Abstract
Epigenetic information, which may affect an organisms’ phenotype, can be stored and stably inherited in the form of cytosine DNA methylation. Changes in DNA methylation can produce meiotically stable epialleles that affect transcription and morphology, but the rates of spontaneous gain or loss of DNA methylation are unknown. We examined spontaneously occurring variation in DNA methylation in Arabidopsis thaliana plants propagated by single-seed descent for 30 generations. 114,287 CG single methylation polymorphisms (SMPs) and 2485 CG differentially methylated regions (DMRs) were identified, both of which show patterns of divergence compared to the ancestral state. Thus, transgenerational epigenetic variation in DNA methylation may generate new allelic states that alter transcription providing a mechanism for phenotypic diversity in the absence of genetic mutation.
Cytosine methylation is a DNA base modification with roles in development and disease in animals as well as in silencing transposons and repetitive sequences in plants and fungi (1). In plants, CG methylation is commonly found within gene bodies (2–5); whereas, non-CG methylation, CHG and CHH (where H = A,C,T) is enriched in transposons and repetitive sequences (1). The RNA-directed DNA methylation (RdDM) pathway targets both CG and non-CG sites for methylation and is commonly associated with transcriptional silencing (6, 7). This pathway can also target and silence protein-coding genes, giving rise to epigenetic alleles or so-called “epialleles” that can be heritable through mitosis and/or meiosis (8, 9) and can be dependent on the methylation of a single CG dinucleotide (10).
Two meiotically heritable epialleles resulting in morphological variation are the peloric (Linaria vulgaris) and colorless non-ripening (Solanum lycopersicum) loci (11, 12). Both show spontaneous epigenetic silencing events within their respective populations (11, 13). However, the frequency at which such spontaneous meiotically heritable epialleles naturally arise in populations is unknown. Although epiallelic variation has been identified between genetically diverse populations within Arabidopsis thaliana (14, 15), it is unclear if these identified epialleles are due to underlying genetic variation. Epialleles have also been artificially generated after mutagenesis or due to mutations in the cellular components required for the maintenance of DNA methylation (14–16).
An Arabidopsis thaliana (Columbia-0) population, “the MA lines”, derived by single-seed descent for 30 generations (17) was used to examine the extent of naturally occurring variation in DNA methylation and the frequency at which spontaneous epialleles emerge over time. We used MethylC-Seq (3) to determine the whole-genome base resolution DNA methylomes for three ancestral MA lines (numbers - 1, 12, 19) and five descendant MA lines (numbers - 29, 49, 59, 69, 119) (fig. S1). We refer to lines 1, 12, 19 as ancestors throughout this study, although they are not direct ancestors as they are three generations removed from the original founder line (fig. S1). These specific descendant lines were selected because their genomes have been sequenced and they have a known level of spontaneous mutation (18). Biological replicates (sibling plants) for each leaf methylome were sequenced to an average of ~34-fold coverage, which allowed for an average per line examination of 39,897,093 (96.35%) uniquely mapped cytosines and 5,307,077 (98.39%) uniquely mapped CGs (table S1).
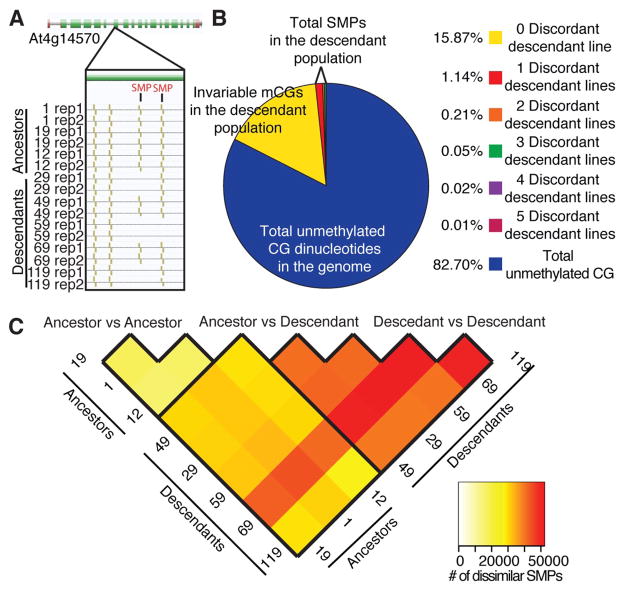
A total of 1,730,761 CGs were methylated (mCGs) in at least one MA line (Fig. 1A) and approximately 91% of the covered mCGs were invariably methylated across all eight lines (19). The variable mCGs revealed a set of 114,287 high-confidence CG single methylation polymorphisms (SMPs) that showed a consensus of the methylation status of CG dinucleotides between biological replicates (Fig. 1A). Next, a reference MA founder DNA methylome was created by pooling the completely conserved mCG site calls for all ancestral MA lines and was used to determine the frequency of discordant CG-SMP sites within the descendant population (Fig. 1B). Within the descendant lines, ~1.62% of the CG methylome shows susceptibility to dynamic acquisitions and losses of mCGs over time (table S2). On average, ~66,000 methylated CG-SMPs (mCG-SMPs) were identified for each ancestral and descendant line (fig. S2). Although the total number of mCG-SMPs was similar between all lines, the conservation of these polymorphisms amongst and between ancestral and descendant populations was different (Fig. 1C, table S3). A pairwise comparison of both populations for methylation conservation, estimated by global similarity of mCG-SMP sites (19), revealed that all of the ancestral lines are highly similar (table S4). Interestingly, descendant lines showed greater similarity in CG-SMPs methylation status to ancestral lines than to other descendant lines (table S4).
