Abstract
For more than thirty years, the dog has been used as a model for human diseases. Despite efforts made to develop canine embryonic stem cells, success has been elusive. Here, we report the generation of canine induced pluripotent stem cells (ciPSCs) from canine adult fibroblasts, which we accomplished by introducing human OCT4, SOX2, c-MYC, and KLF4. The ciPSCs expressed critical pluripotency markers and showed evidence of silencing the viral vectors and normal karyotypes. Microsatellite analysis indicated that the ciPSCs showed the same profile as the donor fibroblasts but differed from cells taken from other dogs. Under culture conditions favoring differentiation, the ciPSCs could form cell derivatives from the ectoderm, mesoderm, and endoderm. Further, the ciPSCs required leukemia inhibitory factor and basic fibroblast growth factor to survive, proliferate, and maintain pluripotency. Our results demonstrate an efficient method for deriving canine pluripotent stem cells, providing a powerful platform for the development of new models for regenerative medicine, as well as for the study of the onset, progression, and treatment of human and canine genetic diseases.
Introduction
Embryonic stem cells (ESCs) were first isolated from preimplantation mouse embryos by Evans and Kaufman in 1981, and subsequently, ESCs were derived from a variety of species including nonhuman primates, humans, rats, and dogs [1–7]. ESCs have the capacity to renew themselves and to differentiate into all cell types found in adult bodies. Although ESC availability has made possible new kinds of developmental and regenerative medicine studies, tissue rejection and immunocompatibility after transplantation remain as obstacles to their clinical use. Researchers have proposed several alternative methods of reprogramming somatic cells to solve this problem, including somatic cell nuclear transfer into unfertilized oocytes and somatic cell fusion with ESCs to attain pluripotency [8,9]. However, a lack of reliable sources of oocytes and the generation of tetraploid cells, respectively, have made their implementation in humans problematic [10]. Success in deriving induced pluripotent stem cells (iPSCs) using a set of transcription factors—such as OCT3/4, SOX2, KLF4, and c-MYC (Yamanaka factors), or OCT4, SOX2, NANOG, and LIN28—into differentiated somatic cells may address the immune rejection problem [11,12]. Induced PSCs are similar to ESCs in morphology, proliferation, and pluripotency. Successful generation of iPSCs has been reported for mice, humans, rats, monkeys, and pigs [11,13–15]. Although the use of iPSCs in basic research is moving forward, their use as a therapeutic tool remains a challenge, mostly because of the lack of appropriate animal models for testing their efficacy and safety.
For more than thirty years, the dog has provided a valuable model for human diseases, particularly in the study and implementation of cell-based therapy protocols [6]. Over 400 dog breeds show a high prevalence of more complex multigenic diseases [16,17]. Approximately 58% of dog genetic diseases resemble the specific human diseases caused by mutations in the same gene [17,18]. Also, dogs share a variety of biochemical and physiological characteristics with humans; their physiologies, disease presentations, and clinical responses often parallel those of humans better than do those of their rodent counterparts [5,17]. This underscores the dog's importance as a reliable preclinical model for testing the feasibility of regenerative medicine and tissue engineering approaches to treat its own diseases and those of man.
Because of dogs' distinct reproductive physiology and embryonic development pattern, the difficulty of deriving their ESCs has blocked the establishment of the canine model for further regenerative medicine studies. The lack of well-defined methods for maturing and fertilizing canine oocytes in vitro has narrowed the choices for harvesting ESCs from natural canine blastocysts [19–21]. Only 1 group has successfully established a bona fide canine ESC line. The scarcity of published data is likely due to poor understanding of canine preimplantation embryonic development and canine embryo culture conditions [21,22]. Recently, a report on the derivation of induced ESC-like cells described the source of donor cells as embryonic fibroblasts [23] and the evidence demonstrating complete reprogramming to pluripotency in such cells is succinct, making the results—while promising—incomplete. We still need an efficient, safe, well-described method for generating canine iPSCs (ciPSCs).
Here, we report the production of iPSCs from adult canine cells using a method like that described for human and mouse iPSCs [11,24,25]. We systematically show the degree of pluripotency of the generated lines, explore their capacity for stable maintenance, and assay their ability to form embryoid bodies (EBs) and to differentiate into multiple cell lineages. We also noticed that the ciPSCs demonstrated dependency on both leukemia inhibitory factor (LIF) and basic fibroblast growth factor (bFGF) to maintain self-renewal. The ciPSC lines described here reveal similarities and differences between canines and other species and reveal ciPSCs as a unique new tool for future application to, and understanding of, analogous conditions in humans.
