′ | 16: 840–856 | 59.55 |
| QUGP-R4 | 5′- CGTACTCCCCAGGCGG-3′ | 16: 856–840 | 59.55 |
| QUGP-F5 | 5′-CCTACGGGAGGCAGCAG-3′ | 17: 326–343 | 54.54 |
| QUGP-R5 | 5′-CTGCTGCCTCCCGTAGG-3′ | 17: 343–326 | 54.30 |
| QUGP-F6 | 5′-GCAGCCGCGGTAATAC-3′ | 16: 481–497 | 54.51 |
| QUGP-R6 | 5′-GTATTACCGCGGCTGC-3′ | 16: 497–481 | 54.30 |
| QUGP-R7 | 5′-CGATTACTAGCGATTCC-3′ | 17:1319–1302 | 47.13 |
| QUGP-R2 | 5′-GACGGGCGGTGTGTAC-3′ | 16: 1376–1392 | 54.85 |
| QUGP-R1 | 5′-TACCTTGTTACGACTTCACCC-3′ | 17: 1468–1447 | 57.90 |
| QUGP-R1b | 5′-TACCTTGTTACGACTTC-3′ | 17: 1468–1451 | 47.90 |
Bold italicized sequences are homologous to Human GENE ID: 100008588 LOC100008588 | 18S ribosomal RNA. The potential products QUGP-Fn6. Rn2, 1096 bp and QUGP-Fn6.Rn1, 1226 bp are unlikely to form at 58°C due to 3′-mismatches to human 18S rDNA when clinical samples contain human DNA. This justifies switching from the short QUGP to the new longer sequences; while QUGP-R2 will match human DNA, QUGP-Rn2 presents a critical mismatch at its 3′-end
aSome of the primers overlapped those used by others, see Table 1. Primer location differ slightly in different bacteria and may be absent from others (QUGP R2/Rn2 is absent from H. pylori, the shown location is for P. fluorescens, some bacteria will mismatch with 5′-end of primer(s)
Table 4.
Al-Quds University 16S general primers new long primers
| Primer | PCR long primers | Oligomer: location | Tm (ºC) |
|---|---|---|---|
| QUGP-Fn3 | 5′-CAGGATTAGATACCCTGGTAGTCC-3′ | 24: 744–768 | 65 |
| QUGP-Fn5 | 5′-ACTCCTACGGGAGGCAGCAG-3′ | 20: 323–343 | 65 |
| QUGP-Fn6 | 5′-CCAGCAGCCGCGGTAATAC-3′ | 19: 479–497 | 62 |
| QUGP-Rn1 | 5′-GGCTACCTTGTTACGACTTC-3′ | 20: 1471–1468 | 58 |
| QUGP-Rn2 | 5′-TGACGGGCGGTGTGTACAAG-3′ | 20: 1406–1386 | 63 |
| QUGP-Rn3 | 5′-GGCGTGGACTACCAGGGTATC-3′ | 21: 775–752 | 65 |
Long form QUGP were used to prepare golden mixtures
Underlined sequences, indicate the short QUGP prior to being re-designed. QUGP (underlined sequences) were extended mostly at their 5′-ends with conserved nucleotides, QUGP-F4/R4 were not modified since their Tm was 59.5°C and are flanked by variable nucleotides. The main differences between the QUGP and redesigned primers are limited to increased primer length and Tm which allowed annealing to be performed at 58°C instead of 50°C
Table 5.
General and Golden Primer Mixtures
| A | B | ||
|---|---|---|---|
| General mixtures | QUGP | Golden mixture | QUGP-primers |
| Level I | |||
| Reaction one | F5, F6, R1b | G1 | Fn3, Fn5, Fn6•Rn1 |
| Reaction two | F5, F6, R3 | G2 | Fn3, Fn5, Fn6•Rn2 |
| Reaction two (b) | F5, F6, R3, R4 | ||
| Reaction three | F3, F4, R1b | G3 | Fn3, Fn5, Fn6•Rn3 |
| Level II | |||
| Reaction four | F1, R1b | G4 | Fn3, Fn5, Fn6•Rn3, Rn2 |
| Reaction five | F1, R3, R4 | G5 | Fn5, Fn6•Rn3, R4 |
| Reaction six | F1, R5. R6 | G6 | Fn3, Fn5, Fn6•Rn1, Rn2 |
| Level III | |||
| Reaction seven | F5, F6, R4 | G7 | Fn3, F4, Fn5, Fn6•Rn1, Rn2, Rn3 |
| Reaction eight | F3, R4 | G8 | Fn3, Fn5, Fn6•Rn1, Rn2, Rn3, R4 |
| Reaction nine | F5, R6 | G9 | Fn5, Fn6•Rn1, Rn2, Rn3, R4 |
| Mq4 | F4, F5, F6, R1b, R2 | G10 | Fn3, F4•Rn1, Rn2 |
| G11 | Fn5, Fn3•Rn1, Rn2 | ||
| G12 | Fn6, F4•Rn1, Rn2 | ||
| G13 | Fn5, Fn6•Rn1, Rn2 | ||
Amplification Parameters
A MiniCycler (MJ Research, Inc., Watertown, MA) heated lid thermocycler was used to amplify DNA; 25-μl reactions were prepared by adding 8 pmol of each primer (0.8 μl of primer mixture), 0.5 μl of DNA sample, 12.5 μl Master Mix from Promega, and pure sterile water to 25 μl. All amplification reactions were hot started at 95°C for 3 min. The PCR protocol used with short General primers was: 94°C 90 s, 48°C 35 s, 50°C 35 s, 72°C 105 s. and 33 cycles, a final extension step at 72°C for 3 min, 4°C for 24 h. When Golden Mixtures (G1–G13) were used, PCR parameters were the same as above except for annealing temperature which was set at 58°C for 1 min. Species specific primers HPU1 and HPU2 [46] were used for detection of H. pylori. The species specific PA-SS-F and PA-SS-R were used for the detection of Pseudomonas aeruginosa. The genus specific PA-GS-F and PA-GS-R primers were used for detection of P. species [18].
Detection and Documentation
Agarose gels containing ethidium bromide (0.1 μg per ml) were used through out the study at concentrations of 1.2 or 1.6% (w/v). LKB power supply (Biochrom,Cambridge, England) and UV Transilluminator (Dinco and Rhenium Industrial Ltd.,), 100-bp ladder was used as molecular weight markers. Gels were photographed using a digital camera (Casio Exilim, Tokyo, Japan) at 3–8 mega pixels with sepia or black and white filter.
