Abstract
Background
Rho kinase signaling plays an important role in the oncogenic process largely through its regulation of F-actin dynamics, and inhibition of this pathway results in reduction in tumor volume and metastasis across a number of tumor types. While the cytoskeletal-regulatory role of Rho kinase has been a topic of in-depth study, the mechanisms linking Rho kinase to altered gene expression are largely unknown.
Materials and Methods
Global gene expression analysis was performed on melanoma tumors treated with sham or the small molecule inhibitor Y27632.
Results
Inhibition of Rho kinase activity in melanoma tumors results in a statistically significant change in gene transcription of 94 genes, many of which are critically involved in tumor initiation and progression.
Conclusion
In addition to regulating tumorigenesis through modulation of the phosphoproteome, Rho kinase signaling also contributes to the regulation of the tumor transcriptome.
Keywords: Melanoma, chorioallantoic membrane tumor assay, Rho kinase, cytoskeleton, microarray, tumor initiation, tumor progression
Rho-associated protein kinases 1 and 2 (ROCK1/2, collectively known as Rho kinase) belong to a family of serine/threonine kinases which serve as key regulators of actin cytoskeletal dynamics and thus control cell migration and motility (1). Rho kinase phosphorylates many protein targets including the catalytic subunit of myosin phosphatase, myosin light chain, intermediate filaments, ezrin/radixin/ moesin family proteins, Lin11/Isl1/Mec3 domain kinase (LIMK), collapsin response mediator protein-2, calponin, and adducin (2). Deregulation of Rho kinase signaling contributes to the metastatic behavior of many tumor types (3–6), and several preclinical and clinical studies have targeted this pathway for anticancer therapeutics in prostate, lung, melanoma, and glioblastoma tumors with good efficacy (7–10).
While the role of Rho kinase in regulating the phosphoproteome is well understood, considerably less attention has been given to the function of Rho kinase signaling in regulating gene transcription. A handful of studies have identified global transcription alterations when Rho kinase is inhibited in epithelial and mesenchymal cell types grown in monolayers on plastic (11–13); however no reports examine global transcription regulation by Rho kinase using in vivo systems such as solid tumors. In this study, we utilized an in ovo melanoma cell xenograft system to perform whole-genome microarray analysis specifically on the melanoma tumor cells, while selectively excluding gene expression changes in cells of non-tumor origin such as endothelial cells, fibroblasts, stromal cells, and immune cells.
Materials and Methods
Cell culture
Mouse B16F1 melanoma (ATCC, Manassas, VA, USA), human NGP neuroblastoma (a generous gift from Dr. Rani George, Harvard Medical School), and human 4T1 breast cancer cells (a generous gift from Dr. Gary Sahagian, Tufts Medical Center) were cultured using standard tissue culture procedures in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 80 U/ml penicillin, and 50 μg/ml streptomycin C.
Melanoma tumors
Tumors were grown using the gelatin sponge-chorioallantoic membrane (CAM) assay according to previously published methods (14). Briefly, a false air-sac was generated using needle aspiration directly over the CAM of fertilized chicken eggs (Charles River Laboratories, North Franklin, CT, USA) at day 8 post-fertilization. Using dissecting scissors, a 10×10 mm window was cut, revealing the underlying embryo and CAM vessels. A hand-cut 1 mm3 gelatin sponge (Harvard Apparatus, Holliston, MA, USA) containing 20,000 dissociated tumor cells was placed onto the CAM and the window was sealed with sterile parafilm. A sham solution of isotonic saline solution or 10 μM solution of trans-4-[(1R)-1-aminoethyl]-N-4-pyridinylcyclohexanecarboxamide dihydrochloride (Y27632) (Enzo Life Sciences, Plymouth Meeting, PA, USA) was added daily directly onto the CAM tumor. At the indicated timepoint, tumors were collected, weighed, photographed on a lightbox, and stored in RNAlater (Ambion, Austin, TX, USA) according to the manufacturer’s directions. In total, greater than 20 tumors per time point were collected for each condition over three independent experiments.
Hematoxylin and eosin staining of tumors
Tumors collected at each timepoint were processed, cryosectioned, and hematoxylin and eosin stained by Excalibur Pathology (Moore, Oklahoma, USA). Images were collected at ×50 magnification using a Carl Zeis Axioscope.
Microarray analysis
Two sham- and two Y27632-treated samples, consisting of at least four tumors pooled per sample, were collected at four days’ treatment and subjected to triplicate microarray analysis per sample. DNA microarray analysis was performed using the Mouse v2 Whole Genome OneArray (Phalanx Biotech, Belmont, CA, USA). RNA quality and integrity were determined utilizing an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA). Only high quality RNA, having a RNA integrity number (RIN) of >7.0, and absorbance ratios A260/A280 >1.8 and A260/A230 >1.6, was utilized for further experimentation. RNA was converted to double-stranded cDNA and amplified using in vitro transcription that included amino-allyl UTP, and the cDNA product was subsequently conjugated with Cy5 NHS ester (GEH Lifesciences, Piscataway, NJ, USA). Fragmented cDNA was hybridized at 42°C overnight using the HybBag mixing system with 1× OneArray Hybridization Buffer (Phalanx Biotech, Belmont, CA, USA), 0.01 mg/ml sheared salmon sperm DNA (Promega, Madison, WI, USA), at a concentration of 0.025 mg/ml labeled target. After hybridization, the arrays were washed according to the OneArray protocol. Raw intensity signals for each microarray were captured using a Molecular Dynamics Axon 4100A scanner, measured using GenePixPro Software, and stored in GPR format. The data from all microarrays in each experimental set was then passed to Rosetta Resolver (Microsoft, Redmond, WA, USA) for analysis. Testing was performed by combining technical replicates and performing statistical analyses using Rosetta Resolver’s proprietary modeling techniques.
Semiquantitative reverse transcriptase polymerase chain reaction (RT-PCR)
RNA was extracted using Trireagent (Molecular Research Center, Cincinnati, OH, USA) according to the manufacturer’s instructions. RNA was converted to cDNA using Verso cDNA kit (Thermo Scientific) according to the manufacturer’s instructions. PCR amplification of specific cDNAs was performed using primers designed by Primer Blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels were used as a control.