Materials and Methods
Derivation of canine fibroblasts and cell culture
Canine testicular fibroblasts (CTFs) were derived from the testicle of a 7-month-old German shorthair pointer from the Small Animal Clinic at Michigan State University (MSU). The testis was minced and incubated in trypsin (Gibco, Carlsbad, CA) at 37°C for 1 h. Then, shredded tissues were spun down, minced again, and subsequently cultured with fibroblast medium (Dulbecco's modified Eagle's medium containing 10% fetal bovine serum) at 37°C with 5% CO2 [24]. We replaced the culture medium every 24 h. All ciPSCs were generated from CTFs older than passage 2.
We maintained ciPSCs on the feeder layer of mitomycin-treated or irradiated mouse embryonic fibroblasts (MEFs) with ciPSC medium, which consisted of Dulbecco's modified Eagle's medium/F-12 (Gibco) supplemented with 15% (v/v) knockout serum (Gibco), 0.1 mM minimal essential medium (MEM) nonessential amino acid solution (Sigma, St. Louis, MO), 1 mM l-glutamine (Invitrogen, Carlsbad, CA), 0.075 mM β-mercaptoethanol, 4 ng/mL human bFGF (Invitrogen), and 10 ng/mL human LIF (Millipore, Billerica, MA). Colonies with compact ES-like cells were mechanically isolated and subcultured onto new MEFs every 4–6 days using glass Pasteur pipettes.
Virus construction and production
We produced and concentrated recombinant OKSIM lentivirus, as previously described [24,25]. Canine fibroblasts were assessed for infection efficiency with recombinant lentivirus using a pSIN-EF1a-YFP reporter gene. We rated lentiviral infection by quantifying the percentage of yellow-fluorescent cells determined to be identical in infectivity to human fibroblasts. Concentrated OKSIM lentivirus was directly titered by infecting canine fibroblasts followed by immunostaining for OCT4 gene product at 72 h. The OKSIM viral titer was ∼3×105 cells/mL, and 0.5 mL (in triplicate) was used to infect 2.5×105 canine cells for iPSC production.
Immunocytochemistry assay
The immunocytochemistry assay protocol was mostly as described in previous reports [24–26]. Supplementary Table S1 (Supplementary Data are available online at www.liebertonline.com/scd) lists details about the primary and secondary antibodies used for some proteins. After washing the cells with phosphate-buffered saline (PBS), we then stained the nuclei by rinsing the cells with PBS containing Hoechst 33342 (1 μg/mL) for 15 min.
RNA extraction and quantitative reverse transcription polymerase chain reaction analysis
RNA was isolated and purified using the NucleoSpin RNA XS Total RNA Isolation Kit (Macherey-Nagle, Bethlehem, PA), following the manufacturer's instructions. We performed the reverse transcription polymerase chain reactions (RT-PCRs) as previously described [24,25]. Supplementary Table S2 lists the primers used.
Bisulfite genome sequencing
Approximately 20,000 cells from ciPSC colonies or CTFs were collected and kept at −80°C until needed. We extracted canine genomic DNA using the ReadyAmp Genomic Kit (Promega, Madison, WI) and conducted bisulfite mutagenesis using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA) according to the manufacturers' instructions. Bisulfited DNA was eluted in 20 μL elution buffer and subjected to 2 rounds of PCR (35 cycles each) with primer pairs for canine OCT4 and NANOG promoters. Primers were designed based on a randomly chosen sequence localized at the OCT4 and NANOG promoters close to the initiators [27–29] (Supplementary Table S3). We verified PCR products on a 2% agarose gel. We ligated PCR products into the pTOPO 10 Vector System (Invitrogen) and randomly chose >10 clones from each cell line to sequence.
Karyotyping analysis
Twenty G-banded metaphase cells were subjected to cytogenetic analysis for each cell line. Cell Line Genetics (Madison, WI) performed standard G-banding karyotype analysis.