The Universal Method Protocol
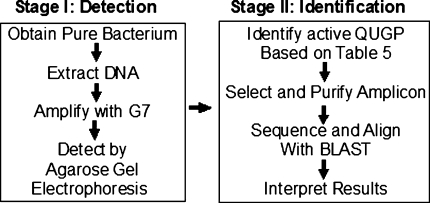
Figure 1 illustrates the two stages of the UM protocol; stage I was designed to allow detection of bacteria followed by stage II which was designed to identify the target bacterium. The two stages are detailed below.
Fig. 1.
A flowchart presentation of the UM protocol; starting from pure bacterial culture or DNA, ending with DNA alignment (BLAST) and bacterial identification
Stage I
Detection of a bacterium: starting with a pure bacterium or its DNA, one or more Golden primer mixtures (Table 5B) is used to amplify the 16S rDNA of the bacterium by PCR. Agarose gel electrophoresis is used to identify the size of the PCR amplicon, the active primer pair is then identified by cross referencing with Table 6. This stage was concluded once detection of amplicons was accomplished.
Table 6.
PCR product size with paired primers based on published rDNA gene H. pylori (HP) HPAG1 gi|108562424:c1140006-1138506 or P. fluorescens (PF) gi|70728250:122811-124349 of Pf-5 NC_004129
| Forward primer | QUGP-F1 | QUGP-Fn3 | QUGP-F4 | QUGP-Fn5 | QUGP-Fn6 |
|---|---|---|---|---|---|
| Reverse primer | HP/PF | HP/PF | HP/PF | HP/PF | HP/PF |
| QUGP-Rn1 | 1463/1503 | 721/743 | 633/639 | 1142/1183 | 995/991 |
| 1277b | |||||
| QUGP-Rn2 | 00/1396 | 623/636 | 532/536 | 00/1073 | 00/893 |
| 1097b | |||||
| QUGP-Rn3 | 764/798 | NPa | NP | 465/473 | 287/298 |
| QUGP-R4 | 847/881 | 112 | NP | 530/561 | 374/379 |
| QUGP-Rn5 | 334/344 | NP | NP | NP | NP |
| QUGP-Rn6 | 488/523 | NP | NP | 171/199 | NP |
The exact PCR product size for each primer pair may vary from one bacterium to another. Some primer sites are missing and their products are indicated as zero (00 bp)
aPrimers that did not form PCR pairs are indicated as no pairing (NP). The exact PCR product size depends on bacterial species. Italicized products indicate size similarity
bHomo sapiens 18S sequence shares homologies with these primers and predicts the possible amplification of 1277 and 1097 bp region
Stage II
To identify the bacterium, the PCR product must be reproduced in a pure form for DNA sequencing, BLAST alignment, and interpretation of results. If unambiguous high identity ≥98% was scored with only a single species, then the results are accepted. If high scores were obtained (usually between 95 and 98%) with several species, this may permit genus identification only. Independent methods must be applied to discriminate between these species and identify the bacterial species. If low identity was obtained (<95%), it is most likely due to unavailability of the sequence for alignment or due to the detection of a new species. This potential for discovering novel species should not be dismissed as an experimental error. Such bacteria must be identified by classical, cultural, biochemical testing, and by whole genome sequencing. The obtained sequence (rDNA or whole genome) should then be deposited into one of the nucleotide data bases. In theory, only 100% matches should be considered as identity. However, due to mutations, gene duplication and copy number, and sequencing errors, it becomes difficult to decide where to draw the identity line.
Results
The Universal Method (UM), described in this work, was applied to 101 different known and unknown bacterial isolates. The method detected all 101 tested isolates by producing one or more PCR products from each isolate. To insure that the 101 isolates represent a wide range of bacteria and to show the utility of the UM in the identification of bacteria; forty isolates of the 101 isolates were subjected to sequencing. A selected PCR products (preferably >500 bp) was purified and sequenced. All 40 isolates were successfully sequenced, aligned with BLAST [43, 44] and bacteria were identified to species or genus level, the results are summarized in Table 7.
Table 7.
Identified bacteria based on sequencing and/or other methods
| Name* | BLAST identification, DNA sequence (%) identity | PCR and sequencing primers | |
|---|---|---|---|
| QUBC1 | P. aeruginosa API 20 NEa | PA-SS | |
| QUBC33 | H. pylori ATCC 43526, confirmed with HP-specific primers HP-F,HP-Rf | QUGP | |
| F6, R3 | |||
| Isolates listed below were sequenced and identified with indicated Primers | QUGP | ||
| QUBC3 |
P. fluorescens NC 004129.61, rhizosphere isolate, PA-GS genus specific primers, API 20NEb |
(98.7) | F3, R1 |
| QUBC4 | Alcaligenes sp. ZB gb|FJ151631.1|, soil isolate | (97)c | F3, R1 |
| QUBC5 | Aeromonas veronii emb|AM937446.1|, spring water pond | (100) | F4, R1 |
| QUBC6 | Serratia marcescens PSB19gb|FJ360759.1|, soil sample | (100) | F3, R1b |
| QUBC7 | Aeromonas salmonicida ref|NC_009348.1|, soil | (94.8)c | F4, R1 |
| QUBC8 | P. aeruginosa gb|EU221384.1|, mosquito | (99.1) | F4, R1 |
| QUBC9 |
Klebsiella oxytoca gb|EU554427.1|, Clinical sample; Al-Maqased Hospital, Jerusalem, Palestine |
(99.7) | F3, R1b |
| QUBC10 | Salmonella enterica NZABEN01000001.1, Fish intestines | (99.3) | F5, R3 |
| QUBC11 | Microbacterium hydrocarbonoxydans emb|FM163607.1|, Soil | (99) | F5, R3 |
| QUBC12 |
Acinetobacter baylyi mcp11c gb|EF419183.1|, A. baumannii API-20E (98.1), wood sawing powder |
(99.7) | F3, R1b |