Results
Small molecule inhibition of Rho kinase signaling in CAM tumor assays
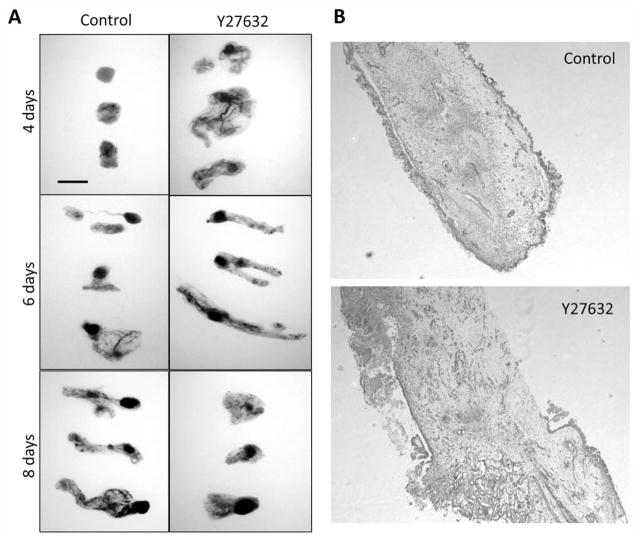
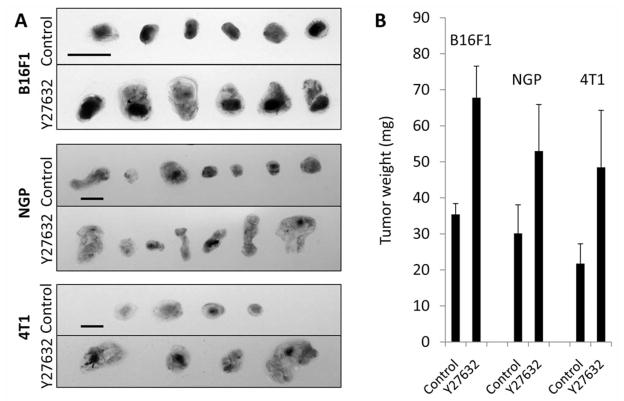
Inhibition of Rho kinase signaling has been shown to decrease tumor progression and metastasis in a number of in vivo models. Indeed our lab has previously demonstrated that systemic treatment of mice harboring subcutaneous B16F1 melanoma tumors with Y27632, a competitive inhibitor of ATP binding to the catalytic site of Rho kinase proteins, resulted in decreased tumor volume and inhibition of oncogenic properties such as tumor cell survival and migration (9). Similarly, B16F1 tumors grown on gelatin sponges using in ovo xenograft CAM tumor assays exhibited reduced tumor size following eight days of Y27632 treatment (Figure 1A), suggesting similar endpoint phenotypes between CAM and mouse tumor assays. Upon visual observation, Y27632-treated tumors appeared spongier and less defined than sham-treated tumors – an observation we previously observed in Y27632-treated B16F1 tumors grown in mice (9). Hematoxylin and eosin staining of cryosections confirmed this observation, revealing significant tissue disorganization and poorly defined tumor edges in Rho kinase inhibited tumors (Figure 1B). While later time points (8 days post-impantation) suggested that Y27632-treated tumors were at a growth disadvantage compared to sham treatments, observations of earlier time points (particularly four days’ post-implantation) revealed Y27632-treated tumors were initially larger in size and weight, albeit greatly disorganized, compared to sham-treated tumors. Similar results were obtained from NGP human neuroblastoma and 4T1 human breast cancer CAM tumors collected after four days of treatment (Figure 2A and B).
Figure 1.
Pharmacological inhibition of Rho kinase disrupts CAM melanoma tumor progression. A: Melanoma tumors were grown using the gelatin sponge-chorioallantoic membrane (CAM) assay according to previously published methods (14). A sham solution of isotonic saline solution or 10 μM solution of Y27632 was added daily directly onto the CAM tumor. At the indicated timepoint after tumor implantation on the CAM, tumors were collected and photographed on a lightbox. At least 20 tumors per time point were collected for each condition over three independent experiments. Scale bar=200 μm. B: Sham and Y27632 treated tumors were cryosectioned, hematoxylin and eosin stained, and photographed at ×50 magnification (data shown is for tumors collected at 6 days post-implantation).
Figure 2.
Y27632 treatment of newly formed CAM tumors results in enlarged initial tumor size and weight. A: B16F1 mouse melanoma, NGP human neuroblastoma, and 4T1 human breast cancer tumors were grown using the gelatin sponge-chorioallantoic membrane (CAM) assay according to previously published methods (14). A sham solution of isotonic saline solution or 10 μM solution of Y27632 was added daily directly onto the CAM tumor. At four days’ treatment, tumors were collected and photographed on a lightbox. B: Average tumor weights at four days’ treatment. At least 20 tumors were collected per condition over three independent experiments. Scale bar=200 μm.
Gene expression analysis
To understand the role of Rho kinase signaling in modulating gene expression in very early stages of tumor formation, we collected sham- and Y27632-treated CAM melanoma tumors four days after tumor inoculation onto the CAM and subjected them to microarray analysis. Out of a total of approximately 26,000 murine genes tested, 94 genes demonstrated a statistically significant fold change of >1.8 in the Y27632-treated melanoma tumors over the sham-treated tumors. Specifically, there were 9 genes that exhibited elevated mean expression levels (1.8- to 3.1-fold) and 85 genes that exhibited reduced mean expression levels (1.8- to 7.1-fold) in Y27632 melanoma tumors compared to sham-treated tumors (Tables I and II).
Table I.
