Abstract
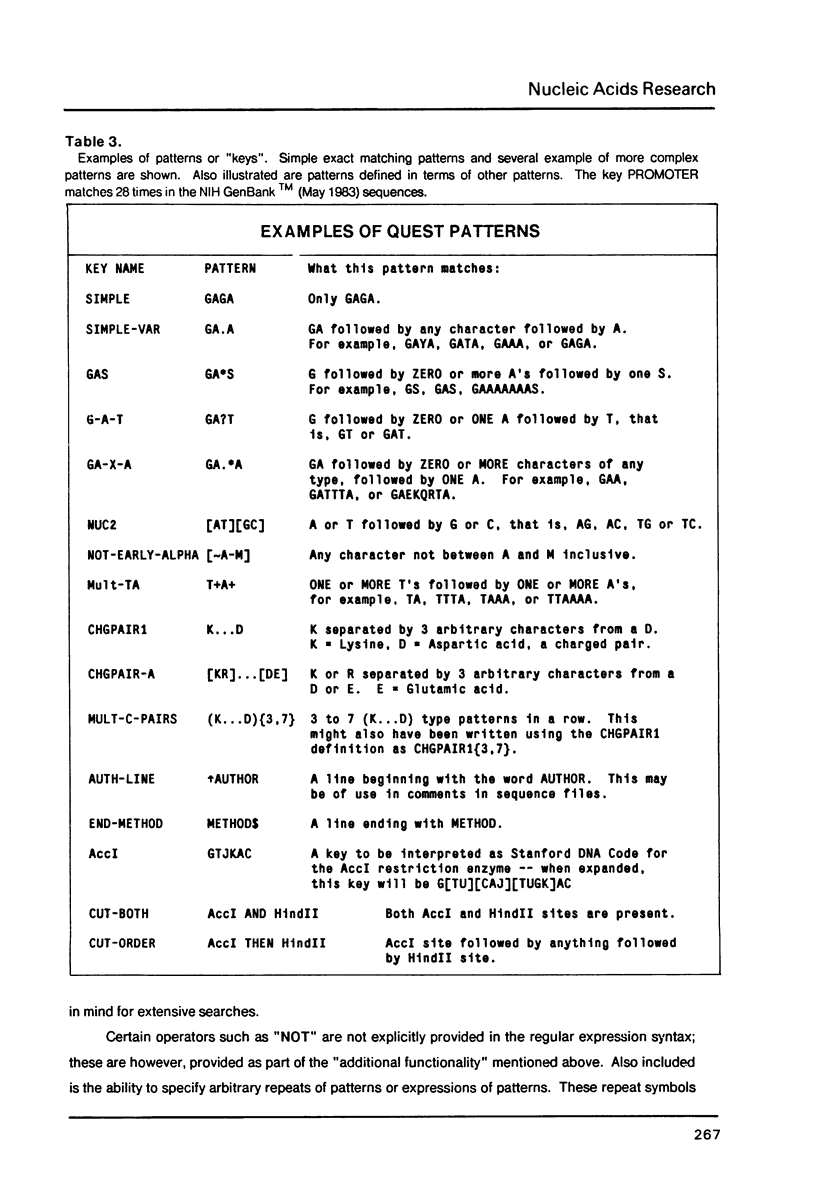
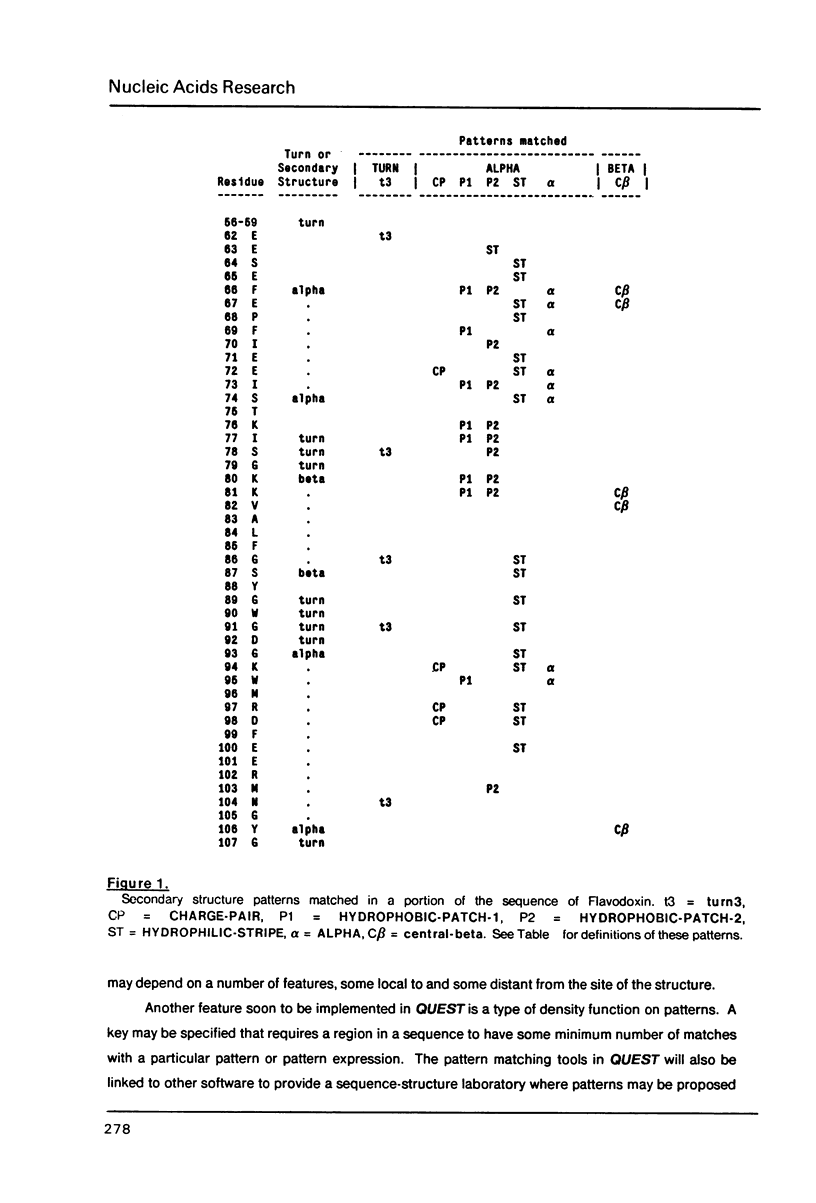
The intrinsic redundancy of genetic information makes searching for patterns in biological sequences a difficult task. We have designed an interactive self-documenting computer program called QUEST that allows rapid searching of large DNA and protein data banks for highly redundant consensus sequences or character patterns. QUEST uses a concise language for specifying character patterns containing several levels of ambiguity and pattern arrangement. Examples of the use of this program for sequence data are given. Details of the algorithm and pattern optimization are explained.
Full text
PDF















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brutlag D. L., Clayton J., Friedland P., Kedes L. H. SEQ: a nucleotide sequence analysis and recombination system. Nucleic Acids Res. 1982 Jan 11;10(1):279–294. doi: 10.1093/nar/10.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor W. R., Thornton J. M. Prediction of super-secondary structure in proteins. Nature. 1983 Feb 10;301(5900):540–542. doi: 10.1038/301540a0. [DOI] [PubMed] [Google Scholar]