| QUBC13 | Ochrobactrum anthropi NC009668.1 Chromosome2 Isolated from decaying broomrape | (99.7) | F3, R1b |
| QUBC14 |
Enterobacter ludwigii T4384 gb|EU999992.1| soil, Enterobacter sp. API-20E 99.8% |
(99.3) | F3, R1b |
| QUBC15 | Enterobacter ludwigii T4384 gb|EU999992.1| soil | (99.5) | F3, R1b |
| QUBC16 | Bacillus atrophaeus gb|EU729737.1|, Autoclave test bag | (99.2) | F3, R1b |
| QUBC18 | Bacillus subtilis ref|NC_000964.2|, fecal sample | (100) | F6, R3 |
| QUBC19 | Kocuria sp. JSM gb|FJ237398.1|, Aphids | (100) | F6, R3 |
| QUBC24 | Aeromonas hydrophila AC-C9gb|DQ865062.1| spring pond | (98.5) | R1 |
| QUBC25 | Bacillus thuringiensis gb|EU162014.1|, lab contaminant | (99) | F1, R6 |
| QUBC32 |
Serratia marcescens gb|FJ360759.1|, soil sample, S.spp. API-20E (99.1%) |
(99.2) | F3, R2 |
| QUBC34 | Staphylococcus aureus ref|NZ_ACJA01000069.1|, a gift, M. Ayesh | (99.3) | F4, R2 |
| QUBC35 |
Citrobacter koseri ref|NC_009792.1|, Citrobacter sp. API (99.9%), a gift from M. Ayesh |
(95.5)c | F3,4R1 |
| QUBC36 | Pseudomonas otitidis gb|AY953147.1|, Clinical sample | (98.5) | F3, R1 |
| QUBC37 | Enterobacter sp gb|FJ025770.1|, a gift from M. Ayesh | (99) | F3, R1 |
| QUBC42 |
Salmonella enterica gb|ABAM02000001.1|, Salmonella sp. API-20E, clinical sample, Al-Maqased Hospital |
(100) | F3, R1b |
| QUBC43 |
Shigella boydii gb|CP001063.1|, Shigella sp. API-20E, clinical sample, Al-Maqased Hospital |
(99.8) | F3, R1b |
| QUBC50 | Streptomyces fragilis gb|EU841657.1, Dove feather | (99.8)e | F3, R2 |
| QUBC60 |
Pantoea agglomerans I10 gb|DQ065752.1|, carrot rot, Pantoea sp. API -20E (97.9%) |
(99.5) | F3, R2 |
| QUBC70 | Phaeospirillum sp., spiral bacterium fecal sample | (95)d2 | F3, R2 |
| QUBC71 | Neisseria meningitides ref|NC_008767.1|, mouth swab | (98.8) | F3, R2 |
| QUBC75 | Dyella marensis gb|FJ535861.1| | (96.4)d | F3, R2 |
| QUBC77 | Alkalilimnicola ehrlichii ref|NC_008340.1|, Sardin fish | (89)d | F3, R2 |
| QUBC80 | Enterococcus faecalis ref|NZ_ACAV01000046.1| Clinical | (98.2) | F3, R2 |
| QUBC81 | Enterococcus faecalis ref|NZ_ACAV01000046.1| Clinical | (98.9) | F3, R2 |
| QUBC82 | Clavibacter michiganensis ref|NC_010407.1| Clinical | (93)d | F3, R2 |
| QUBC83 | Enterococcus faecalis ref|NZ_ACAV01000046.1| Clinical | (99.3) | F3, R2 |
| QUBC91 | Listeria monocytogenes ref|NZ_AARP03000051.1| ATCC 19115 | (99) | F3, R2 |
| QUBC102 | Streptomyces griseus ref|NC_010572.1| Pigeon feather | (98.9) | F3, R2 |
| QUBC103 | Streptomyces griseus ref|NC_010572.1| Pigeon feather | (99)e | F3, R2 |
| QUBC104 | Streptomyces coelicolor A3 ref|NC_003888.3| Soil | (99)e | F3, R2 |
| QUBC106 | Streptomyces mediolani gb|FJ486429.1| Soil | (99)e | F3, R2 |
* These bacteria are stored frozen at −70°C as part of Al-Quds University Bacterial Collection
aThis test was performed with API 20NE by the Microbiology Laboratory, The Karitas Hospital, Bethlehem
bThe Microbiology laboratory, Ministry of Health, Ramallah, Palestine
cPoor sequencing results
d2Good sequencing results but poor BLAST alignment result, 2 indicates an independent repeat of sample processing on a different day
eDifferent colony morphologies
fH. pylori specific primers were used to confirm isolate, according to reference [8], while PCR product was generated with QUGP-F6.R3, no sequencing was attempted for this bacterium
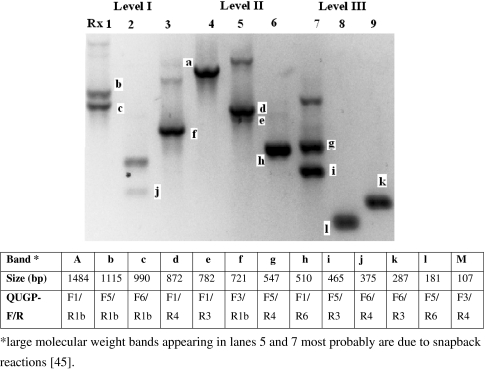
Predicted PCR Amplicons were Obtained with QUGP Primers
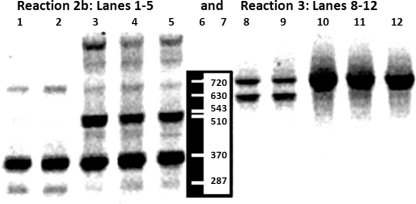
Table 6 predicts the possible PCR amplicons that may be generated with each pair of Al-Quds University Primers (QUGPs). Table 6 was constructed based on published 16S rDNA sequences of H. pylori and Pseudomonas fluorescens. It shows that some primer sites are missing; (e.g. QUGP-Fn5 with QUGP-Rn2 will result in no amplification when applied to H. pylori, but will produce 1073 bp PCR product if applied to P. fluorescens). When the designed QUGPs were mixed (Table 5A) and applied to DNA extracted from Streptococcus sp. QUBC26, the predicted PCR amplicons were obtained as illustrated in Fig. 2. These PCR products were consistent with those predicted in Table 6, demonstrating that all short QUGPs were functional. The produced amplicons were also consistent with primer distribution shown in Table 2 for the bacterium Streptococcus pyogenes. Reaction 2b and 3 (Table 5A) were applied to five additional bacterial isolates (Shigella sp. QUBC43, Salmonella sp. QUBC42, Klebsiella sp. QUBC41, Proteus sp. QUBC40, and Staphylococcus aureus QUBC34). All isolates were detected as illustrated in Fig. 3. Amplicons were consistent with those predicted by Table 6. PCR product at 630 bp indicating the activity of primer pair QUGP-F4.R1 (of reaction 3, Table 5A) was detected with two isolates (lanes 8 and 9). The same amplicon was missing from lanes 10, 11, and 12 indicating sequence mismatch with QUGP-F4 or suboptimal reaction conditions that favoured the formation of the larger product (720 bp) amplified with QUGP-F3.R1b; in lanes 8–12 of Fig. 3. The large molecular weight bands appearing in lanes 5 and 7 of Fig. 2 and lanes 3, 4 and 5 of Fig. 3 are non-specific bands; most probably due to snapback mediated amplification associated with suboptimal annealing temperature for QUGP-R4 primer [45].