Genes up-regulated by more than 1.8-fold in Y27632-treated melanoma tumors.
| Gene symbol | Gene name | Accession number | Fold change | P-value |
|---|---|---|---|---|
| Fcgbp | Fc fragment of IgG binding protein | NM_001122603.1 | 3.1 | 0.002 |
| Gmps | Guanine monophosphate synthetase | NM_001033300.2 | 2.6 | 0.002 |
| Agxt2l1 | Alanine glyoxylate-aminotransferase 2-like 1 | NM_001163587.1 | 2.3 | 0.009 |
| Capzb | Capping protein (actin filament) muscle Z-line beta | NM_001037761.1 | 2.1 | 0.008 |
| Prps1l1 | Phosphoribosyl pyrophosphate synthetase 1-like 1 | NM_029294.2 | 2.1 | 0.006 |
| Dmd | Dystrophin, muscular dystrophy | NM_007868.5 | 2.0 | 0.007 |
| Gm10876 | Predicted gene 10876 | XM_003085005.1 | 1.9 | 0.0003 |
| Grk4 | G-Protein-coupled receptor kinase 4 | NM_001080743.1 | 1.9 | 0.004 |
| Slc26a9 | Solute carrier family 26, member 9 | NM_177243.4 | 1.8 | 0.01 |
Table II.
Genes down-regulated by more than 1.8-fold in Y27632-treated melanoma tumors.
| Gene symbol | Gene name | Accession number | Fold change | P-value |
|---|---|---|---|---|
| Rnu12 | RNA U12, small nuclear | NR_004432.2 | −7.1 | 0.00001 |
| LOC100503583 | Fibrinogen silencer-binding protein-like | XM_003084562.1 | −3.7 | 0.006 |
| Etnk1 | Ethanolamine kinase 1 | NM_029250.2 | −3.2 | 0.001 |
| Atf3 | Activating transcription factor 3 | NM_007498.3 | −2.9 | 0.0002 |
| Dscc1 | Defective in sister chromatid cohesion 1 | NM_183089.2 | −2.9 | 0.005 |
| Lig3 | Ligase III, DNA, Atp-depedent | NM_010716.2 | −2.9 | 0.002 |
| Gm10825 | Predicted gene 10825 | NR_028580.1 | −2.7 | 0.008 |
| Mospd2 | Motile sperm domain containing 2 | NM_029730.3 | −2.7 | 0.0004 |
| Maf | Musculoaponeurotic fibrosarcoma | NM_001025577.2 | −2.5 | 0.003 |
| Mylip | Myosin regulatory light chain interactin protein | NM_153789.3 | −2.5 | 0.005 |
| Slc44a1 | Solute carrier family 44, member 1 | NM_001159633.1 | −2.5 | 0.0009 |
| Lbr | Lamin B receptor | NM_133815.2 | −2.4 | 0.003 |
| Tnks2 | TRF1-interacting ankyrin-related ADP ribose | NM_001163635.1 | −2.4 | 0.002 |
| Fam84b | Family with sequence similarity 84, member B | NM_001162926.1 | −2.4 | 0.0007 |
| Plxnc1 | Plexin C1 | NM_018797.2 | −2.4 | 0.0002 |
| Pde4d | Phosphodiesterase 4D cAMP specific | NM_011056.2 | −2.3 | 0.0003 |
| Med1 | Mediator complex subunit 1 | NM_134027.2 | −2.3 | 0.009 |
| 4632427E13Rik | RIKEN cDNA 4632427E13 gene | NR_015510.1 | −2.3 | 0.001 |
| Cdkl3 | Cyclin dependent kinase-like 3 | NM_001166657.1 | −2.3 | 0.003 |
| Unc93b1 | Unc-93 homolog B1 | NM_019449.2 | −2.3 | 0.0004 |
| Tcf12 | Transcription factor 12 | NM_011544.3 | −2.3 | 0.0003 |
| Heatr3 | HEAT repeat containing 3 | NM_172757.3 | −2.2 | 0.009 |
| Slc38a9 | Solute carrier family 38, member 9 | NM_178746.4 | −2.2 | 0.0001 |
| Mbnl2 | Muscleblind-like 2 | NM_207515.2 | −2.2 | 0.003 |
| Tyr | Tyrosinase | NM_011661.4 | −2.1 | 0.0005 |
| 5031439G07Rik | RIKEN cDNA 5031439G07 gene | NM_001033273.2 | −2.1 | 0.0001 |
| Serinc5 | Serine incorporator 5 | NM_172588.2 | −2.1 | 0.006 |
| Ggps1 | Geranylgeranyl diphosphate synthase 1 | NM_010282.2 | −2.1 | 0.001 |
| Pcdh17 | Protocadherin 17 | NM_001013753.2 | −2.1 | 0.007 |
| Zfp618 | Zing finger protein 618 | NM_028326.1 | −2.1 | 0.007 |
| Erbb3 | Erythroblastic leukemia viral oncogene homolog 3 | NM_010153.1 | −2.1 | 0.002 |
| Zdhhc2 | Zinc finger, DHHC domain containing 20 | NM_029492.4 | −2.1 | 0.001 |
| Thop1 | Thimet oligopeptidase 1 | NM_022653.4 | −2.0 | 0.0005 |
| 1700066M21Rik | RIKEN cDNA 1700066M21 gene | NM_028546.1 | −2.0 | 0.001 |
| Tob1 | Transducer of ErbB-2.1 | NM_009427.2 | −2.0 | 0.0002 |
| Decr2 | 2-4-Dienoyl-coenzyme A reductase 2, peroxisomal | NM_011933.2 | −2.0 | 0.004 |
| Trim23 | Tripartite motif-containing 23 | NM_030731.3 | −2.0 | 0.002 |
| Stt3a | Subunit oligosaccharyltransferase complex homolog-A | NM_008408.4 | −2.0 | 0.006 |
| Glrx2 | Glutaredoxin 2 (thioltransferase) | NM_001038593.1 | −1.9 | 0.009 |
| Enpp2 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 | NM_015744.2 | −1.9 | 0.003 |
| Pknox1 | Pbx/knotted 1 homeobox | NR_027493.1 | −1.9 | 0.001 |
| Tec | Tec protein tyrosine kinase | NM_001113464.1 | −1.9 | 0.005 |
| 2310035C23Rik | RIKEN cDNA 2310035C23 gene | NM_029349.1 | −1.9 | 0.0002 |
| Osbp | Oxysterol binding protein | NM_001033174.1 | −1.9 | 0.006 |
| Cyr61 | Cysteine rich protein 61 | NM_010516.2 | −1.9 | 0.0009 |
| Pgf | Placental growth factor | NM_008827.2 | −1.9 | 0.002 |
| 4930579G24Rik | RIKEN cDNA 4930579G24 gene | NM_029482.1 | −1.9 | 0.0001 |
