Abstract
Extrahepatic replication has important implications for the transmission and treatment of hepatitis C virus (HCV). We analyzed longitudinal HCV diversity in peripheral-blood mononuclear cells (PBMCs) and serum during HCV monoinfection and HCV/HIV coinfection to determine whether distinct amino acid signatures characterized HCV replicating within PBMCs. Analysis of E1-HVR1 sequences demonstrated higher serum genetic distances among HCV/human immunodeficiency virus (HIV)–coinfected persons. Moreover, consensus PBMC sequences were rarely identical to those in the corresponding serum, suggesting divergence in these 2 compartments. Three of 5 HCV/HIV-coinfected participants showed evidence of HCV compartmentalization in PBMCs. Additionally, signature sequence analysis identified PBMC-specific amino acids in all HCV/HIV-coinfected persons. To our knowledge, this is the first study to identify specific amino acids that may distinguish HCV variants replicating in PBMCs. It is provocative to speculate that extrahepatic HCV diversity may be an important determinant of treatment response and thus warrants additional study, particularly during HCV/HIV coinfection.
Because of their shared routes of transmission, hepatitis C virus (HCV) and HIV coinfection occurs in ~150,000–300,000 people in the United States [1]. Multiple studies have demonstrated the adverse impact of HIV coinfection on HCV RNA levels, liver disease progression, and response rates to HCV treatment [2–5].
Although the mechanisms by which these 2 viruses interact remain unclear, peripheral-blood mononuclear cells (PBMCs) may serve as important sites for interaction between HIV and HCV; thus, evaluating this and other potential reservoirs of HCV replication is important. Evidence of HCV replication in granulocytes, monocytes/macrophages, dendritic cells, B lymphocytes, and other extrahepatic tissues has been suggested (reviewed in [6]). Furthermore, several studies suggest that extrahepatic replication of HCV may be an important predictor of HCV treatment outcome [7–12]. To date, most analyses of HCV in PBMCs have involved a small number of patients and generally included only HCV-monoinfected persons [13–17]. No longitudinal studies have assessed replication of HCV in PBMCs during coinfection with HIV, nor have specific viral amino acid sequences associated with replication in PBMCs been identified.
Within an individual, HCV exists as a population of closely related, yet distinct, variants termed “the viral quasispecies.” This heterogenous nature allows HCV to adapt quickly to a variety of changing selection pressures that may exist within a given host. The majority of HCV quasispecies studies to date have focused on the envelope glycoproteins, E1 and E2, because of their putative role in receptor recognition and binding and their interactions with the adaptive immune system. Several studies have analyzed HCV variants from various tissues to examine HCV tropism for a given cell type [18–24]. Thus far, the data have been obtained from small, cross-sectional studies and have not generally included HCV/HIV-coinfected persons. Furthermore, viral diversity in the PBMC compartment has not been compared directly in HCV-monoinfected and HCV/HIV-coinfected persons from the same study population. Therefore, we sought to (1) investigate longitudinal HCV diversity during HCV mono-infection versus HCV/HIV coinfection and (2) use a robust phylogenetic approach to identify amino acids that may be associated with extrahepatic replication of HCV.
SUBJECTS, MATERIALS, AND METHODS
Study population
From 1993 until 1999, a prospective natural history study of HIV infection—the HIV Epidemiologic Research (HER) Study—was conducted in US women who had clinic visits at 6-month intervals [25]. By study design, one-half reported intravenous drug use (IVDU), whereas the other half reported only sexual risk behavior. Overall, the prevalence of HCV was 56.5%, with rates of 48.0% and 60.8% among HIV-uninfected and HIV-infected women, respectively [26].
Participants were included in the present study if they (1) had paired serum and PBMC samples available from at least 2 (HCV monoinfected) or 3 (HCV/HIV coinfected) consecutive study visits (separated by ~6 months between visits) conducted at the Providence, Rhode Island, site; (2) were not receiving antiretroviral therapy at the study onset for HIV-infected women; (3) had quantifiable HCV RNA in all serum samples tested; and (4) were infected with HCV genotype 1, based on previous reports that suggested genotype-specific differences in variability [27, 28].
Polymerase chain reaction (PCR) amplifications
Serum samples and cells were processed and stored in accordance with standard laboratory procedures to ensure viral preservation. Real-time PCR conditions to quantify strand-specific HCV RNA levels in the serum and PBMC compartments have been described in detail elsewhere [29]. Briefly, RNA was extracted from 140 μL of serum using the QIAamp Viral RNA Kit (Qiagen) and eluted in 60 μL of diethylpyrocarbonate (DEPC)–treated dH2O. For PBMC samples, the number of cells available was limited and varied per sample (1.4 × 106 to 7.6 × 106 cells/mL). Therefore, we normalized quantitative HCV RNA data in the PBMC compartment to the copy number of a housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Cells (500 μL) were washed with DEPC-treated dH2O, and cellular RNA was extracted with TRIzol. The resulting RNA was resuspended in 40 μL of DEPC-treated dH2O and treated 2 times with DNase I (Ambion).