Fig. 2.
All nine Level reactions (Table 5A) were positive with isolate QUBC26. Lanes represent Level reactions in the same order (1–9). Interpreted results are shown in the table
Fig. 3.
Reaction 2b (lanes1–5) and Reaction 3 (lanes 8–12 (Table 5A). Lanes 1 and 8 QUBC43 Shigella (API), lanes 2 and 9 QUBC42 Salmonella (API), lanes 3 and 10 QUBC41 Klebsiella (API), lanes 4 and 11 QUBC40 Proteus, and lanes 5 and 12 QUBC34 Staphylococcus aureus. Notice the presence of the 510 bp in lanes3–5 and its absence from lanes 1 and 2 and the presence of a 630 bp band in lanes 8 and 9 and its absence in lanes 10, 11, and 12
Selection of Golden Mixtures for Efficient Bacterial Detection
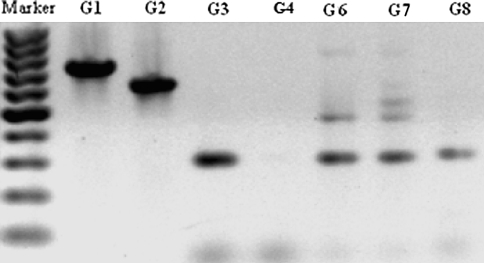
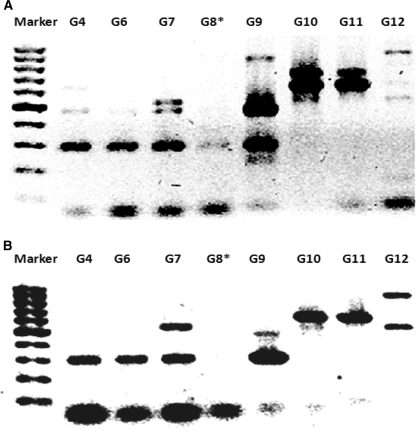
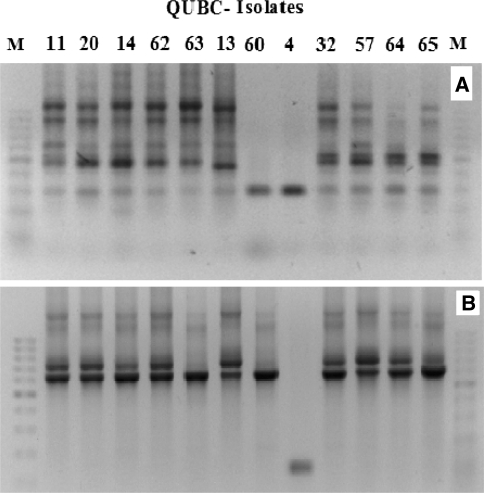
Different Golden Mixtures were prepared (Table 5B) and their efficacy to detect bacteria was tested. The mixtures G1, G2, and G3 were considered too simple and were only included as control mixtures. G4, G6, G8, and G9 had failed even when tested against few bacterial isolates. G4 had failed with Streptomyces sp. QUBC50 which was detected by G1, G2, and G3 (Fig. 4), yet G4 was productive with both QUBC56 and 58 (Fig. 5). G8, G9, and G13 had several failures when tested against few (≤5) bacterial isolates (not shown). G6 and G9 showed bias to producing small PCR products (Figs. 4, 5). On preliminary screening, G5, G7, G10, G11, and G12 performance was acceptable. Further investigation of G7 and G10 showed that G7 was capable of producing positive PCR reactions with all tested bacterial isolates (67/67; 100%). 12 bacterial isolates were randomly selected for detection with G7 (Fig. 6a) which detected one band from isolate QUBC4. The same isolates were tested with G10 (Fig. 6b) which detected all except QUBC4, G10 produced positive PCR results with a total of 25 bacterial isolates (>96%). Both G11 and G5 performed well, they detected all 23 and 11 tested isolates respectively; QUBC50 was clearly detected with G5 but poorly with G11.
Fig. 4.
G4 has failed to detect this bacterium; Streptomyces sp.(QUBC50), notice the absence of the 630 bp which appeared with G2, G4 showed very week band at 287/298 bp that was clearly present with G3
Fig. 5.
a (QUBC56) and b (QUBC58) tested with different Golden mixtures; G4–G12, showing poor performance of G8 (asterisks)
Fig. 6.
Detection of bacterial rDNA from 12 different QUBC isolates with G7 (a) or with G10 (b), notice that isolate QUBC4 was negative with G10
Validation of Bacterial Detection and Bacterial Identification (Stage II)
Amplicon Sequences are Those of 16S rDNA Genes
To confirm that PCR amplicons obtained from Stage I of the UM (Fig. 1) represent target DNA sequences, it was necessary to show that the produced PCR amplicons represent the 16S rDNA gene, and to show that these amplicons can be used in the identification of the source bacterium.
Having the 40 amplicons sequenced and aligned using BLAST, only 16S rDNA was detected indicating 100% specificity of amplification of rDNA from those 40 different isolates and most likely from the rest of the 101 tested isolates (Table 7). The results are highly supportive of the conclusion that the UM-QUGPs can only amplify and detect 16S rDNA sequences.
DNA Sequence Alignment and Bacterial Identification
Identification of Pseudomonas spp.
The isolate (QUBC1) was used as a control bacterium to test the concurrent validity of the PCR and sequencing system, published P. aeruginosa specific PCR primers PA-SS-F and PA-SS-R [18] were utilized with this isolate. The primers were used to amplify a segment of the 16S rDNA gene, the product was sequenced (Automated Sequencing, Bethlehem University, Bethlehem, Palestine) [19]. Alignment of the sequenced amplicon showed 98.5% identity to the 16S gene of P. aeruginosa (Table 7).
The isolate QUBC3 obtained from the rhizosphere of a garden weed, was identified with API 20NE as P. fluorescens/putida. The UM was applied to this bacterium. A 720 bp segment of the 16S rDNA of QUBC3 was amplified with QUGP-F3 and QUGP-R1, upon sequencing of the PCR amplicon and BLAST alignment, QUBC3 was confirmed to be P. fluorescens (98.7%) offering additional validation of the UM capacity to detect 16S rDNA and to identify the source bacterium. This process of validation was repeated with a total of 40 isolates (Table 7). Another suspected Pseudomonas isolate QUBC37 was identified (98.5% identity) by the UM as P. otitidis. The isolate QUBC8 obtained from a mosquito larva was identified as P. aeruginosa (99.1%; Table 7) as a result of sequencing and alignment a 640 bp amplicon produced with QUGP-F4.R1.