| Itgb5 | Integrin beta 5 | NM_001145884.1 | −1.9 | 0.0008 |
| Atg4a | Autophagy-related 4A | NM_174875.3 | −1.9 | 0.0007 |
| Gm10786 | Predicted gene 10786 | XM_003084921.1 | −1.9 | 0.0001 |
| Slmo1 | Slowmo homolog 1 | NM_144867.2 | −1.9 | 0.009 |
| Etf1 | Eukaryotic translation termination factor 1 | NM_144866.3 | −1.9 | 0.0003 |
| Usp31 | Ubiquitin specific peptidase 31 | NM_001033173.1 | −1.9 | 0.0001 |
| Srsf2 | Serine/arginine-rich splicing factor 2 | NM_011358.2 | −1.9 | 0.0001 |
| Mdfi | MyoD family inhibitor | NM_001109973.1 | −1.9 | 0.000005 |
| Foxn3 | Forkhead box N3 | NM_183186.2 | −1.9 | 0.0008 |
| Fam46a | Family with sequence similarity 46, member A | NM_001160379.1 | −1.9 | 0.00009 |
| Smad7 | MAD homolog 7 | NM_001042660.1 | −1.9 | 0.001 |
| Dimt1 | DIM1 dimethyladenosine transferase 1-like | NM_025447.4 | −1.8 | 0.002 |
| Rock1 | Rho-associated coiled-coil containing protein kinase 1 | NM_009071.2 | −1.8 | 0.0002 |
| Acox2 | Acyl-coenzyme A oxidase 2, branched chain | NM_001161667.1 | −1.8 | 0.0007 |
| Lta4h | Leukotriene A4 hydrolase | NM_008517.2 | −1.8 | 0.001 |
| Dync1li1 | Dynein cytoplasmic 1 light intermediate chain 1 | NM_146229.2 | −1.8 | 0.00005 |
| Fam161a | Family with sequence similarity 161, member A | NM_028672.2 | −1.8 | 0.008 |
| Ube2d3 | Ubiquitin-conjugating enzyme E2D 3 | NM_025356.4 | −1.8 | 0.008 |
| Mrps10 | Mitochondrial ribosomal protein S10 | NM_001146211.1 | −1.8 | 0.001 |
| Ppp2r3c | Protein phosphatase 2, regulatory subunit B, gamma | NM_021529.3 | −1.8 | 0.0005 |
| Mtch2 | Mitochondrial carrier homolog 2 | NM_019758.2 | −1.8 | 0.001 |
| Gm4769 | Predicted gene 4769 | NM_177596.3 | −1.8 | 0.004 |
| Klhl24 | Kelch-like 24 | NM_029436.3 | −1.8 | 0.002 |
| Cyp20a1 | Cytochrome P450, family 20, subfamily A, polypeptide 1 | NM_030013.2 | −1.8 | 0.000009 |
| Ankrd26 | Ankyrin repeat domain 26 | NM_001081112.1 | −1.8 | 0.00007 |
| Zfp385b | Zinc finger protein 385B | NM_178723.6 | −1.8 | 0.00007 |
| Prosc | Proline synthetase co-transcribed | NM_001039078.2 | −1.8 | 0.00008 |
| Idh1 | Isocitrate dehydrogenase 1 (NADP+), soluble | NM_001111320.1 | −1.8 | 0.0003 |
| Slc25a36 | Solute carrier family 25, member 36 | NM_138756.4 | −1.8 | 0.00009 |
| Asnsd1 | Asparagine synthetase domain containing 1 | NM_133728.2 | −1.8 | 0.0001 |
| 1110018J18Rik | RIKEN cDNA 1110018J18 gene | NM_025370.2 | −1.8 | 0.0007 |
| Bc030336 | cDNA sequence BC030336 | NM_001164580.1 | −1.8 | 0.0001 |
| Strada | STE20-related kinase adaptor alpha | NM_028126.2 | −1.8 | 0.0001 |
| Cdk2 | Cyclin-dependent kinase 2 | NM_016756.4 | −1.8 | 0.001 |
| Sypl | Synaptophysin-like protein | NM_013635.3 | −1.8 | 0.0008 |
| Stc1 | Stanniocalcin 1 | NM_009285.3 | −1.8 | 0.002 |
| Mzt1 | Mitotic spindle organizing protein 1 | NM_175245.4 | −1.8 | 0.0004 |
| Tars2 | Threonyl-tRNA synthetase 2, mitochondrial (putative) | NM_001163617.1 | −1.8 | 0.00003 |
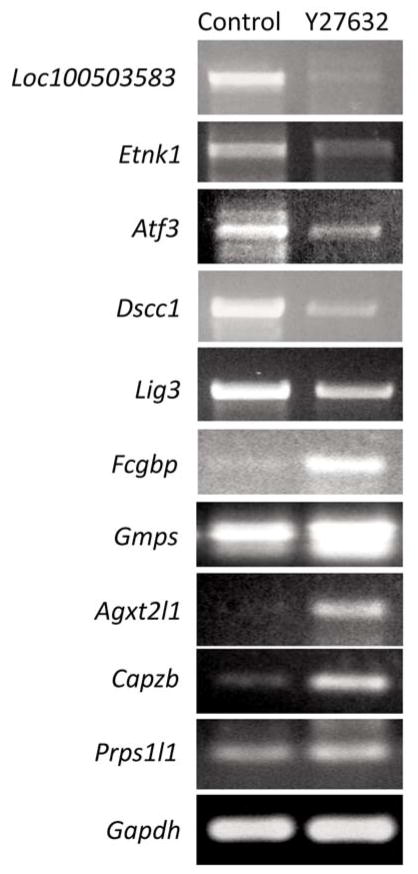
The molecular and cellular functions of both up- and down-regulated genes included cell cycle regulation (Strada, Cdk2, Smad7, Tob1, Cdkl3), DNA replication (Lig3, Dscc1), extracellular matrix/cell adhesion (Fcgbp, Itgb5, Cyr61, Pcdh17), cytoskeletal dynamics (Capzb, Dmd, Mzt1, Lbr, Mylip), cell death (Atg4a), transcriptional regulation (Foxn3, Pknox1, Zfp618, Tcf12, Med1, Maf, Atf3), growth factors and hormones (Stc1, Pgf), cell signaling (Smad7, Rock1, Tec, Pde4d), biogenesis and metabolism (Gmps, Agxt2l1, Prps1l1, Cyp20a1, Idh1, Tars2, Lta4h, Acox2, Dimt1, Enpp2, Glrx2, Decr2, Ggps1, Serinc5, Tyr, Tnks2, Etnk1), post-translational protein modification (Ube2d3, Ppp2r3c, Usp31, Stt3a, Trim23, Thop1, Zdhhc2), vesicle transport (Dync1li1), receptor proteins and receptor regulators (Grk4, Klhl24, Erbb3, Plxnc1), transport proteins (Slc26a9, Mtch2, Slc25a36, Slc38a9, Unc93b1, Slc44a1), nuclear localization (Osbp, Mdfi), mRNA splicing (Srsf2, Rnu12, Mbnl2), translational regulation (Mrps10, Etf1) and of unknown/presumed function (Gm10876, Fam161a, Gm4769, Ankrd26, Zfp385b, Prosc, Asnsd1, 1110018J18Rik, Bc030336, Sypl, Fam46a, Slmo1, Gm10786, 4930579G24Rik, 2310035C23Rik, 1700066M21Rik, 5031439G07Rik, Heatr3, 4632427E13Rik, Fam84b, Mospd2, Gm10825, Loc100503583). The genes whose expression was most significantly altered following Y27632 treatment were independently verified by semi-quantitative RT-PCR on cDNA synthesized from sham-and Y27632-CAM melanoma tumor samples collected four days-post-implantation (Figure 3).