Microsatellite assay
We used the following tetranucleotide microsatellite markers, each located on a separate autosome, for genotype analysis: FH2054, FH2165, FH2233, FH2313, and FH2324. We obtained primer sequences for these markers from Mellersh et al. [30], and the allele frequencies, derived from over 1,000 dogs from 28 dog breeds, were obtained from Irion et al. [31]. Amplified fragments were fluorescently labeled with 6-FAM using chimeric primers and a labeled M13 primer [32]. We amplified all markers in 25 μL reactions under the following conditions: 50 mM KCl, 10 mM Tris (pH 8.3 at 20°C), 1.5 mM MgCl2, 100 μM dNTPs, 0.1 μM M13 and reverse primers, 0.01 μM chimeric primer, 10–100 ng DNA, and 0.5 U Taq DNA polymerase (Invitrogen). Reactions were cycled under the following conditions: 1 min, 94°C; 2 min, 59°C; and 3 min, 72°C (for 50 cycles). Amplification was verified by imaging agarose gels on a Typhoon scanner (Amersham Biosciences, Piscataway, NJ), and high-resolution fragment analysis was performed using an ABI PRISM 3130 Genetic Analyzer at the Michigan State University Research Technology Support Facility. We calculated the probability that the samples derived from an unrelated dog genome, by chance, had identical allele sizes with the CTF-derived cell lines, using the allele frequencies obtained from Irion et al. (taking into account the size of the M13 tail for the comparisons) [31]. To produce a conservative probability, we assumed that the allele size between our data and that of Irion et al. could be one repeat unit off, so we used the most frequent allele of the 3 possible alleles (the determined allele size, plus or minus 1 repeat unit) from Irion et al. for each calculation [31].
EB formation
We isolated ciPSC colonies from the MEF and transferred them to ciPSC medium without bFGF or human LIF in 35×10 mm Petri dishes. After 5 days in suspension, we transferred the EBs to tissue culture dishes coated with 0.1% gelatin (Sigma), culturing them using the same medium without growth factors but with 5% fetal bovine serum (Gemini, West Sacramento, CA) and 10% serum replacement. The culture medium for suspension and subsequent spontaneous differentiation was partially changed daily. We cultured the attached EBs in the differentiation media for at least 3 weeks.
Terminal deoxynucleotidyl transferase dUTP nick-end labeling assay
We washed cells with PBS and fixed them in 4% paraformaldehyde for 15 min. We performed terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assays using the In Situ Cell Death Detection Kit (Roche Applied Science, Indianapolis, IN) following manufacturer's instructions. As positive control, the cells were treated with RQ1 DNase (10 IU/mL; Promega). After washing in PBS, we counterstained all nuclei with Hoechst 33342 (1 μg/mL) for 10 min at room temperature.
5-Bromo-2-deoxyuridine incorporation assay
We cultured cells overnight with 30 μg/mL of 5-bromo-2-deoxyuridine (BrdU) before immunostaining. We have described the BrdU incorporation assay protocol in a previously published report [26]. The nuclei were counterstained with Hoechst 33342 (1 μg/mL) for 5 min at room temperature.
Results
Generation of ciPSCs
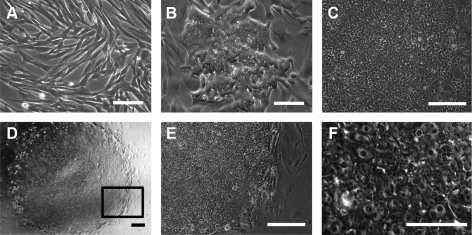
We derived CTFs from canine testicular tissue, as described earlier (Fig. 1A). The infection efficiency of recombinant lentivirus was initially examined in CTFs and canine skin fibroblasts (CSFs) from an old (>10 passages) canine fibroblast line derived from another dog, using a yellow fluorescent protein (YFP) reporter vector. Infection efficiency, shown by YFP, was over 75% in both CTFs and CSFs (Supplementary Fig. S1). The CTFs and CSFs were then infected by lentivirus OKSIM, which had been previously used to generate human iPSC lines [24]. We confirmed successful introduction of OKSIM at 72 h postinfection by immunostaining for OCT4 and SOX2 transgenes; 40% of the target cells carried the virus (Supplementary Fig. S2). To understand the best conditions for reprogramming, we added different concentrations of LIF (1 or 10 ng/mL) or bFGF (0.4 or 4 ng/mL). No ESC-like colonies were observed when using LIF or bFGF alone at 10 days postinfection (Supplementary Fig. S3). However, when both LIF and bFGF were supplied, we observed ESC-like colonies on days 6–8 postinfection (Fig. 1B and Supplementary Fig. S4F). From 2 independent infections, 2 (DI-A1 and DI-A2) and 5 (DI-B1, DI-B2, DI-B3, DI-B4, and DI-B5) cell lines were derived and passed to new MEFs (Fig. 1C, D). Three to four days after the first passage, the morphology of the colonies in all cell lines resembled human ESCs (Fig. 1D–F and Supplementary Fig. S4A–E). All 7 cell lines proliferated at similar rates and required subculturing at 1:6 dilution ratios every 5 days. We chose the DI-B2 iPSC line to characterize growth rate. The ciPSC doubling time at passage 5 (P5) took 27 h, compared with the CTFs at P5, which doubled in 43 h. The ciPSC line DI-B3, which was selected for further studies on growth factor dependency, was maintained and expanded for >20 passages. The other ciPSC lines including DI-B1, 2, 4, and 5 were continuously cultured for over 10 passages. Beyond that ciPSCs require both LIF and bFGF, these results demonstrate that ciPSC can be generated and maintained using a protocol similar to the one used to derive human iPSCs.