Fig. 1.
Epigenetic variation of CG-SMPs. (A) An example of a CG-SMP. (Gold lines = CG methylation, maroon rectangle indicates the untranslated regions and green rectangles indicated exons). (B) A breakdown of the methylation distribution of CG dinucleotides amongst all samples. (C) A heatmap indicating the number of CG-SMPs that differ between two samples (see table S3).
We calculated an estimate of the “epimutation rate” per generation in this population using linear regression and TREE PUZZLE, which revealed 704 and 2876 methylation changes each generation, respectively (19). We estimated a lower bound of the epimutation rate with the linear regression results, which revealed 4.46 × 10−4 methylation polymorphisms per CG site per generation (P <0.0000216, table S5). This finding contrasts with the previously reported spontaneous genetic mutation rate of 7 × 10−9 base substitutions per site per generation for these same MA lines (18). It is noteworthy that the TREE PUZZLE analysis revealed higher estimated epimutation rates in earlier generations (19). One possible source of this variation could be due to seed age, storage and/or selection for seed survival. Therefore, although DNA methylation is predominantly static over relatively long periods of time, changes in cytosine methylation do occur, and at a frequency greater than that of mutation observed at the DNA sequence level.
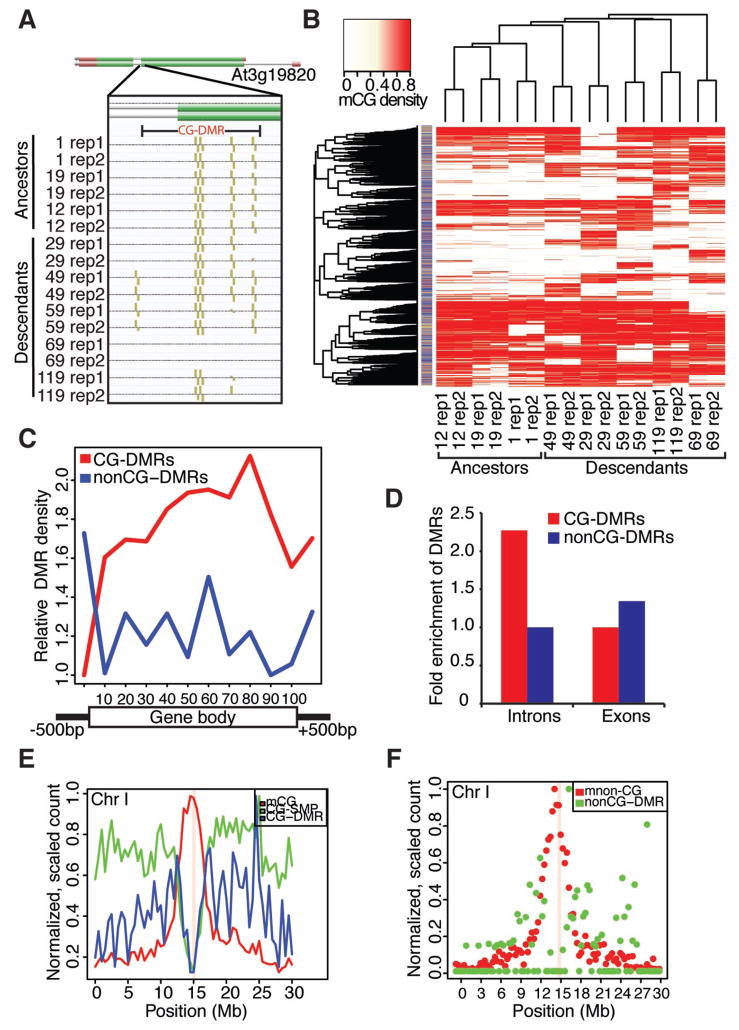
Using CG-SMPs derived from both ancestral and descendant populations, we carried out a genome-wide analysis of differentially methylated regions (DMRs) and identified 2,485 CG-DMRs that ranged in size from 11 bp to 1,110 bp (Fig. 2A, table S6). Hierarchical clustering of CG-DMRs in this population, calculated solely on the basis of the methylation density, revealed that the ancestral lines segregate as an independent cluster from the descendant lines (Figs. 2B, S3). Multivariate distance-based regression (MDMR) (20, 21) confirmed this finding, indicating a statistically significant (P < 0.00005) association between ancestor/descendant status and methylation density of the CG-DMR profiles. The ancestor/descendant status explained 47% of the variance in the dissimilarity in methylation density of CG-DMRs between pairs of samples, indicating that, over time, there is a divergence of DNA methylation patterns in both the formation and elimination of CG-DMRs. Furthermore, the genome-wide locations of these CG-DMRs were not uniformly distributed (P < 2.20e−16) as 60.5% (1,504/2,485) were found in genic regions compared to 3.3% (82/2,485) and 36.2% (899/2,485) located in intergenic regions and transposons, respectively (Fig. 2B).
Fig. 2.