Figure 3.
Semi-quantitative RT-PCR confirmation of microarray results. cDNA collected from sham- and Y27632-treated tumors at four days’ treatment were subjected to semi-quantitative RT-PCR analysis using primers specific for the five most up- and down-regulated protein coding genes identified in the microarray analysis.
Discussion
Rho kinase regulation of the phosphoproteome has been extensively studied over the past decade, yet how these cellular changes regulate the global transcriptome is poorly understood. An extensive literature search primarily reveals reports mentioning singular instances of gene regulation by altering Rho kinase signaling, but only a handful of large scale genomic approaches have addressed global Rho kinase mediated transcriptional changes. Global analysis of gene regulation in Rho kinase-inhibited fibroblast monolayers revealed that 2.3% of genes exhibited significantly altered expression levels to greater or less than 20% those of the sham (11). Pharmacological ROCK2 inhibition in endothelial cells, smooth muscle cells, and fibroblasts using the small molecule inhibitor SLx-2119 revealed changes in the low hundreds of genes per cell type, with only a very minimal overlap of common gene changes between cell types (12), suggesting that transcriptional alterations in response to Rho kinase inhibition are uniquely cell type specific. Lastly, a total of 66 genes were altered by Y27632 treatment in cultured human corneal stromal cells (13). These three studies utilized in vitro cell culture systems where cells were grown in monolayers on plastic – a system that is convenient, but hardly represents the complex microenvironment of an in vivo tissue. To date, no genomics-based study has been performed on Rho kinase-inhibited cancer lines or solid tumors. The findings reported in this manuscript are the first of their kind to describe the global transcriptome regulation by Rho kinase signaling in solid tumors.
Distinct disadvantages often exist when performing microarray analysis on tumors. Namely, in addition to the cancer cells, a mixture of various host cell types including immune cells, stromal cells, endothelial cells, and fibroblasts, are present which contaminate and complicate the analysis. Therefore gene expression profiles using mRNA isolated from total tumor tissue are very unlikely to reflect the genomic changes that occur specifically in the cancer cells themselves. To overcome this issue, we utilized an in ovo xenograph system whereby melanoma cancer cells were solely of mouse origin, while all other associated infiltrating cells were of chicken origin. Thus we could address the global gene changes specific for the melanoma cells in response to Rho kinase inhibition without non-tumor cell contamination. We established a cutoff for significant gene changes at greater or less than 1.8-fold, with a stringent p-value of less than 0.01. Under these conditions, a total of 94 genes were identified, with 9 genes exhibiting elevated mean expression levels and 85 genes exhibiting reduced mean expression levels in Y27632-treated melanoma tumors.
Among the many genes identified in this study, approximately 20% of these have been suggested to have roles as tumor suppressors/oncogenes, angiogenesis regulators, and/or serve as molecular markers of cancer. Plxnc1 (−2.4-fold) is a receptor for the GPI-anchored semaphorin Sema7A and serves as a tumor suppressor by opposing melanocyte dendricity and melanoma progression, as well as inactivating cofilin and R-Ras GAP-mediated cell migration (15, 16). Moreover, Plxnc1 expression is lost during melanoma metastasis and is down-regulated across a panel of melanomas (15–17). A deficiency in the Pknox1 gene (−1.9-fold), encoding an atypical homeobox transcription factor that binds DNA cooperatively with E2A/myogenic transcription factors, leads to decreased levels of the anti-apoptotic protein Bcl-Xl and spontaneous tumor formation in mice (18, 19). Additionally, 70% of tumors have been reported to exhibit reduced or a lack of expression of Pknox1 (18). Maf (−2.5-fold) is considered a potent oncogene with strong transforming activity, and functions as a transcriptional activator of cyclin D2 and other proliferative genes (20). Maf is translocated or up-regulated in multiple myelomas and T-cell lymphomas (21, 22). Erbb3 (−2.1-fold) is a member of the epidermal growth factor receptor family of proteins. This strong oncogene regulates the phosphoinositol 3-kinase/AKT pathway to promote survival and proliferation, and recent results link high ERBB3 activity with escape from chemotherapy targeting other ERBBs in lung and breast cancer (23). Erbb3 has been implicated in the initiation and progression of tumors in a very large number of tissue types (23). Smad7 (−1.9-fold) is an inhibitor of the TGFbeta signaling pathway, and under normoxic conditions functions as a strong tumor suppressor; however, its expression is greatly up regulated in response to hypoxia and contributes to the resistance of TGFbeta growth suppression in hypoxic tumors (24, 25). Moreover, particular polymorphisms in the Smad7 gene have been shown to increase an individual’s risk to colon cancer (26). Cyr61 (−1.9-fold) is a strongly pro-angiogenic secreted extracellular matrix (ECM) protein that regulates cell survival, apoptosis, inflammation, cell adhesion, migration, and metastasis, and is believed to influence the disease course of many cancers through binding with integrins (27). Not only does Cyr61 promote in vitro and in vivo angiogenesis, but it also serves a role in the recruitment of endothelial progenitor cells to tumors (28). Expression of Cyr61 is regulated in response to hypoxia (29) and is up-regulated in a variety of cancer types (29–31). Interestingly, Rho kinase inhibition down-regulates the expression of Rho kinase 1 (ROCK1; −1.8-fold). ROCK1 plays a major role in the regulation of tumor cell invasion and metastasis (32). Moreover, pharmacological inhibition of Rho kinase activity effectively inhibits solid tumor growth and spreading across numerous cancer types (32). Unfortunately, small molecule inhibition of Rho kinase in cisplatin treated neuroblastomas leads to acquisition of a chemoresistant phenotype through altered cell cycle regulation, up regulation of cisplatin influx/efflux genes, and up regulation of nucleotide excision repair genes (33), suggesting that Rho kinase may also play a role in chemotherapy resistance. Rho kinase is also an essential regulator of endothelial cell fate determination and in vitro and in vivo blood vessel formation (34). Other interesting genes identified in this study are the tumor suppressors Pcdh17 (35), Zdhhc2 (36), and Tob1 (37), the cell type variable tumor suppressor/oncogene Atf3 (38, 39), the angiogenic regulators Pgf (40) and Itgb5 (41), and the tumor biomarkers Stc1 (42, 43), Fcgbp (44), and Med1 (45–47).