In the present study, a qualitative reverse-transcriptase (RT)–PCR was used in which cDNA corresponding to E1-HVR1 was generated using 1–2 μL of serum or PBMC RNA and the antisense primer 5′-AGG CTT TCA TTG CAG TTC AAG GCC TTG CTA TTG ATG TGC C-3′ (nt 1639–1600 according to the numbering of H77 [GenBank number AF009606]). First-round PCR amplifications using the sense primer 5′-GCG TCC GGG TTC TGG AAG ACG GCG TGA ACT ATG CAA CAG G-3′ (nt 802–841) were performed as follows: 94°C for 2 min, followed by 35 cycles at 94°C for 1 min, 65°C for 2 min, and 72°C for 2 min, with a final elongation step at 72°C for 8 min. Second-round PCR conditions were identical to first-round conditions and used the antisense primer 5′-AGT TCA AGG CCG TGC TAT TGA TGT GCC AAC TGC CGT TGG T-3′ (nt 1626–1587) and the sense primer 5′-GGC ATG GGA TAT GAT GAT GAA CTG GTC CCC TAC-3′ (nt 1295–1327). To minimize PCR-generated errors, the high-fidelity pfu polymerase (Stratagene) was used. To avoid potential cross-contamination, samples from the same individual were handled separately. Additionally, all RT-PCR amplifications included one reaction containing no RT and a separate reaction containing no template as negative controls.
PCR products were gel purified and ligated into the PCR2.1-TOPO vector (Invitrogen). Plasmids were propagated and purified before sequencing with dye terminator chemistry. A minimum of 10 plasmids per time point per compartment were sequenced in the forward and reverse directions and edited using Sequencher software (version 4.2; GeneCodes). A previous study has validated that sequencing 10 clones reliably and reproducibly represents the viral quasispecies circulating within an individual [30].
Phylogenetic analyses
By aligning the compartment-specific clones at each time point, consensus amino acid sequences were generated using Clustal X [31]. The database references used to confirm HCV genotype included D10749 (1A), D11355 (1B), D14853 (1C), AF177039 (2A), AB030907 (2B), D50409 (2C), AF046866 (3A), Y11604 (4A), AB031663 (5A), and D84262 (6B), as well as relevant consensus genotype references and the laboratory reference strain H77 (AF009606). Separate minimum evolution trees were generated for the E1-HVR1 (258 nt) by use of the MEGA program [32]. Positions with gaps were excluded from analysis, and corrections for multiple substitutions were performed for all trees. The statistical robustness and reliability of the branching order within each phylogenetic tree were confirmed by bootstrap analysis using 100 replicates [33]. Only bootstrap values >70% were considered statistically significant. Sequences have been submitted to GenBank under accession numbers EF193810–EF193846.
Quasispecies parameters
HCV quasispecies variability was analyzed according to HIV status, compartment, and time point. Intrapatient genetic distances were calculated by pairwise comparison of E1-HVR1 nucleotide sequences at each time point using the Kimura method of MegAlign (DNASTAR). Shannon entropy was also calculated as follows: Sn =−Σ(pilnpi)/lnN, where pi is the frequency of each distinct amino acid sequence and N is the total number of sequences analyzed [34]. Nonsynonymous (dN):synonymous (dS) ratios were calculated using the Nei-Gojobori method in MEGA [32].
Signature sequence analysis
Signature amino acid patterns that distinguish PBMC viral variants from those in the corresponding serum sample were identified using the VESPA program [35]. Only amino acid signatures above a 70% threshold were considered significant.
Statistical analyses
Baseline demographics and virologic variables were compared using Fisher’s exact test for categorical variables and the Wilcoxon rank sum test for continuous variables. Because of the limited sample size, the exact Wilcoxon rank sum test was used to compare differences between the HCV-monoinfected and HCV/HIV-coinfected groups, whereas the exact Wilcoxon signed rank test was used to compare differences between PBMCs and serum samples within the HCV/HIV-coinfected group. All P values are 2-tailed; P <.05 was considered statistically significant. All analyses were performed using SAS software (SAS Institute).
RESULTS
Patient characteristics
Four HCV-monoinfected and 5 HCV/HIV-coinfected women were selected for the current phylogenetic analysis of HCV diversity in PBMCs. The median age was 31.4 (range, 21.6–39.0) and 34.1 (33.5–38.3) years for HCV-monoinfected and HCV/HIV-coinfected women, respectively. Median CD4 cell counts were 1322 cells/mm3 at visit A and 1192 cells/mm3 at visit B among HCV-monoinfected persons. Median CD4 cell counts were 377, 356, and 337 cells/mm3 at visits A, B, and C, respectively, among HCV/HIV-coinfected women. Serum HCV RNA levels were 2.55–5.77 and 5.21–6.34 log10 copies/μL for HCV-monoinfected and HCV/HIV-coinfected persons, respectively. No study participant received antiviral therapy for HCV at any study visit. All 4 HCV-monoinfected and 3 of 5 HCV/HIV-coinfected women reported heterosexual exposure as their main risk factor for HCV; 2 of 5 HCV/HIV-coinfected women reported IVDU as their main risk factor. HIV treatment for coinfected women (after visit A, at which time they were HIV-treatment naive) consisted of 2 nucleoside RT inhibitors (NRTIs; n =1) or 2 NRTIs plus 1 protease inhibitor (n =4).