Identification of Three Bacillus Species
Three Bacillus species defined as aerobic spore forming Gram positive rods. Isolates QUBC16, 18, and 25 were isolated accordingly. DNA extracted from these isolates was amplified using the UM (QUGP multiplex mixtures; Table 5A) for identification.
QUBC16: Reaction 1 was negative, while two bands were obtained with Reaction 2; a 450 bp/QUGP-F5.R3 and a 287 bp/QUGP-F6.R3. Reaction 3 produced a 720 bp/QUGP-F3.R1b which was sequenced with QUGP-F3 and QUGP-R1b. BLAST analysis of the 611 bp identified the bacterium as Bacillus atrophaeus with 100% identity. QUBC18 isolate was positive with all Level I reactions, it was identified as B. subtilis with 100% identity. The 287 bp product was obtained and sequenced with QUGP-F6.R3. The third isolate, QUBC25 was identified as B. cereus with 99% identity using the primer pair QUGP-F1.R6. Although, other primers can be used since the bacterium was positive for all Level II reactions 4, 5 and 6. The intentional application of different primer pairs with these isolates was aimed at showing the flexibility of the UM in accommodating the length and location of the amplicon for identification purposes, Table 7. Therefore, it may not be critical as to which segment and length should be utilized, but for the purpose of rigorousness segments >500 bases are preferable.
Identification of Two Different Colonies as Enterobacter ludwigii
Two morphologically different isolates, QUBC14 and 15, were obtained from a soil sample as pink colonies on the same MacConkey agar plate. When amplified and sequenced, both isolates were identified as Enterobacter ludwigii (>99%) and confirmed with API 20E as Enterobacter sp. (99.8%). Both were PCR positive with Mq10 (Table 5A). A 720 bp PCR product obtained from each isolate and sequenced with QUGP-F3 and QUGP-R1b.
Identification of Ochrobactrum anthropi
Unknown soil isolate QUBC13 was PCR-sequenced with QUGP-F3 and QUGP-R1b. Alignment with BLAST [43, 44], identified the bacterium as Ochrobactrum anthropi, chromosome2 with 99.7% identity. Biochemical reactions were consistent with this identification, the bacterium was Gram negative, aerobic, non-fermenter rod, urease and oxidase positive, and indole negative [47]. Neither API 20 NE nor API 20 E could identify this isolate.
Identification of Acinetobacter
API 20NE system identified QUBC12 as A. baumannii (98.1%). The isolate was identified by PCR-sequencing as A. baylyi (99.7%). Both methods identified the bacterium at the genus level but disagreed at the species level, sequence identity to A. baumannii was 99.1%, the sequence was 100% identical to uncultured bacterium gb|EF121340.1|.
Identification of Citrobacter Isolate
A putative Citrobacter isolate QUBC35, was subjected to PCR-sequencing with QUGP-F3 and QUGP-R1b followed by BLAST analysis. The bacterium was identified as Citrobacter koseri (95.5%) ref|NC_009792.1|. API 20E system identification indicated Citrobacter sp. (99.9%); C. freundii (92.5%) or C. youngae (7.4%).
Identification of Additional Isolates
Additional examples of identified bacteria are presented in Table 7, the sequencing primers are indicated for each isolate as well. Isolate QUBC32 produced deep pink colonies characteristic of Serratia marcescens and was identified as such (95.9%) or 99.8% as Serratia sp. with API 20E. PCR-sequencing with QUGP-F3 and QUGP-R2 confirmed its identity as S. marcescens gb|FJ360759.1| (99.2%). QUBC50 sequenced with QUGP-F3 and R2 was identified as Streptomyces fragilis (99.8%). The result was in agreement with characteristic odor and morphology of Streptomyces spp. Isolate QUBC42 was API 20E indentified as Salmonella sp., PCR-sequencing identified it to species level as S. enterica (100%). Isolate QUBC43 was identified as Shigella sp. with API 20E or S. boydii (99.8%) by PCR-sequencing (Table 7). Poor sequencing may result in poor identification as indicated for QUCB4, QUBC7, and QUBC35.
Primer Specificity and Sensitivity
The sequenced amplicons obtained from 40 different isolates were found to be those of 16S rDNA, they were aligned with BLAST which identified the bacterium to species or genus level; 24 genera and 34 species were represented. Among the 101 detected bacteria, eight additional genera were represented; Agrobactrum sp., Campylobacter sp., H. pylori ATCC 43526, Lactobacillus sp., Proteus sp., Staphylococcus epidermidis, Stenotrophomonas sp., and Streptococcus sp. bringing the total number of detected bacterial to exceed 32 genera. The ability of the UM to detect and discriminate between several genera and species makes it highly specific and sensitive provided that good DNA sequences are obtained and that matching sequences exist in nucleotide data bases for alignment analysis.
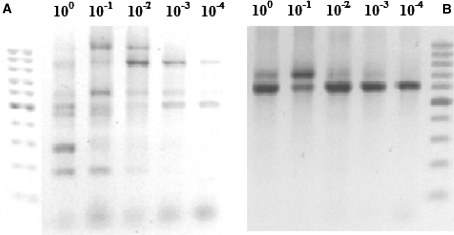
To test the sensitivity of detection of bacterial DNA, a diluted sample containing 1 μl of bacterial DNA diluted by a factor of 10−4 folds, was detected with both G7 and G10 (Fig. 7). Some PCR amplicons disappeared as DNA concentration was reduced, possibly due to imperfect annealing of some primers (Table 2). One of the amplified bands continued to show strong presence even when sample DNA was diluted 10−4 folds, higher dilutions were not tested.
Fig. 7.
Ten-fold serially diluted DNA samples from clinical Escherichia coli isolate QUBC62; API 20E, were tested with G7 (a) or G10 (b). 100-bp ladder markers were included
Validation of the UM was supported by the obtained results; only 16S sequences were amplified. Two methods of bacterial identification were in agreement; the UM and another valid method. The obtained amplicons had similar sizes to predicted amplicons shown in Table 6. The detection process was successful with 67 isolates when G7 was applied. A total of 101 isolates were detected (100%). Sequencing and alignment were performed on 40 different isolates i.e. the system was tested repeatedly (40 times) and isolates were identified.