CG-DMRs diverge over time and are enriched in gene bodies. (A) Example CG-DMR present in an unmethylated state in both replicates of line 69. (B) A heatmap representation of a two-dimensional hierarchical clustering based on DMRs. Columns represent samples. Rows indicate DMRs. The column to the left of the heatmap indicates the genomic location of the DMR (blue – gene body, gold – transposon, gray – intergenic, red – transposon in gene body). (C) The average distribution of CG-DMRs (red) and nonCG-DMRs (blue) across gene bodies (from the start of the 5′ UTR to the end of the 3′ UTR, including 500 bp up/downstream). (D) CG gene-body DMRs are specifically depleted in exons. (E) Genome-wide distributions of mCG (red), CG-SMPs (green), and CG-DMRs (blue) across chromosome I. (F) Genome-wide distributions of methylated non-CGs (mnonCG - red) and nonCG-DMRs (green) across chromosome I. The centromere is indicated by the pink vertical bar for (E) and (F).
Next, we performed a genome-wide survey for nonCG-DMRs and uncovered a total of 284 among all eight lines (table S7). In general, the nonCG-DMRs were largely localized to intergenic regions (141/284) of the genome as only 57/284 overlapped with genes and 86/284 overlapped with transposons. The size ranges of the nonCG-DMRs were similar to the CG-DMRs as the vast majority occurred in smaller segments of the genome (10 bp to 682 bp). Therefore, variation in DNA methylation appears to occur in all three methylation sequence contexts.
CG methylation is present within gene bodies and is enriched towards the 3′ end (2–5) whereas CG and nonCG methylation is associated with heterochromatin, transposons and repetitive sequences (1). In agreement with these findings, we observed that the 3′ portion of genes contained the greatest source of CG-DMRs and that the majority of nonCG-DMRs were enriched outside of the gene bodies (Fig. 2C). Furthermore, we observed a ~2-fold depletion of CG-DMRs in exons compared to introns (Fig. 2D). The genome-wide distributions of CG-SMPs, CG-DMRs and nonCG-DMRs were depleted in heterochromatic regions in the genome (Fig. 2E and F). This is mostly observed at the pericentromeres and centromeres (Figs. 2E, F; S4, S5). It is noteworthy that CG-DMRs are enriched in transposons located in euchromatin, but depleted in transposons present near the centromere. As the centromeric regions of the genome contain the highest density of DNA methylation (Fig. 2E, F), these observations combined with the observations that CG-DMRs are enriched in intron sequences, may indicate that DNA methylation that is associated with nucleosomes (22) (i.e., exons or tightly packaged chromatin in the pericentromeres and centromeres) may be maintained at a higher fidelity and DNA methylation not associated with nucleosomes may undergo greater epigenetic drift.