Interpatient variability
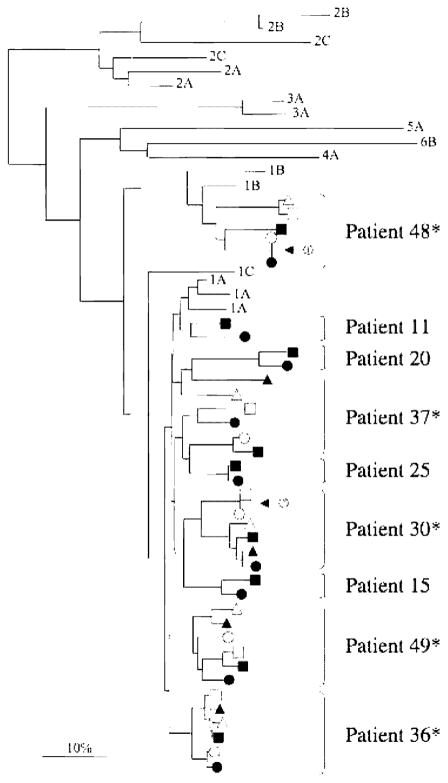
Alignment of all E1-HVR1 clones demonstrated highly significant clustering of sequences by patient (data not shown). Consensus amino acid sequences for HCV-monoinfected and HCV/HIV-coinfected persons were then compared by phylogenetic analysis to database reference sequences (figure 1). All 4 HCV-monoinfected participants and 4 of 5 HCV/HIV-coinfected participants were infected with HCV genotype 1A, whereas 1 HCV/HIV-coinfected participant was infected with HCV genotype 1B. There was no apparent clustering of sequences by HIV status. Importantly, E1-HVR1 consensus amino acid sequences were nearly always distinct over time (cf. visits A, B, and C) and by compartment (cf. serum to PBMCs). However, one HCV/HIV-coinfected woman had an identical consensus sequence in her serum and PBMC samples at visit A, whereas another HCV/HIV-coinfected woman had an identical PBMC consensus sequence at visits A and B. Despite repeated attempts, we could only consistently amplify E1-HVR1 from the PBMCs of 1 HCV-monoinfected person, even though the housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), was routinely amplified for all PBMC samples (data not shown).
Figure 1.
Phylogenetic tree of consensus E1-HVR1 amino acid sequences from serum (black symbols) and peripheral-blood mononuclear cell (PBMC; white symbols) compartments at visits A (circles), B (squares), and C (triangles). Database reference sequences are indicated by their genotype. Arrow 1 indicates a time point at which the consensus amino acid sequences from the serum and PBMC compartments are identical for patient 48 (hepatitis C virus [HCV]/HIV coinfected). Arrow 2 indicates 2 time points at which the consequence amino acid sequences in the PBMC compartment are identical for patient 30 (HCV/HIV coinfected). *HCV/HIV-coinfected individuals.
Quasispecies evaluation
Because analysis of consensus amino acid sequences masks subtle shifts in the viral quasispecies over time, longitudinal intrapatient variability was also evaluated in serum and PBMCs. Multiple measures of HCV diversity and complexity were assessed in each compartment at each time point, including genetic distance, entropy, and the dN:dS ratio. The genetic distance reflects the similarity among all sequences within an individual at a given time point. In HCV-monoinfected participants, the median intrapatient genetic distance in serum samples was 0.7% (range, 0.3%–2.2%) and 1.5% (range, 0.6%–2.5%) at visits A and B, respectively (table 1, upper panel). In HCV/HIV-coinfected participants, the median intrapatient genetic distance in serum was 4.4% (range, 0.8%–7.6%), 3.3% (range, 2.3%–7.7%), and 5.1% (range, 1.6%–6.8%) at visits A–C (table 1, middle panel). The difference in serum genetic distance between the HCV-mono-infected and HCV/HIV-coinfected groups approached statistical significance at visit A (P =.06) and was significant at visit B (P =.03). In the PBMCs of HCV/HIV-coinfected participants, the median intrapatient genetic distance was 0.8% (range, 0.0%–3.2%) at visit A, 2.4% (range, 2.0%–5.2%) at visit B, and 2.8% (range, 0.4%–7.3%) at visit C (table 1, lower panel). Among HCV/HIV-coinfected persons, the genetic distance in serum was not significantly different from that in the corresponding PBMCs for any visit.
Table 1.
E1-HVR1 intrapatient diversity for the serum and peripheral-blood mononuclear cell (PBMC) quasispecies from hepatitis C virus (HCV)–monoinfected and HCV/HIV-coinfected participants.