The data presented in this study examine for the first time the global transcriptional regulation controlled by Rho kinase in melanoma tumors. These findings identified many Rho kinase-responsive genes involved directly in the tumorigenic process, and further suggest that targeting Rho kinase activity may not only disrupt the tumor phosphoproteome, but also disturb the tumor transcriptome. Future experiments should address temporal regulation of global gene expression during tumor progression and identify commonalities and differences in genes regulated by Rho kinase between different tumor cell types.
Acknowledgments
We would like to thank Dr. Ellen Fynan (Worcester State University) for technical knowledge and assistance with tumor CAM assays, Dr. Rani George (Harvard Medical School) and Dr. Gary Sahagian (Tufts Medical Center), respectively, for generous use of the NGP and 4T1 cell lines, and Dr. Patricia D’Amore (Harvard Medical School) for use of the Axioscope. The project described was supported by a National Heart, Lung, and Blood Institute Grant (HL098931) awarded to BAB.
References
- 1.Riento K, Ridley AJ. Rocks: multifunctional kinases in cell behaviour. Nat Rev Mol Cell Biol. 2003;4:446–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- 2.Takuwa Y. Rho-Rho kinase pathway. Nippon Rinsho. 2004;62:43–48. [PubMed] [Google Scholar]
- 3.Kamai T, Arai K, Sumi S, Tsujii T, Honda M, Yamanishi T, Yoshida KI. The rho/rho-kinase pathway is involved in the progression of testicular germ cell tumour. BJU Int. 2002;89:449–453. doi: 10.1046/j.1464-4096.2001.01920.x. [DOI] [PubMed] [Google Scholar]
- 4.Kamai T, Tsujii T, Arai K, Takagi K, Asami H, Ito Y, Oshima H. Significant association of Rho/ROCK pathway with invasion and metastasis of bladder cancer. Clin Cancer Res. 2003;9:2632–2641. [PubMed] [Google Scholar]
- 5.Kaneko K, Satoh K, Masamune A, Satoh A, Shimosegawa T. Expression of ROCK-1 in human pancreatic cancer: its down-regulation by morpholino oligo antisense can reduce the migration of pancreatic cancer cells in vitro. Pancreas. 2002;24:251–257. doi: 10.1097/00006676-200204000-00007. [DOI] [PubMed] [Google Scholar]
- 6.Kleer CG, van Golen KL, Zhang Y, Wu ZF, Rubin MA, Merajver SD. Characterization of RhoC expression in benign and malignant breast disease: a potential new marker for small breast carcinomas with metastatic ability. Am J Pathol. 2002;160:579–584. doi: 10.1016/S0002-9440(10)64877-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amine A, Rivera S, Opolon P, Dekkal M, Biard DS, Bouamar H, Louache F, McKay MJ, Bourhis J, Deutsch E, Vozenin-Brotons MC. Novel anti-metastatic action of cidofovir mediated by inhibition of E6/E7, CXCR4 and Rho/ROCK signaling in HPV tumor cells. PLoS One. 2009;4:e5018. doi: 10.1371/journal.pone.0005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rattan R, Giri S, Singh AK, Singh I. Rho/ROCK pathway as a target of tumor therapy. J Neurosci Res. 2006;83:243–255. doi: 10.1002/jnr.20707. [DOI] [PubMed] [Google Scholar]
- 9.Routhier A, Astuccio M, Lahey D, Monfredo N, Johnson A, Callahan W, Partington A, Fellows K, Ouellette L, Zhidro S, Goodrow C, Smith A, Sullivan K, Simone P, Le L, Vezuli B, Zohni M, West E, Gleason D, Bryan B. Pharmacological inhibition of Rho-kinase signaling with Y-27632 blocks melanoma tumor growth. Oncol Rep. 2010;23:861–867. [PubMed] [Google Scholar]
- 10.Somlyo AV, Bradshaw D, Ramos S, Murphy C, Myers CE, Somlyo AP. Rho-kinase inhibitor retards migration and in vivo dissemination of human prostate cancer cells. Biochem Biophys Res Commun. 2000;269:652–659. doi: 10.1006/bbrc.2000.2343. [DOI] [PubMed] [Google Scholar]
- 11.Berenjeno IM, Bustelo XR. Identification of the Rock-dependent transcriptome in rodent fibroblasts. Clin Transl Oncol. 2008;10:726–738. doi: 10.1007/s12094-008-0279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boerma M, Fu Q, Wang J, Loose DS, Bartolozzi A, Ellis JL, McGonigle S, Paradise E, Sweetnam P, Fink LM, Vozenin-Brotons MC, Hauer-Jensen M. Comparative gene expression profiling in three primary human cell lines after treatment with a novel inhibitor of Rho kinase or atorvastatin. Blood Coagul Fibrinolysis. 2008;19:709–718. doi: 10.1097/MBC.0b013e32830b2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harvey SA, Anderson SC, SundarRaj N. Downstream effects of ROCK signaling in cultured human corneal stromal cells: microarray analysis of gene expression. Invest Ophthalmol Vis Sci. 2004;45:2168–2176. doi: 10.1167/iovs.03-1218. [DOI] [PubMed] [Google Scholar]
- 14.Ribatti D, Nico B, Vacca A, Presta M. The gelatin sponge- chorioallantoic membrane assay. Nat Protoc. 2006;1:85–91. doi: 10.1038/nprot.2006.13. [DOI] [PubMed] [Google Scholar]