Discussion
As predicted in the hypothesis and illustrated in Figs. 1, 2, 3, 4, 5, 6, and 7, the appearance of different PCR amplification profiles for different bacterial isolates (Fig. 3) supports the hypothesis in that more than one general primer pair was required to detect all bacteria, the hypothesis predicted the shortcoming of “Universal Primers”. The results showed that G4, G6, G8, G9, and G10 had failed with one or more isolates. The inevitable failure of universal primer pairs (or general primer pairs) can be further predicted based on primer distributions; Table 2 shows that no single primer could react with all 44 listed genera, meaning that any given primer pair will at least fail once (2.2%). The “Universal” primer pair 909F.R (Table 1) [17] will fail with 22 different genera or 50% of those listed in Table 2. This renders the 909-primer pair neither specific nor universal. The Primer pair QUGP-F3.R2 (which proved useful in this study, Table 7) will fail with three genera that lack the QUGP-R2 sequence (Aquifex, Rickettsia, Thermotoga; Table 2) and it will fail with nine genera that lack QUGP-F3 (Anabaena, Aquifiex, Bordetella, Campylobacter, Chlamedophila1, Desulfovibrio, Mycoplasma, Prochlorococcus, and Thermotoga). In applying this PCR primer pair to the 44 genera listed in Table 2, the primer pair will fail with ten genera allowing the detection of 34 genera only (72%). Similarly, the primer pair QUGP-F5.R4 will detect 84% of the listed genera; QUGP-F5 will fail with two genera (Chlamydophila1,Rhodopirellula Table 2) while QUGP-R4 will fail with Chlamydophila1 and five additional genera (Mycoplasma, Syntrophobacter, Thermus, Treponema, and Wolbachia). Accordingly, QUGP-F5.R4 will only detect 37 (84%) genera of those listed in Table 2. Mathematically, the two pairs QUGP-F3.R2 and QUGP-F5.R4 will detect (95.4%) all 44 but (Chlamydophila1 and Mycoplasma). However, mixing the two pairs will have the added advantage of generating a new pair (QUGP-F5.R2) which will detect Mycoplasma but not Chlamydophila1. Therefore mixing the four primers will result in the detection of 43 genera (97.8%). Now, it is clear how G7 with its 10 primer pairs could detect bacteria. G7 will detect all 44 genera shown in Table 2 which may be a representative sample of all bacteria. According to the present study, additional genera (not listed in Table 2) had been detected by G7 (Acinetobacter, Alkaligenes, Serratia, Klebsiella and others; Table 7). These calculations strongly support the potential of the UM approach over the “Universal Primer” approach.
The limiting step in the identification of a bacterium using the UM resides in the detection step (Stage I). The UM solved this limitation by providing an array of primer mixtures (Table 5A, B). According to the UM, Golden Mixtures G7 and G5 can potentially produce 12 different primer pairs (or PCR amplicons). Additional primer pairs can be generated by incorporation of QUGP-F1 and QUGP-R7 in Golden mixtures. Incorporation of QUGP-F1 generates 6 additional primer pairs (Table 6). The 20 possible primer pairs shown in Table 6 are likely to detect any bacterium. G7 allowed for the formation of 10 different primer pairs responsible for the amplicons observed in Fig. 6a (QUGP-Fn3 neither pairs with QUGP-Rn3 nor with QUGP-F4). Two additional primer pairs (QUGP-Fn5.R4 and QUGP-Fn6.R4) can be generated with G5. Therefore, applying G7 and G5 to any bacterium will enhance the chances of detecting that bacterium by 12 folds over any single PCR primer pair, or by 20 folds if all primer pairs of Table 6 are to be used.
The detection of all 101 tested bacterial isolates utilizing the different QUGP mixtures with zero failure strongly supports the capacity of the UM to detect any bacterium. Golden Mixtures were developed for practical reasons and to simplify the process relative to utilizing single primer pairs; G7 alone capacity to produce positive PCR amplicons with each of (67/67) tested bacterial isolates, testifies to its potential to detect most if not all bacteria.
Both G7 and G10 were able to detect bacterial DNA when diluted 10,000 folds, this implies that samples should be sufficiently diluted when casual contamination is suspected; dilution of blood samples contaminated with Staphylococcus epidermidis or other bacteria can help avoiding detection of such contaminants, this area needs extensive investigation before it can be put to any practical use. G7 can be applied to test for the presence or absence of bacteria in a variety of samples including clinical samples.
Application of G7 is probably most suitable in the analysis of bacterial communities since it is expected to amplify the vast majority, if not all species, within the tested sample. Qualitative analysis of mixed PCR products with available techniques such as microarrays [12], C0t analysis, cloning and sequencing, RFLP, denaturing gradient gel electrophoresis analysis [14, 39] or any combination of these methods will improve our understanding of bacterial communities.
A critical requirement for DNA sequencing is that a sequencing primer should only anneal to a single site on the template DNA. Therefore, samples containing mixed DNA species obtained with General primers are not suitable for direct DNA sequencing. Another obstacle associated with sequencing based ribotyping is its dependency on the availability of rDNA sequences for alignment analysis (BLAST). For example, BLAST analysis identified isolate QUBC35 as Citrobacter koseri (95.5%), it was also identified as Citrobacter freundii (92.5%) or C. youngae (7.4%) with API 20E system, the unavailability of some species for BLAST analysis may have biased the identification of the isolate as C. koseri. Another example was obtained when the spiral bacterium QUBC70 was PCR-sequenced but was poorly identified. It was 95% identical to Phaeospirillum sp. most probably as a result of absence of the matching sequence from nucleotide data bases. The isolate QUBC70 is being identified using biochemical profiling, the obtained sequence will be deposited in the gene bank after bacterium identification is accomplished and confirmed. Having a complete bacterial nucleotide/gene bank (especially for ribosomal nucleotides sequences) will certainly improve the reliability of the UM and any other sequence-based methods.
Isolates showing BLAST homologies less than 98% were not accepted unless verified by another method. Two main reasons contributed to poor identification; poor sequencing and absence of corresponding sequence from the nucleotide database. Additional threats to the validity of the UM come from strong associations between bacterial species as in case of the parasitic bacteria (Bdellovibrio sp.), gene copy number, and polymorphism.
When highly conserved genes such as 16S genes are used for bacterial identification, long sequences (≥500 bases) may provide sufficient discriminatory power amongst bacteria than would shorter sequences. Therefore, sequences ≥500 bases long are preferable for identification although species specific shorter sequences can be applied [9, 20–22, 24].