| Sample, patient, time point | Intrapatient genetic distance | Entropy | dN:dS |
|---|---|---|---|
| Serum compartment of HCV monoinfected | |||
| 11 | |||
| A | 2.19 | 0.47 | 0.43 |
| B | 1.41 | 0.59 | 0.40 |
| 15 | |||
| A | 0.51 | 0.14 | 0.15 |
| B | 2.51 | 0.35 | 0.67 |
| 20 | |||
| A | 0.88 | 0.41 | 0.35 |
| B | 0.60 | 0.59 | a |
| 25 | |||
| A | 0.32 | 0.41 | 1.00 |
| B | 1.50 | 0.56 | 9.50 |
|
| |||
| Serum compartment of HCV/HIV coinfected | |||
| 30 | |||
| A | 4.74 | 0.56 | 0.69 |
| B | 3.33 | 0.58 | 0.55 |
| C | 4.39 | 0.56 | 0.55 |
| 36 | |||
| A | 0.80 | 0.52 | 1.50 |
| B | 2.27 | 0.64 | 1.15 |
| C | 1.57 | 0.61 | 2.00 |
| 37 | |||
| A | 7.56 | 0.60 | 1.01 |
| B | 7.66 | 0.54 | 0.90 |
| C | 6.75 | 0.55 | 0.48 |
| 48 | |||
| A | 4.04 | 0.39 | 1.00 |
| B | 5.40 | 0.64 | 1.74 |
| C | No sample | No sample | No sample |
| 49 | |||
| A | 4.35 | 0.54 | 0.96 |
| B | 2.72 | 0.60 | 2.67 |
| C | 5.81 | 0.50 | 1.07 |
|
| |||
| PBMC compartment of HCV/HIV coinfected | |||
| 30 | |||
| A | 1.01 | 0.28 | 0.10 |
| B | 3.68 | 0.39 | 0.56 |
| C | 2.11 | 0.51 | 0.47 |
| 36 | |||
| A | 1.27 | 0.35 | 3.00 |
| B | 3.12 | 0.94 | 2.12 |
| C | 3.14 | 1.00 | 2.12 |
| 37 | |||
| A | 5.61 | 0.94 | 0.86 |
| B | 1.87 | 0.70 | 0.21 |
| C | 8.13 | 0.70 | 0.60 |
| 48 | |||
| A | 3.40 | 0.47 | 1.03 |
| B | 3.47 | 0.56 | 0.48 |
| C | 0.40 | 0.14 | 0.22 |
| 49 | |||
| A | 0.19 | 0.27 | a |
| B | 2.44 | 0.80 | 2.42 |
| C | 4.23 | 0.51 | 1.48 |
NOTE. dN, nonsynonymous; dS, synonymous.
Indicates that dS =0; therefore, this time point could not be defined.
As an additional measure of genetic complexity, entropy was calculated. Because entropy takes into account the number of distinct viral variants as well as the relative frequency of each variant within the quasispecies [34], it is less susceptible than genetic distance to variation as a result of hypermutation. In HCV-monoinfected participants, the median serum HCV entropy was 0.41 (range, 0.14–0.47) at visit A and 0.57 (range, 0.35–0.59) at visit B (table 1, upper panel). In HCV/HIV-coinfected participants, the median serum HCV entropy was 0.54 (range, 0.39–0.60) at visit A, 0.60 (range, 0.54–0.64) at visit B, and 0.55 (range, 0.50–0.61) at visit C (table 1, middle panel). The difference in entropy between the HCV-monoinfected and HCV/HIV-coinfected groups was marginally significant for visit A (P =.09) but not for visit B (P =.26). In the PBMCs of HCV/HIV-coinfected participants, median entropy was 0.35 (range, 0.28–0.94) at visit A, 0.70 (range, 0.39–0.94) at visit B, and 0.51 (range, 0.14–1.0) at visit C (table 1, lower panel). Among HCV/HIV-coinfected persons, entropy in the serum samples was not significantly different from that in the corresponding PBMCs for any visit.
As a measure of immunologic selection pressure, dN:dS ratios were also calculated. Ratios >1 are consistent with positive immune selection pressure and have previously been used to explore HCV evolution [36, 37]. Among HCV-monoinfected individuals, the dN:dS ratio was >1 in serum for only 1 of 7 (14.3%) time points; dS was 0 for 1 time point; therefore, this sample could not be analyzed further for this measure of quasispecies diversity. In contrast, among HCV/HIV-coinfected individuals, the dN:dS ratio was >1 in serum for 7 of 14 (50%) time points. Among PBMCs from HCV/HIV-coinfected individuals, the dN:dS ratio was >1 for 6 of 14 (42.9%) time points, suggesting that virus present within this compartment may also be subject to immunologic selection pressures.