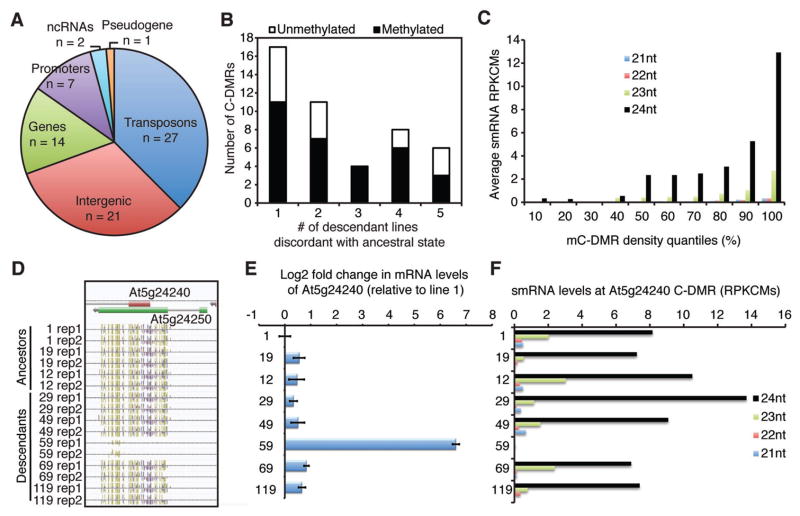
A genome-wide screen for DMRs simultaneously occurring in all three methylation sequence contexts (C-DMRs = CG, CHG and CHH) was performed to assess the extent of epiallelic variation that is characteristic of RdDM across the MA population. In total, 72 C-DMRs were identified of which functional categorization revealed that two-thirds overlapped with transposon and intergenic sequences while approximately one-third overlapped with gene bodies and promoters (Fig. 3A, table S8). To determine if transposition-induced methylation could potentially give rise to the methylated C-DMRs (mC-DMRs) (23), genomic DNA encompassing all C-DMRs was amplified and compared in all ancestral and descendant lines. In every case, the observed amplicon size was identical for all MA lines and was equal to the expected size of the locus (table S8), indicating that these C-DMRs are unlinked to cis-genetic variation located within 500 bp; a distance that would be expected to reveal methylation induced by transposon insertions at these loci (23). Additionally, none of the genetic variants identified by genome resequencing of this population (18) overlapped with any of these C-DMRs. Lastly, restriction enzyme digestion and Southern blot analyses were performed to rule out the possibility that copy number variants were the cause of spontaneous epiallele formation as is the case for the PAI and BAL epialleles (24, 25). In all cases examined, the observed hybridization pattern and gene copy number were identical for each of the MA lines (fig. S6). Therefore, we conclude that the 72 C-DMRs represent a set of spontaneously occurring epialleles within the MA lines, as they were not associated with any genetic variation.
Fig. 3.
Epiallelic variation at protein-coding loci is associated with transcriptional variation. (A) Classification of C-DMRs and their genomic locations. (B) The number of descendant lines discordant with the ancestral C-DMR state and the C-DMR methylation status. The black portions of the bar indicate the descendant C-DMRs that become methylated whereas the white portions indicate regions that become unmethylated compared to the ancestral population. (C) 24nt smRNA levels are associated with increasing methylation density. 24nt smRNA RPKCMs for all 576 C-DMRs (8 MA lines × 72 C-DMRs) were ranked and binned into 10% quantiles and then the average mC densities were plotted. (D) A representative C-DMR at At5g24240 in which both biological replicates of descendant line 59 were unmethylated. (E) qRT-PCR analysis of At5g24240 reveals >50-fold increase in mRNA abundance in unmethylated line 59. Error bars indicate standard error of the mean (s.e.m.). (F) 24nt smRNAs are enriched specifically in the MA lines that are transcriptionally silenced in (E) for the At5g24240 locus with the exception of line 59 which is abundantly expressed in (E).
Using a set of C-DMRs that exhibited an identical methylation status (fig. S7), we determined the frequency of discordance of the ancestral state with the descendant lines and found that 29 of the C-DMRs were highly variable (>1 descendant line was discordant with the ancestral state) (Fig. 3B). C-DMRs discordant in only one of the five descendant lines were the most frequent class, but there were a surprisingly high number of C-DMRs (63%) that were discordant in more than one descendant (Fig. 3B). Interestingly, within the set of 576 C-DMRs identified (8 lines × 72 C-DMRs), seven were discordant between the biological replicates (table S8). These data suggest that although many C-DMRs represent the formation of spontaneous epialleles, a small subset may reflect the presence of “hotspots” (metastable epialleles).