- 15.Scott GA, McClelland LA, Fricke AF. Semaphorin 7a promotes spreading and dendricity in human melanocytes through beta1-integrins. J Invest Dermatol. 2008;128:151–161. doi: 10.1038/sj.jid.5700974. [DOI] [PubMed] [Google Scholar]
- 16.Scott GA, McClelland LA, Fricke AF, Fender A. Plexin C1, a receptor for semaphorin 7a, inactivates cofilin and is a potential tumor suppressor for melanoma progression. J Invest Dermatol. 2009;129:954–963. doi: 10.1038/jid.2008.329. [DOI] [PubMed] [Google Scholar]
- 17.Uesugi K, Oinuma I, Katoh H, Negishi M. Different requirement for Rnd GTPases of R-Ras GAP activity of Plexin-C1 and Plexin-D1. J Biol Chem. 2009;284:6743–6751. doi: 10.1074/jbc.M805213200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longobardi E, Iotti G, Di Rosa P, Mejetta S, Bianchi F, Fernandez-Diaz LC, Micali N, Nuciforo P, Lenti E, Ponzoni M, Doglioni C, Caniatti M, Di Fiore PP, Blasi F. Prep1 (pKnox1)-deficiency leads to spontaneous tumor development in mice and accelerates EmuMyc lymphomagenesis: a tumor suppressor role for Prep1. Mol Oncol. 2010;4:126–134. doi: 10.1016/j.molonc.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Micali N, Ferrai C, Fernandez-Diaz LC, Blasi F, Crippa MP. Prep1 directly regulates the intrinsic apoptotic pathway by controlling Bcl-XL levels. Mol Cell Biol. 2009;29:1143–1151. doi: 10.1128/MCB.01273-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishizawa M, Kataoka K, Vogt PK. MafA has strong cell transforming ability but is a weak transactivator. Oncogene. 2003;22:7882–7890. doi: 10.1038/sj.onc.1206526. [DOI] [PubMed] [Google Scholar]
- 21.Chang H, Qi Q, Xu W, Patterson B. c-Maf nuclear oncoprotein is frequently expressed in multiple myeloma. Leukemia. 2007;21:1572–4. doi: 10.1038/sj.leu.2404669. [DOI] [PubMed] [Google Scholar]
- 22.Murakami YI, Yatabe Y, Sakaguchi T, Sasaki E, Yamashita Y, Morito N, Yoh K, Fujioka Y, Matsuno F, Hata H, Mitsuya H, Imagawa S, Suzuki A, Esumi H, Sakai M, Takahashi S, Mori N. c-Maf expression in angioimmunoblastic T-cell lymphoma. Am J Surg Pathol. 2007;31:1695–1702. doi: 10.1097/PAS.0b013e318054dbcf. [DOI] [PubMed] [Google Scholar]
- 23.Sithanandam G, Anderson LM. The ERBB3 receptor in cancer and cancer gene therapy. Cancer Gene Ther. 2008;15:413–448. doi: 10.1038/cgt.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heikkinen PT, Nummela M, Jokilehto T, Grenman R, Kahari VM, Jaakkola PM. Hypoxic conversion of SMAD7 function from an inhibitor into a promoter of cell invasion. Cancer Res. 2010;70:5984–5993. doi: 10.1158/0008-5472.CAN-09-3777. [DOI] [PubMed] [Google Scholar]
- 25.Nakahata S, Yamazaki S, Nakauchi H, Morishita K. Down regulation of ZEB1 and overexpression of SMAD7 contribute to resistance to TGF-beta1 mediated growth suppression in adult T-cell leukemia/lymphoma. Oncogene. 2010;29:4157–4169. doi: 10.1038/onc.2010.172. [DOI] [PubMed] [Google Scholar]
- 26.Slattery ML, Herrick J, Curtin K, Samowitz W, Wolff RK, Caan BJ, Duggan D, Potter JD, Peters U. Increased risk of colon cancer associated with a genetic polymorphism of SMAD7. Cancer Res. 2010;70:1479–1485. doi: 10.1158/0008-5472.CAN-08-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhar A, Ray A. The CCN family proteins in carcinogenesis. Exp Oncol. 2010;32:2–9. [PubMed] [Google Scholar]
- 28.Grote K, Salguero G, Ballmaier M, Dangers M, Drexler H, Schieffer B. The angiogenic factor CCN1 promotes adhesion and migration of circulating CD34+ progenitor cells: potential role in angiogenesis and endothelial regeneration. Blood. 2007;110:877–885. doi: 10.1182/blood-2006-07-036202. [DOI] [PubMed] [Google Scholar]
- 29.Hirschfeld M, zur Hausen A, Bettendorf H, Jager M, Stickeler E. Alternative splicing of Cyr61 is regulated by hypoxia and significantly changed in breast cancer. Cancer Res. 2009;69:2082–2090. doi: 10.1158/0008-5472.CAN-08-1997. [DOI] [PubMed] [Google Scholar]
- 30.Chen PP, Li WJ, Wang Y, Zhao S, Li DY, Feng LY, Shi XL, Koeffler HP, Tong XJ, Xie D. Expression of CYR61, CTGF, and WISP-1 correlates with clinical features of lung cancer. PLoS One. 2007;2:e534. doi: 10.1371/journal.pone.0000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lv H, Fan E, Sun S, Ma X, Zhang X, Han DM, Cong YS. CYR61 is up-regulated in prostate cancer and associated with the p53 gene status. J Cell Biochem. 2009;106:738–744. doi: 10.1002/jcb.22075. [DOI] [PubMed] [Google Scholar]
- 32.Narumiya S, Tanji M, Ishizaki T. Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 2009;28:65–76. doi: 10.1007/s10555-008-9170-7. [DOI] [PubMed] [Google Scholar]