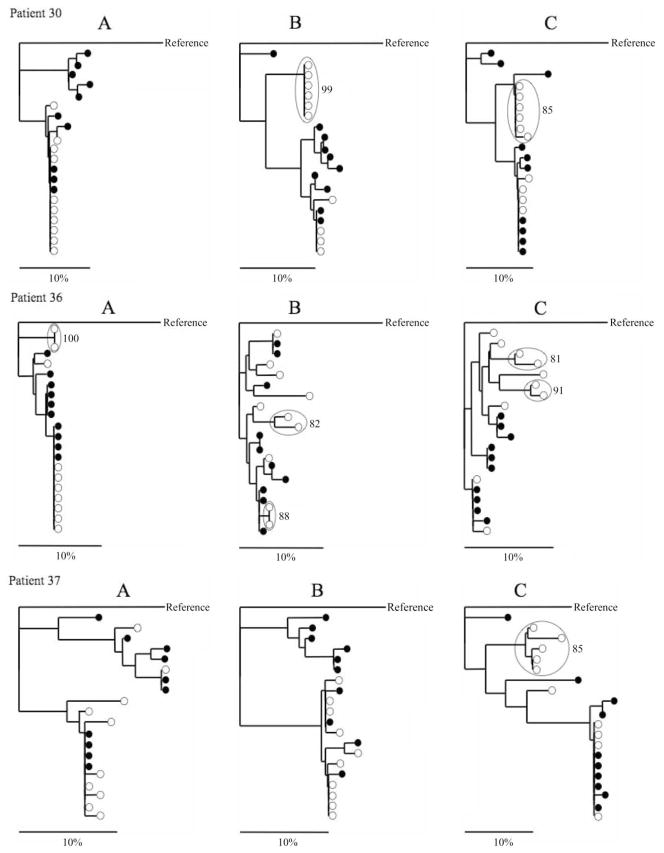
Further phylogenetic analysis was performed to investigate the potential for compartmentalization of viral variants between PBMCs and serum samples from the 5 HCV/HIV-coinfected persons by analyzing each time point independently (figure 2). Participant 30 showed no compartmentalization of E1-HVR1 variants between PBMCs and serum at visit A; however, a distinct population of 6 PBMC-specific variants was observed at visits B and C. In participant 36, 2 PBMC-specific variants were observed at visit A. Two distinct PBMC populations, each consisting of 2 viral variants, were also observed at visits B and C. In participant 37, a population of 5 PBMC-specific variants was identified at visit C but not at other visits. Participants 48 and 49 showed no evidence of PBMC compartmentalization of E1-HVR1 variants at any available time point. Thus, 3 of 5 (60%) HCV/HIV-coinfected participants and 6 of 14 (43%) time points showed obvious phylogenetic evidence of HCV compartmentalization in PBMCs.
Figure 2.
Phylogenetic tree analysis of quasispecies E1-HVR1 amino acid sequences from the serum (black circles) and peripheral-blood mononuclear cell (PBMC; white circles) compartments of hepatitis C virus (HCV)/HIV–coinfected women. Relevant bootstrap values >70 (of 100) are shown for PBMC-specific viral variants (highlighted by large, open circles). HCV genotype 2 was used as the reference sequence.
Signature sequence analysis
To identify specific amino acid differences associated with HCV replication in PBMCs, signature sequence analysis was performed at each time point. Amino acids that distinguish viral variants in the PBMCs from those in the corresponding serum samples were identified in all 5 HCV/HIV-coinfected individuals (figure 3); amino acid positions at which the distribution of E1-HVR1 clones are significantly different between the PBMC and serum compartment are underlined. Using this approach, PBMC-specific signature sequences were identified at 8 of 14 (57%) time points analyzed. Among these 8 time points, the mean number of PBMC-specific signature amino acids was 6.25 (range, 1–11). Forty-eight (96%) of 50 signature amino acids were located within HVR1, whereas only 2 (4%) were located within E1, suggesting that HVR1 could play an important role in HCV compartmentalization within PBMCs.
Figure 3.
Signature sequence analysis. Amino acid positions at which the peripheral-blood mononuclear cell (PBMC) and serum clonal distributions are significantly different at a threshold of 70% are underlined. “S30A” and “P30A” indicate, for example, a serum sample and a PBMC sample, respectively, from patient 30 at time point A. Nos. in parentheses indicate the no. of amino acid signatures present in PBMCs at that time point. Shown are the consensus amino acids at each position. In several instances, a given amino acid may appear to be identical between the 2 compartments; however, the frequency distribution of the 10 viral variants that make up that consensus is different between the 2 compartments. The bar highlights the 27 amino acids of HVR1.
Among the HVR1 sequences from the 4 HCV/HIV-coinfected women infected with HCV genotype 1A (visit A), positions 2 (T), 4 (V), 6 (G), 7 (G), 23 (G), and 26 (Q) were conserved in all variants from the serum and PBMC compartments, whereas the frequency of amino acid residues at other positions was highly variable (data not shown).
DISCUSSION
We have previously investigated HCV quasispecies diversity in the serum of HCV/HIV-coinfected persons [37]; however, evaluation of HCV diversity in the PBMC compartment has not been adequately explored in HCV/HIV-coinfected persons. Because PBMCs could serve as a critical site for direct interactions between HIV and HCV, further examination of this compartment is warranted.
In the present study, we analyzed the HCV quasispecies in the serum and PBMC of 4 HCV-monoinfected and 5 HCV/HIV-coinfected women. Several lines of evidence suggested PBMC-specific compartmentalization of HCV. First, consensus PBMC sequences were rarely identical to those in the corresponding serum sample, suggesting divergence of the viral quasispecies in these 2 compartments (figure 1). Second, 3 of 5 HCV/HIV-coinfected participants and 6 of 14 time points showed obvious evidence of HCV compartmentalization in PBMCs by phylogenetic analysis (figure 2). However, it is likely that this approach underestimates the true probability of detecting PBMC-specific variants, because this is a very conservative approach that included only a small portion of the viral genome and a limited follow-up period. Nonetheless, signature sequence analysis identified PBMC-specific amino acids in all 5 HCV/HIV-coinfected women. Third, additional analysis demonstrated a distribution of viral variants that appeared to be compartment dependent (figure 3). Although a similar non-random distribution of HCV sequences in hepatic and extra-hepatic compartments has been reported previously in cross-sectional studies of HCV monoinfection [18, 20, 22, 24], this phenomenon has not been described previously during HCV/HIV coinfection. These data also suggest that specific amino acids within HVR1 may contribute to HCV replication within PBMCs and altered cell tropism. The increased intrapatient genetic distances observed over time in the PBMC compartment also suggest that this data was not simply due to long-term sequestration of HCV debris in PBMCs or long-lived antibody complexes—both of which would likely result in decreased or stable diversity (and dN:dS ratios <1) over time. Nonetheless, the exact mechanisms by which HIV may influence extrahepatic replication of HCV require additional investigation.