FIG. 1.
Induction of canine induced pluripotent stem cells (ciPSCs) from adult canine testicular fibroblasts (CTFs). (A) Input (CTFs); (B) a typical first-observed ciPSC colony on day 6 after lentiviral-mediated transduction; (C) ciPSC colony on day 9 after viral transduction; (D) ciPSC colony (DI-A2) after being passaged on the feeder layer of mouse embryonic fibroblasts (MEFs); the frame represents the location of image in (E); (E) ciPSC colony on MEF with 10×objective; (F) ciPSCs with 40×objective. Scale bar: 100 μm for A and B; 250 μm for C–E; 25 μm for F.
Immunocytochemistry assay
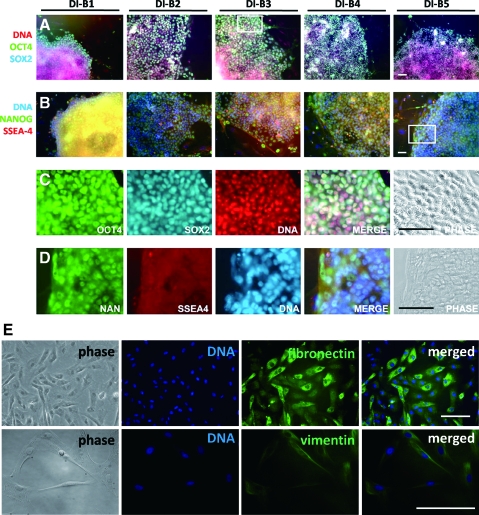
The expression of pluripotency-associated transcription factors OCT4, SOX2, NANOG, and LIN28 was positively displayed in ciPSC colonies; they were also positive for carbohydrate antigens TRA-1-60 and SSEA-4 (Fig. 2A–D and Supplementary Fig. S5). In contrast, the parental CTF cells expressed fibroblast markers, including fibronectin and vimentin, but pluripotency markers were not detected (Fig. 2E and Supplementary Fig. S5).
FIG. 2.
Immunocytochemistry of ciPSCs. (A–D) Immunofluorescent staining of pluripotent cell markers OCT4, SOX2 (A), NANOG, and SSEA-4 (B) in 5 cell lines cultured on MEFs (A and B, from left to right: DI-B1, DI-B2, DI-B3, DI-B4, and DI-B5). Localizations of nuclei were visualized by staining with propidium iodide (A and C) and DAPI (B and D). Localizations of representative cells in C and D were chosen, respectively, from the frames in A and B. (E) CTFs express fibroblast markers, including fibronectin (upper) and vimentin (lower). Scale bars: 100 μm for A–D; 250 μm for E. Color images available online at www.liebertonline.com/scd
Pluripotency gene expression and epigenetics
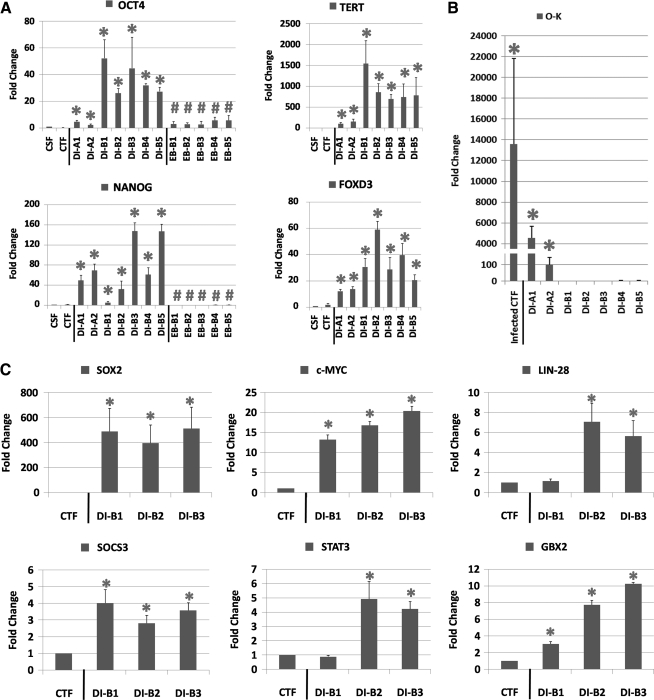
We examined the expression of pluripotency genes in ciPSCs by quantitative RT-PCR (qRT-PCR) assay. Canine-specific pluripotency genes (OCT4, NANOG, TERT, and FOXD3) were robustly expressed in all ciPSC lines but not in CSFs or CTFs (P<0.05; Fig. 3A). However, the levels of OCT4, TERT, and FOXD3 in DI-B1 to B5 were significantly higher than in DI-A1 and DI-A2. Also, fold change of NANOG expression in DI-B1 was comparatively lower than in other ciPSC lines (P<0.05). To confirm the specificity of canine gene amplification, primers for canine OCT4 were used in qRT-PCR for human H9 ESCs; no PCR products were detected (Supplementary Fig. S6). To confirm the silence of viral vectors, we compared transgene expression in ciPSCs to CTFs harvested at 2 days after viral transduction (Fig. 3B). Forward and reverse primers were designed for the intersection between viral OCT4 and KLF4 (O-K). The result indicated that DI-B1 to B5 