We sequenced smRNAs populations for all eight lines and found that smRNAs (represented as RPKCMs (Reads Per Kilobase of each C-DMR per Million reads) in table S9-12) were associated with an increase in the average methylation density of C-DMRs (Fig. 3C). Furthermore, this association resembled a binary switch as the most densely methylated C-DMRs contained abundant 24nt smRNAs (Fig. 3C).
Of the nine previously documented plant epialleles resulting in phenotypic variation, all affected transcriptional output of the differentially methylated locus (10–12, 23–28). mRNA abundance was measured in all eight lines with quantitative RT-PCR at eight C-DMRs that overlapped with protein-coding regions. In four of these genes the gain or loss of DNA methylation was correlated with a large decrease or increase in mRNA abundance, respectively and with the presence of 24nt smRNAs at each silenced epiallele (Fig. 3D to F and fig. S8). These findings reveal that changes in epiallelic state can lead to major effects on transcriptional output (fig. S9).
We also observed that the methylation status of one C-DMR resulted in alternative promoter usage of ACTIN RELATED PROTEIN 9 (At5g43500) (fig. S10C). Interestingly, the loss of DNA methylation within the 5′UTR of the At5g43500.1 isoform led to an increase in mRNA expression, whereas expression of a second isoform At5g43500.2 with a transcriptional start site located further downstream was unaffected (fig. S10D and E).
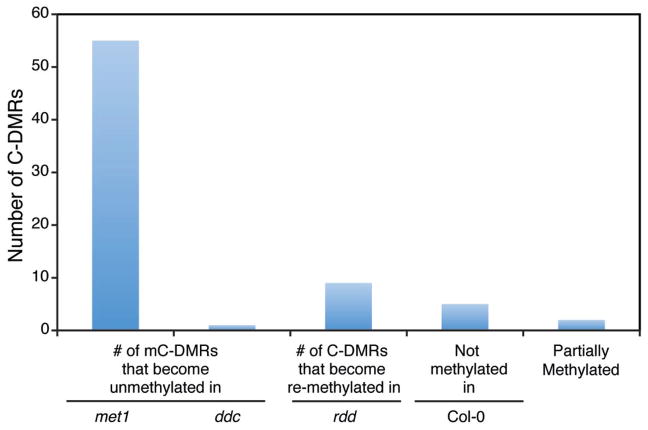
Although epialleles can have major impacts on phenotypic diversity, until now their identification was not trivial. Even more puzzling is the origin of “pure” alleles, which are defined by their formation in absence of any genetic variation in cis or trans (9). One route to epiallele formation may be the failure to correctly maintain the proper methylation status throughout epigenetic reprogramming that occurs postfertilization (29, 30). It is noteworthy that 63 of the 72 C-DMRs overlap with regions previously shown to have altered methylation patterns in methylation enzyme mutants (Fig. 4) (3). Of the 14 C-DMRs that overlap with genes, five become re-expressed in met1-3 and one transcript becomes silenced in rdd (3). Therefore, these results suggest that a failure to faithfully maintain genome-wide methylation patterns by MET1 and/or RDD is likely one source of spontaneous epiallele formation.
Fig. 4.
Methylation status of all 72 epialleles in methylation and demethylation mutant backgrounds. Most of the epialleles become unmethylated in met1-3 while a smaller number become re-methylated in the DNA demethylase triple mutant rdd.
Regardless of their origin, the majority of epialleles identified in this study are meiotically stable and heritable across many generations in this population. Understanding the basis for such transgenerational instability and the mechanism(s) that trigger and/or release these epiallelic states will be of great importance for future studies.
Supplementary Material
Acknowledgments
We thank M. White, R. Lister, M. Galli and R. Amasino for discussions and R. Shaw and E. Darmo for seeds and M. Axtell for his Southern blot protocol. RJS was supported by an NIH NRSA postdoctoral fellowship (F32-HG004830). MDS was supported by a National Science Foundation IGERT training grant (DGE-0504645). MGL was supported by an EU FP7 Marie Curie International Outgoing Fellowship (project 252475). OL and NJS are supported by NIH/NCRR Grant Number UL1 RR025774. This work was supported by the Mary K. Chapman Foundation, the National Science Foundation (MCB-0929402 and MCB1122246), the Howard Hughes Medical Institute and the Gordon and Betty Moore foundation to JRE. JRE is a HHMI-GBMP Investigator. Analyzed datasets can be viewed at http://neomorph.salk.edu/30_generations/browser.html. Sequence data can be downloaded from NCBI SRA (SRA035939.1).