To our knowledge, the present study is the first to identify specific amino acid sequences that distinguish viral variants in PBMCs from those in the serum of HCV/HIV-coinfected persons. However, several limitations should be noted. First, because of the extensive sequencing conducted in each participant, a limited number of women were included; thus, generalizing our results to other populations requires caution. Second, because E1-HVR1 was not consistently amplified from the PBMCs of the HCV-monoinfected women, direct comparison of HCV sequences in PBMCs during HCV monoinfection versus HCV/HIV coinfection was not possible. However, it should be reiterated that several previous studies have suggested that extrahepatic replication of HCV occurs more frequently in the presence of HIV coinfection [29, 38]. Furthermore, a housekeeping gene could be amplified from all PBMC samples tested. Additional methodologic considerations that could explain different abilities to detect HCV RNA in PBMCs include the quality and quantity of viral RNA, the genomic region targeted for amplification, and the sensitivity of the qualitative or quantitative assay employed. Third, although it is possible that minority viral variants may not be represented, multiple measures of viral diversity/complexity were examined here. Moreover, a previous study has shown that sequencing 10 viral variants is sufficient for quasispecies evaluation [30]. Additionally, we have demonstrated that multiple amplifications of the same template yielded similar estimates of quasispecies diversity [37]. Fourth, given the limited number of PBMCs available, we were unable to sort the cell population to precisely define the cell type(s) that supports HCV replication. Nonetheless, there is compelling evidence to suggest that HCV can infect several peripheral-blood cell types, including B lymphocytes, granulocytes, monocytes/macrophages, and dendritic cells [13, 39, 40]. Importantly, the current study is unique because it permitted longitudinal evaluation of PBMC compartmentalization in HCV/HIV-coinfected individuals and is restricted to HCV genotype 1, thereby controlling for potential genotype-specific differences in diversity [27, 28].
The detection of HCV RNA in hidden reservoirs, such as PBMCs, suggests that higher doses of current HCV medications and/or combination therapy with antiviral agents specific for HCV may be warranted to completely eradicate HCV from all reservoirs of replication. Furthermore, the presence of PBMC-specific variants with particular amino acid signatures implies that subtle changes in the viral quasispecies could have a profound effect on cell tropism and replication of HCV and that diversity in the PBMC compartment may be an important of determinant of treatment response and the duration of therapy in some individuals.
HIV EPIDEMIOLOGIC RESEARCH (HER) STUDY GROUP
The HER Study Group consists of Robert S. Klein, Ellie Schoenbaum, Julia Arnsten, Robert D. Burk, Penelope Demas, and Andrea Howard (Montefiore Medical Center and the Albert Einstein College of Medicine); Paula Schuman, Jack Sobel, Suzanne Ohmit, William Brown, Michael Long, Wayne Lancaster, and Jose Vazquez (Wayne State University School of Medicine); Anne Rompalo, David Vlahov, and David Celentano (Johns Hopkins University School of Medicine); Charles Carpenter, Kenneth Mayer, Susan Cu-Uvin, Timothy Flanigan, Joseph Hogan, Valerie Stone, Karen Tashima, and Josiah Rich (Brown University School of Medicine); Ann Duerr, Lytt I. Gardner, Chad Heilig, Scott D. Holmberg, Denise J. Jamieson, Janet S. Moore, Ruby M. Phelps, Dawn K. Smith, and Dora Warren (Centers for Disease Control and Prevention); and Katherine Davenny (National Institute of Drug Abuse).
Acknowledgments
We thank Dr. Julie A. E. Nelson for helpful discussions and review of the manuscript. We also thank the HIV Epidemiologic Research Study staff and participants.
Financial support: Partners/Fenway/Shattuck Center for AIDS Research (NIH P30-AI42851); National Institute on Drug Abuse (R21 DA022148-01); Lifespan/Tufts/Brown Center for AIDS Research (NIH P30 AI42853); Ministry of Education, Culture, Sports, Science, and Technology of Japan (15790350); Centers for Disease Control and Prevention (U64/CCU106795 to fund data collection at Brown University).
Footnotes
Potential conflicts of interest: none reported.