The validity of the UM was established; all 40 sequenced PCR amplicons were those of 16S rDNA. The UM detected and identified 24 genera and 34 species. This wide range of species detection and identification justifies the application of the method to any other bacterium. The UM was further validated; since it was in agreement with other identification methods such as API, morphological and biochemical profiles, and species specific PCR primers. The reproducibility of the results obtained by the UM testify for its stability; detection of all Enterococcus faecalis isolates (QUBC80,81, and 83). The ability of the UM to discriminate between Streptomyces spp. QUBC50, 102, 103, 104, and 106 was illustrated as well. Repetition of sequencing and BLAST analysis with the same isolate as in QUBC70 revealed the same exact results.
In conclusion, G7 together with G5 may represent a true substitution to universal primers; as predicted in the hypothesis, these two mixtures contained a total of 8 general primers (7 primers in G7 and 4 primers in G5). Potentially, they may detect all bacteria since they generate a total of 12 primer pairs. G7 and G5 can be utilized to serve a number of other applications such as the detection of bacterial presence in samples, analyses of bacterial communities, and bacterial identification.
Acknowledgments
This work was partially supported by DFG grant Number: 885-2341. Special thanks to Mr. Mohamed Hassan Qabajah for volunteer work, and Mr. Mohamed Ayesh for provision of some bacterial isolates. Department of Medical Laboratory Sciences and professor Samira Barghouthi, Dean of Research, Al-Quds University, for suggestions and support.
References
- 1.Bergeron MG, Ouellette M. Preventing antibiotic resistance through rapid genotypic identification of bacteria and of their antibiotic resistance genes in the clinical microbiology laboratory. J Clin Microbiol. 1998;36:2169–2172. doi: 10.1128/jcm.36.8.2169-2172.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felske A, Wolterink A, Lis R, Akkermans ADL. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands) Appl Envir Microbiol. 1998;64:871–879. doi: 10.1128/aem.64.3.871-879.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mareckova MS, Cermak L, Novotna J, Plhackova K, Forstova J, Kopecky J. Innovative methods for soil DNA purification tested in soils with widely differing characteristics. Appl Environ Microbiol. 2008;74:2902–2907. doi: 10.1128/AEM.02161-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ott SJ, Musfeldt M, Ullmann U, Hampe J, Schreiber S. Quantification of intestinal bacterial populations by real-time pcr with a universal primer set and minor groove binder probes: a global approach to the enteric flora. J Clin Microbiol. 2004;422:566–2572. doi: 10.1128/JCM.42.6.2566-2572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sigal N, Senez JC, Le Gall J, Sebald M. Base composition of the deoxyribonucleic acid of sulfate-reducing bacteria. J Bacteriol. 1963;85:1315–1318. doi: 10.1128/jb.85.6.1315-1318.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perry D, Slade H. Transformation of streptococci to streptomycin resistance. J Bacteriol. 1961;83:443–449. doi: 10.1128/jb.83.3.443-449.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inácio J, Flores O, Martins IS. Efficient identification of clinically relevant Candida yeast species by use of an assay combining panfungal loop-mediated isothermal DNA amplification with hybridization to species-specific oligonucleotide probes. J Clin Microbiol. 2008;46:713–720. doi: 10.1128/JCM.00514-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nilsson H, Taneera J, Castedal M, Glatz E, Olsson R, Wadström T. Identification of Helicobacter pylori and Other Helicobacter Species by PCR, hybridization and partial DNA sequencing in human liver samples from patients with primary sclerosing cholangitis or primary biliary cirrhosis. J Clin Microbiol. 2000;38:1072–1076. doi: 10.1128/jcm.38.3.1072-1076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owen RH, Dymock D, Booth V, Weightman AJ, Wade WG. Detection of unculturable bacteria in periodontal health and disease by PCR. J Clin Microbiol. 1999;37:1469–1473. doi: 10.1128/jcm.37.5.1469-1473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lund M, Nordentoft S, Pedersen K, Madsen M. Detection of Campylobacter spp. in chicken fecal samples by real-time PCR. J Clin Microbiol. 2004;42:5125–5132. doi: 10.1128/JCM.42.11.5125-5132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards GP, Watson MA, Fankhauser RL, Monroe SS. Genogroup I and II noroviruses detected in stool samples by real-time reverse transcription-PCR using highly degenerate universal primers. Appl Envir Microbiol. 2004;70:7179–7184. doi: 10.1128/AEM.70.12.7179-7184.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitterer G, Huber M, Leidinger E, Kirisits C, Lubitz W, Mueller MW, Schmidt WM. Microarray-based identification of bacteria in clinical samples by solid-phase PCR amplification of 23S ribosomal DNA sequences. J Clin Microbiol. 2004;42:1048–1057. doi: 10.1128/JCM.42.3.1048-1057.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasuoka MO. A multiplex polymerase chain reaction–based diagnostic method for bacterial vaginosis. Obst Gynecol. 2002;100:759–764. doi: 10.1016/S0029-7844(02)02201-9. [DOI] [PubMed] [Google Scholar]
- 14.Lelie D, Lesaulnier C, McCorkle S, Geets J, Taghavi S, Dunn J. Use of single-point genome signature tags as a universal tagging method for microbial genome surveys. Appl Environ Microbiol. 2006;72:2092–2101. doi: 10.1128/AEM.72.3.2092-2101.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson M, Bollinger D, Hagler A, Hartwell H, Rivers B, Ward K, Steck TR. Viable but Nonculturable Bacteria are Present in Mouse and Human Urine Specimens. J Clin Microbiol. 2004;42:753–758. doi: 10.1128/JCM.42.2.753-758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haas JH, Moore LW, Ream W, Manulis S. Universal PCR primers for detection of phytopathogenic Agrobacterium strains. Appl Environ Microbiol. 1995;61:2879–2884. doi: 10.1128/aem.61.8.2879-2884.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng X, Luo W, Zhang J, Wang S, Lin S. Rapid detection of Shigella species in environmental sewage by an immunocapture PCR with universal primers. Appl Environ Microbiol. 2002;68:2580–2583. doi: 10.1128/AEM.68.5.2580-2583.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spilker T, Coenye T, Vandamme P, LiPuma JJ. PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol. 2004;42:2074–2079. doi: 10.1128/JCM.42.5.2074-2079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bethlehem University, DNA sequencing http://hrl.bethlehem.edu/services.shtml