Footnotes
References and Notes
- 1.Law JA, Jacobsen SE. Nat Rev Genet. 2010;11:204. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cokus SJ, et al. Nature. 2008;452:215. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lister R, et al. Cell. 2008;133:523. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, et al. Cell. 2006;126:1189. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Nat Genet. 2006;39:61. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 6.Chan SW, Zhang X, Bernatavichute YV, Jacobsen SE. PLoS Biol. 2006;4:e363. doi: 10.1371/journal.pbio.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan SW, et al. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- 8.Paszkowski J, Grossniklaus U. Curr Opin Plant Biol. 2011 doi: 10.1016/j.pbi.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Richards EJ. Nat Rev Genet. 2006;7:395. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- 10.Shibuya K, Fukushima S, Takatsuji H. Proc Natl Acad Sci U S A. 2009;106:1660. doi: 10.1073/pnas.0809294106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cubas P, Vincent C, Coen E. Nature. 1999;401:157. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- 12.Manning K, et al. Nat Genet. 2006;38:948. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 13.Thompson AJ, et al. Plant Physiol. 1999;120:383. doi: 10.1104/pp.120.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johannes F, et al. PLoS Genet. 2009;5:e1000530. doi: 10.1371/journal.pgen.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teixeira FK, et al. Science. 2009;323:1600. doi: 10.1126/science.1165313. [DOI] [PubMed] [Google Scholar]
- 16.Vongs A, Kakutani T, Martienssen RA, Richards EJ. Science. 1993;260:1926. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- 17.Shaw RG, Byers DL, Darmo E. Genetics. 2000;155:369. doi: 10.1093/genetics/155.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ossowski S, et al. Science. 2010;327:92. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Additional experiments and descriptions of methods used to support our conclusions are presented as supporting material on Science Online.
- 20.Nievergelt CM, et al. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:234. doi: 10.1002/ajmg.b.30252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zapala MA, Schork NJ. Proc Natl Acad Sci U S A. 2006;103:19430. doi: 10.1073/pnas.0609333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chodavarapu RK, et al. Nature. 2010;466:388. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, He Y, Amasino R, Chen X. Genes Dev. 2004;18:2873. doi: 10.1101/gad.1217304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bender J, Fink GR. Cell. 1995;83:725. doi: 10.1016/0092-8674(95)90185-x. [DOI] [PubMed] [Google Scholar]
- 25.Yi H, Richards EJ. Genetics. 2009;183:1227. doi: 10.1534/genetics.109.105569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobsen SE, Meyerowitz EM. Science. 1997;277:1100. doi: 10.1126/science.277.5329.1100. [DOI] [PubMed] [Google Scholar]
- 27.Saze H, Kakutani T. EMBO J. 2007 doi: 10.1038/sj.emboj.7601788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soppe WJ, et al. Mol Cell. 2000;6:791. doi: 10.1016/s1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- 29.Mosher RA, et al. Nature. 2009;460:283. doi: 10.1038/nature08084. [DOI] [PubMed] [Google Scholar]
- 30.Slotkin RK, et al. Cell. 2009;136:461. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langmead B, Trapnell C, Pop M, Salzberg SL. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lister R, et al. Nature. 2009;462:315. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. Bioinformatics. 2002;18:502. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 34.Nievergelt CM, Libiger O, Schork NJ. PLoS Genet. 2007;3:e51. doi: 10.1371/journal.pgen.0030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McArdle BH, Anderson MJ. Ecology. 2001;82:290. [Google Scholar]
- 36.Lister R, et al. Nature. 2011;471:68. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewsey MG, et al. Mol Plant Microbe Interact. 2010;23:835. doi: 10.1094/MPMI-23-7-0835. [DOI] [PubMed] [Google Scholar]
- 38.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Neurosci Lett. 2003;339:62. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 39.Ruijter JM, et al. Nucleic Acids Res. 2009;37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinoshita T, et al. Science. 2003;303:521. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.