References
- 1.Sherman K, Rouster S, Chung R, Rajicic N. Hepatitis C virus prevalence among patients infected with human immunodeficiency virus: a cross-sectional analysis of the US adult AIDS clinical trials groups. Clin Infect Dis. 2002;34:831–7. doi: 10.1086/339042. [DOI] [PubMed] [Google Scholar]
- 2.Sulkowski M, Thomas D. Hepatitis C in the HIV-infected person. Ann Intern Med. 2003;138:197–207. doi: 10.7326/0003-4819-138-3-200302040-00012. [DOI] [PubMed] [Google Scholar]
- 3.Chung R, Andersen J, Volberding P, et al. Peginterferon alpha-2a plus ribavirin versus interferon alpha-2a plus ribavirin for chronic hepatitis C in HIV-coinfected persons. N Engl J Med. 2004;351:451–9. doi: 10.1056/NEJMoa032653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perez-Olmeda M, Nunez M, Romero M, et al. Pegylated IFN-alpha2b plus ribavirin as therapy for chronic hepatitis C in HIV-infected patients. AIDS. 2003;17:1023–8. doi: 10.1097/00002030-200305020-00011. [DOI] [PubMed] [Google Scholar]
- 5.Torriani F, Rodriguez-Torres M, Rockstroh J, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection in HIV-infected patients. N Engl J Med. 2004;351:438–50. doi: 10.1056/NEJMoa040842. [DOI] [PubMed] [Google Scholar]
- 6.Blackard JT, Kemmer N, Sherman KE. Extrahepatic replication of HCV: insights into clinical manifestations and biological consequences. Hepatology. 2006;44:15–22. doi: 10.1002/hep.21283. [DOI] [PubMed] [Google Scholar]
- 7.Pham T, MacParland S, Mulrooney P, Cooksley H, Naoumov N, Michalak T. Hepatitis C virus persistence after spontaneous or treatment-induced resolution of hepatitis C. J Virol. 2004;78:5867–74. doi: 10.1128/JVI.78.11.5867-5874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radkowski M, Gallegos-Orozco J, Jablonska J, et al. Persistence of hepatitis C virus in patients successfully treated for chronic hepatitis C. Hepatology. 2005;41:106–14. doi: 10.1002/hep.20518. [DOI] [PubMed] [Google Scholar]
- 9.Watkins-Riedel T, Ferenci P, Steindl-Munda P, Gschwantler M, Mueller C, Woegerbauer M. Early prediction of hepatitis C virus (HCV) infection relapse in nonresponders to primary interferon therapy by means of HCV RNA whole-blood analysis. Clin Infect Dis. 2004;39:1754–60. doi: 10.1086/425614. [DOI] [PubMed] [Google Scholar]
- 10.Saleh M, Tibbs C, Koskinas J, et al. Hepatic and extrahepatic hepatitis C virus replication in relation to response to interferon therapy. Hepatology. 1994;20:1399–404. doi: 10.1002/hep.1840200604. [DOI] [PubMed] [Google Scholar]
- 11.Gong G, Lai L, Jiang Y, He Y, Su X. HCV replication in PBMC and its influence on interferon therapy. World J Gasterenterol. 2003;9:291–4. doi: 10.3748/wjg.v9.i2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taliani G, Badolato M, Lecce R, et al. Hepatitis C virus RNA in peripheral blood mononuclear cells: relation with response to interferon treatment. J Med Virol. 1995;47:16–22. doi: 10.1002/jmv.1890470105. [DOI] [PubMed] [Google Scholar]
- 13.Boisvert J, He X, Cheung R, Keeffe E, Wright T, Greenberg H. Quantitative analysis of hepatitis C virus in peripheral blood and liver: replication detected only in liver. J Infect Dis. 2001;184:827–35. doi: 10.1086/323391. [DOI] [PubMed] [Google Scholar]
- 14.Lin L, Fevery J, Yap S. A novel strand-specific RT-PCR for detection of hepatitis C virus negative-strand RNA (replicative intermediate): evidence of absence or very low level of HCV replication in peripheral blood mononuclear cells. J Virol Methods. 2002;100:97–105. doi: 10.1016/s0166-0934(01)00399-8. [DOI] [PubMed] [Google Scholar]
- 15.Lerat H, Berby F, Trabaud MA, et al. Specific detection of hepatitis C virus minus strand RNA in hematopoietic cells. J Clin Invest. 1996;97:845–51. doi: 10.1172/JCI118485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laskus T, Radkowski M, Wang L, Cianciara J, Vargas H, Rakela J. Hepatitis C virus negative strand RNA is not detected in peripheral blood mononuclear cells and viral sequences are identical to those in serum: a case against extrahepatic replication. J Gen Virol. 1997;78:2747–50. doi: 10.1099/0022-1317-78-11-2747. [DOI] [PubMed] [Google Scholar]
- 17.Kao J, Chen P, Lai M, Wang T, Chen D. Positive and negative strand of hepatitis C virus RNA sequences in peripheral blood mononuclear cells in patients with chronic hepatitis C: no correlation with viral genotypes 1b, 2a, and 2b. J Med Virol. 1997;52:270–4. [PubMed] [Google Scholar]