- 20.Kirchman DL, Yu L, Cottrell MT. Diversity and abundance of uncultured Cytophaga-like bacteria in the Delaware Estuary. Appl Environ Microbiol. 2003;69:6587–6596. doi: 10.1128/AEM.69.11.6587-6596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kox LFF, Leeuwen J, Knijper S, Jansen HM, Kolk HJ. PCR assay based on DNA coding for 16S rRNA for detection and identification of Mycobacteria in clinical samples. J Clin Microbiol. 1995;33:3225–3233. doi: 10.1128/jcm.33.12.3225-3233.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verma A, Sampla AK, Tyagi JS. Mycobacterium tuberculosis rrn promoters: differential usage and growth rate-dependent control. J Bacteriol. 1999;181:4326–4333. doi: 10.1128/jb.181.14.4326-4333.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barghouthi S. Helicobacter pylori: a theoretical background with a practical approach to growth and behavior. Ramallah, Palestine: Acknaton Design and Print; 2009. [Google Scholar]
- 24.Tyler SD, Strathdee CA, Rozee KR, Johnson WM. Oligonucleotide primers designed to differentiate pathogenic Pseudomonads on the basis of the sequencing of genes coding for 16S–23S rRNA internal transcribed spacers. Clin Diag Lab Immunol. 1995;2:448–453. doi: 10.1128/cdli.2.4.448-453.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aakra A, Utaker JB, Nes IF. RFLP of rRNA genes and sequencing of the 16S–23S rDNA intergenic spacer region of ammonia-oxidizing bacteria: a phylogenetic approach. Int J Sys Bacteriol. 1999;49:123–130. doi: 10.1099/00207713-49-1-123. [DOI] [PubMed] [Google Scholar]
- 26.Bosshard PP, Zbinden R, Abels S, Boddinghaus B, Altwegg M, Bottger EC. 16S rRNA gene sequencing versus the API 20 NE system and the VITEK 2 ID-GNB card for identification of nonfermenting gram-negative bacteria in the clinical laboratory. J Clin Microbiol. 2006;44:1359–1366. doi: 10.1128/JCM.44.4.1359-1366.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall SM, Melito PL, Woodward DL, Johnson WM, Rodgers FG, Mulvey MR. Rapid identification of Campylobacter, Arcobacter and Helicobacter isolates by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene. J Clin Microbiol. 1999;37:4158–4160. doi: 10.1128/jcm.37.12.4158-4160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raut AD, Kapadnis BP, Shashidhar R, Bandekar JR, Vaishampayan P, Shouche YS. Letter to the editor “nonspecific pcr amplification of the 16s rrna gene segment in different bacteria by use of primers specific for Campylobacter, Arcobacter and Helicobacter spp”. J Clin Microbiol. 2007;45:1376–1377. doi: 10.1128/JCM.02559-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexeeva I, Elliott EJ, Rollins S, Gasparich GE, Lazar J, Rohwer RG. Absence of Spiroplasma or other bacterial 16S rRNA genes in brain tissue of hamsters with scrapie. J Clin Microbiol. 2006;44:91–97. doi: 10.1128/JCM.44.1.91-97.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ben-Dov E, Shapiro OH, Siboni N, Kushmaro A. Advantage of using inosine at the 3-termini of 16S rRNA gene universal primers for the study of microbial diversity. Appl Envir Microbiol. 2006;72:6902–6906. doi: 10.1128/AEM.00849-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Hara CM. Manual and Automated instrumentation for identification of Enterobacteriaceae and Other Aerobic Gram-negative Bacilli. Clin Microbiol Rev. 2005;18:147–162. doi: 10.1128/CMR.18.1.147-162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pujol BS, Vabret A, Legrand L, Dina J, Gouarin S, Lecherbonnier PJ, Pozzetto B, Ginevra C, Freymuth F. Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J Virol Methods. 2005;126:53–63. doi: 10.1016/j.jviromet.2005.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hendolin PH, Markkanen A, Ylikoski J, Wahlfors JJ. Use of multiplex PCR for simultaneous detection of four bacterial species in middle ear effusions. J Clin Microbiol. 1997;35:2854–2858. doi: 10.1128/jcm.35.11.2854-2858.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daly K, Sharp RJ, McCarthy AJ. Development of oligonucleotide probes and PCR primers for detecting phylogenetic subgroups of sulfate-reducing bacteria. Microbiol. 2000;146:1693–1705. doi: 10.1099/00221287-146-7-1693. [DOI] [PubMed] [Google Scholar]
- 35.Han J, Swan DC, Smith SJ, Lum SH, Sefers SE, Unger ER, Tang YW. Simultaneous amplification and differentiation of 25 human papillomaviruses with Templex technology. J Clin Microbiol. 2006;44:4157–4162. doi: 10.1128/JCM.01762-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daims H, Bru¨hl A, Amann R, Schleifer KH, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 37.Hathaway LJ, Brugger S, Martynova A, Aebi S, Mühlemann K. Use of the Agilent 2100 Bioanalyzer for Rapid and Reproducible Molecular Typing of Streptococcus pneumoniae. J Clin Microbiol. 2007;45:803–809. doi: 10.1128/JCM.02169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luna RA, Fasciano LR, Jones SC, Boyanton BL, Jr, Ton TT, Versalovic J. DNA pyrosequencing-based bacterial pathogen identification in a pediatric hospital setting. J Clin Microbiol. 2007;45:2985–2992. doi: 10.1128/JCM.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muyzer G, Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stull TL, LiPuma JJ, Edlind TD. A broad-spectrum probe for molecular epidemiology of bacteria: ribosomal RNA. Infect Dis. 1988;157:280–285. doi: 10.1093/infdis/157.2.280. [DOI] [PubMed] [Google Scholar]
- 41.Eubacterial Species http://www.speciesaccounts.org/SPECIES%20LISTS.htm
- 42.Barghouthi S (2008) A UM for the identification of bacteria based on general PCR primers. In: Abstract Annual General Meeting American Society for Microbiology, K-085, p 150
- 43.BLAST: http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi
- 44.BLAST (Nucleotide collection): http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Nucleotides&PROGRAM=blastn&MEGABLAST
- 45.Maniatis T, Fritsch EF, Sambrook J. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 46.Clayton CL, Kleanthous H, Coates PJ, Morgan DD, Tabaqchali S. Sensitive detection of Helicobacter pylori by using polymerase chain reaction. J Clin Microbiol. 1992;30:192–200. doi: 10.1128/jcm.30.1.192-200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teyssier C, Marchandin H, Jean-Pierre H, Diego I, Darbas H, Jeannot JL, et al. Molecular and phenotypic features for identification of the opportunistic pathogens Ochrobactrum spp. J Med Microbiol. 2005;54:945–953. doi: 10.1099/jmm.0.46116-0. [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto S, Harayama S. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of pseudomonas putida strains. Appl Envir Microbiol. 1995;61:1104–1109. doi: 10.1128/aem.61.3.1104-1109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]