- 33.Street CA, Routhier AA, Spencer C, Perkins AL, Masterjohn K, Hackathorn A, Montalvo J, Dennstedt EA, Bryan BA. Pharmacological inhibition of Rho-kinase (ROCK) signaling enhances cisplatin resistance in neuroblastoma cells. Int J Oncol. 2010;37:1297–1305. doi: 10.3892/ijo_00000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bryan BA, Dennstedt E, Mitchell DC, Walshe TE, Noma K, Loureiro R, Saint-Geniez M, Campaigniac JP, Liao JK, D’Amore PA. RhoA/ROCK signaling is essential for multiple aspects of VEGF-mediated angiogenesis. Faseb J. 2010;24:3186–3195. doi: 10.1096/fj.09-145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haruki S, Imoto I, Kozaki K, Matsui T, Kawachi H, Komatsu S, Muramatsu T, Shimada Y, Kawano T, Inazawa J. Frequent silencing of protocadherin 17, a candidate tumour suppressor for esophageal squamous cell carcinoma. Carcinogenesis. 2010;31:1027–1036. doi: 10.1093/carcin/bgq053. [DOI] [PubMed] [Google Scholar]
- 36.Planey SL, Keay SK, Zhang CO, Zacharias DA. Palmitoylation of cytoskeleton associated protein 4 by DHHC2 regulates antiproliferative factor-mediated signaling. Mol Biol Cell. 2009;20:1454–1463. doi: 10.1091/mbc.E08-08-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helms MW, Kemming D, Contag CH, Pospisil H, Bartkowiak K, Wang A, Chang SY, Buerger H, Brandt BH. TOB1 is regulated by EGF-dependent HER2 and EGFR signaling, is highly phosphorylated, and indicates poor prognosis in node-negative breast cancer. Cancer Res. 2009;69:5049–5056. doi: 10.1158/0008-5472.CAN-08-4154. [DOI] [PubMed] [Google Scholar]
- 38.Hackl C, Lang SA, Moser C, Mori A, Fichtner-Feigl S, Hellerbrand C, Dietmeier W, Schlitt HJ, Geissler EK, Stoeltzing O. Activating transcription factor-3 (ATF3) functions as a tumor suppressor in colon cancer and is up-regulated upon heat-shock protein 90 (Hsp90) inhibition. BMC Cancer. 2010;10:668. doi: 10.1186/1471-2407-10-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang A, Arantes S, Yan L, Kiguchi K, McArthur MJ, Sahin A, Thames HD, Aldaz CM, Macleod MC. The transcription factor ATF3 acts as an oncogene in mouse mammary tumorigenesis. BMC Cancer. 2008;8:268. doi: 10.1186/1471-2407-8-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carmeliet P, Moons L, Luttun A, Vincenti V, Compernolle V, De Mol M, Wu Y, Bono F, Devy L, Beck H, Scholz D, Acker T, DiPalma T, Dewerchin M, Noel A, Stalmans I, Barra A, Blacher S, Vandendriessche T, Ponten A, Eriksson U, Plate KH, Foidart JM, Schaper W, Charnock-Jones DS, Hicklin DJ, Herbert JM, Collen D, Persico MG. Synergism between vascular endothelial growth factor and placental growth factor contributes to angiogenesis and plasma extravasation in pathological conditions. Nat Med. 2001;7:575–583. doi: 10.1038/87904. [DOI] [PubMed] [Google Scholar]
- 41.Leifheit-Nestler M, Conrad G, Heida NM, Limbourg A, Limbourg FP, Seidler T, Schroeter MR, Hasenfuss G, Konstantinides S, Schafer K. Overexpression of integrin beta 5 enhances the paracrine properties of circulating angiogenic cells via Src kinase-mediated activation of STAT3. Arterioscler Thromb Vasc Biol. 2010;30:1398–1406. doi: 10.1161/ATVBAHA.110.206086. [DOI] [PubMed] [Google Scholar]
- 42.Law AY, Ching LY, Lai KP, Wong CK. Identification and characterization of the hypoxia-responsive element in human stanniocalcin-1 gene. Mol Cell Endocrinol. 2010;314:118–127. doi: 10.1016/j.mce.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Liu G, Yang G, Chang B, Mercado-Uribe I, Huang M, Zheng J, Bast RC, Lin SH, Liu J. Stanniocalcin 1 and ovarian tumorigenesis. J Natl Cancer Inst. 2010;102:812–827. doi: 10.1093/jnci/djq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gazi MH, He M, Cheville JC, Young CY. Down-regulation of IgG Fc-binding protein (Fc gammaBP) in prostate cancer. Cancer Biol Ther. 2008;7:70–75. doi: 10.4161/cbt.7.1.5131. [DOI] [PubMed] [Google Scholar]
- 45.Howard JH, Frolov A, Tzeng CW, Stewart A, Midzak A, Majmundar A, Godwin A, Heslin M, Bellacosa A, Arnoletti JP. Epigenetic down-regulation of the DNA repair gene MED1/MBD4 in colorectal and ovarian cancer. Cancer Biol Ther. 2009;8:94–100. doi: 10.4161/cbt.8.1.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lucci-Cordisco E, Neri G. Silent beginning: early silencing of the MED1/MBD4 gene in colorectal tumorigenesis. Cancer Biol Ther. 2009;8:192–193. doi: 10.4161/cbt.8.2.7647. [DOI] [PubMed] [Google Scholar]
- 47.Ndong Jde L, Jean D, Rousselet N, Frade R. Down-regulation of the expression of RB18A/MED1, a cofactor of transcription, triggers strong tumorigenic phenotype of human melanoma cells. Int J Cancer. 2009;124:2597–2606. doi: 10.1002/ijc.24253. [DOI] [PubMed] [Google Scholar]