- 18.Roque-Afonso A, Jiang J, Penin F, et al. Nonrandom distribution of hepatitis C virus quasispecies in plasma and peripheral blood mono-nuclear cell subsets. J Virol. 1999;73:9213–21. doi: 10.1128/jvi.73.11.9213-9221.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laskus T, Radkowski M, Wang L, Nowicki M, Rakela J. Uneven distribution of hepatitis C virus quasispecies in tissues from subjects with end-stage liver disease: confounding effect of viral adsorption and mounting evidence for the presence of low-level extrahepatic replication. J Virol. 2000;74:1014–7. doi: 10.1128/jvi.74.2.1014-1017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roque-Afonso A, Ducoulombier D, Di Liberto G, et al. Compartmentalization of hepatitis C virus genotypes between plasma and peripheral blood mononuclear cells. J Virol. 2005;79:6349–57. doi: 10.1128/JVI.79.10.6349-6357.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okuda M, Hino K, Korenaga M, Yamaguchi Y, Katoh Y, Okita K. Differences in hypervariable region 1 quasispecies of hepatitis C virus in human serum, peripheral blood mononuclear cells, and liver. Hepatology. 1999;29:217–22. doi: 10.1002/hep.510290117. [DOI] [PubMed] [Google Scholar]
- 22.Maggi F, Fornai C, Morrica A, et al. Divergent evolution of hepatitis C virus in liver and peripheral blood mononuclear cells of infected patients. J Med Virol. 1999;57:57–63. [PubMed] [Google Scholar]
- 23.Navas S, Martin J, Quiroga J, Castillo I, Carreno V. Genetic diversity and tissue compartmentalization of the hepatitis C virus genome in blood mononuclear cells, liver, and serum from chronic hepatitis C patients. J Virol. 1998;72:1640–6. doi: 10.1128/jvi.72.2.1640-1646.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maggi F, Fornai C, Vatteroni M, et al. Differences in hepatitis C virus quasispecies composition between liver, peripheral blood mononuclear cells and plasma. J Gen Virol. 1997;78:1521–5. doi: 10.1099/0022-1317-78-7-1521. [DOI] [PubMed] [Google Scholar]
- 25.Smith D, Warrne D, Vlahov D, et al. Design and baseline participant characteristics of the Human Immunodeficiency Virus Epidemiology Research (HER) Study: a prospective cohort of human immunodeficiency virus infection in US women. Am J Epidemiol. 1997;146:459–69. doi: 10.1093/oxfordjournals.aje.a009299. [DOI] [PubMed] [Google Scholar]
- 26.Thomas D, Rich J, Schuman P, et al. Multicenter evaluation of hepatitis C RNA levels among female injection drug users. J Infect Dis. 2001;183:973–6. doi: 10.1086/319256. [DOI] [PubMed] [Google Scholar]
- 27.Winters MA, Welles SL, Holodniy M. Hepatitis C virus protease gene diversity in patients coinfected with human immunodeficiency virus. J Virol. 2006;80:4196–9. doi: 10.1128/JVI.80.8.4196-4199.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roque-Afonso A, Robain M, Simoneau D, et al. Influence of CD4 cell counts on the genetic heterogeneity of hepatitis C virus in patients coinfected with human immunodeficiency virus. J Infect Dis. 2002;185:728–33. doi: 10.1086/339297. [DOI] [PubMed] [Google Scholar]
- 29.Blackard J, Smeaton L, Hiasa Y, et al. Detection of hepatitis C virus (HCV) in serum and peripheral blood mononuclear cells of HCV-monoinfected and HIV/HCV-coinfected persons. J Infect Dis. 2005;192:258–65. doi: 10.1086/430949. [DOI] [PubMed] [Google Scholar]
- 30.Torres-Puente M, Bracho M, Jiminez N, Garcia-Robles I, Moya A, Gonzalez-Candelas F. Sampling and repeatability in the evaluation of hepatitis C virus genetic variability. J Gen Virol. 2003;84:2343–50. doi: 10.1099/vir.0.19273-0. [DOI] [PubMed] [Google Scholar]
- 31.Thompson J, Gibson T, Plewniak F, Jeanmougin F, Higgins D. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S, Tamura K, Jakobsen I, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–5. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 33.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 34.Wolinksy S, Korber B, Neumann A, et al. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science. 1996;272:537–42. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]
- 35.Korber B, Myers G. Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res Hum Retroviruses. 1992;8:1549–60. doi: 10.1089/aid.1992.8.1549. [DOI] [PubMed] [Google Scholar]
- 36.Ray S, Wang Y, Laeyendecker O, Ticehurst J, Villano S, Thomas D. Acute hepatitis C virus structural gene sequences as predictors of persistent viremia: hypervariable region 1 as a decoy. J Virol. 1999;73:2938–46. doi: 10.1128/jvi.73.4.2938-2946.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blackard J, Yang Y, Bordoni P, et al. Hepatitis C virus (HCV) diversity in HIV-HCV coinfected subjects initiating highly active antiretroviral therapy. J Infect Dis. 2004;189:1472–81. doi: 10.1086/382959. [DOI] [PubMed] [Google Scholar]
- 38.Laskus T, Radkowski M, Wang L, Vargas H, Rakela J. Search for hepatitis C virus extrahepatic replication sites in patients with acquired immunodeficiency syndrome: specific detection of negative-strand viral RNA in various tissues. Hepatology. 1998;28:1398–401. doi: 10.1002/hep.510280531. [DOI] [PubMed] [Google Scholar]
- 39.Lerat H, Rumin S, Habersetzer F, et al. In vivo tropism of hepatitis C virus genomic sequences in hematopoietic cells: influence of viral load, viral genotype, and cell phenotype. Blood. 1998;91:3841–9. [PubMed] [Google Scholar]
- 40.Goutagny N, Fatmi A, De Ledinghen V, et al. Evidence of viral replication in circulating dendritic cells during hepatitis C virus infection. J Infect Dis. 2003;187:1951–8. doi: 10.1086/375350. [DOI] [PubMed] [Google Scholar]