expressed transgenes negligibly, compared with infected CTFs, which displayed 13,000-fold higher transgene expression (P<0.05). DI-A1 and DI-A2 had higher transgene expression (4,000-fold and 100-fold, respectively) than DI-B1, suggesting that the vectors were not shut down in DI-A1 and DI-A2. We further evaluated the expression of other canine pluripotency genes (including SOX2, c-MYC, LIN-28, SOCS3, STAT3, and GBX2) in CTFs and in DI-B1, DI-B2, and DI-B3 cell lines. Except for LIN-28 and STAT3 in the DI-B1 cell line, we found significantly higher gene expressions in ciPSCs than in CTFs (Fig. 3C).
FIG. 3.
Gene expression of ciPSCs. (A) Quantitative reverse transcription–polymerase chain reaction (qRT-PCR) analysis of relative transcript amounts of pluripotency-associated genes in canine skin fibroblast (CSF), CTF, all 7 ciPSC lines, and all 5 cell lines from embryoid bodies (EBs) (OCT4 and NANOG only). Pluripotency-associated genes include canine OCT4, NANOG, TERT, and FOXD3. Values in the y-axis represent fold changes relative to canine RPL13 expression. The gene expression in CTF and ciPSC lines is relative to that in CSF (*P<0.05), and the expression in EB cells is relative to their ciPSC lines, respectively (#P<0.05). (B) qRT-PCR analysis of relative transcript amounts of the transgene sequence in CSF, CTF, and all 7 ciPSC lines. The transcripts of transgenes are represented by amplification of the intersection between hOCT4 and hKLF4 within the transgene. The y-axis stands for fold changes relative to canine RPL13 expression. (C) qRT-PCR analysis of relative transcript amount of pluripotency-associated genes in CTF, DI-B1, DI-B2, and DI-B3. Values in the y-axis represent fold change relative to canine RPL13 expression (*P<0.05).
We further investigated the CpG dinucleotide methylation status in one canine NANOG regulatory region and 2 OCT4 regulatory regions (regions 1 and 2) by bisulfite genomic sequencing. We selected ciPSCs DI-A1, DI-A2, DI-B1, and DI-B5 to compare with CTFs. Results showed demethylated NANOG promoters in DI-A2 and DI-B5, whereas DI-A1 and DI-B1 maintained the same level as CTFs. However, OCT4 methylation status in ciPSCs maintained at the same level as CTFs or even increased (Supplementary Fig. S7). These results indicate that, at least for the residues investigated, the DNA methylation level for the OCT4 gene does not always correlate with the gene expression observed.
Karyotype analysis
We randomly chose DI-A1, DI-A2, DI-B2, and DI-B5 for karyotype analysis. Results indicated that all ciPSC lines had normal karyotypes (Supplementary Fig. S8). Specifically, ciPSCs with normal karyotypes among all the G-banded ciPSCs had ratios of 17/17 (DI-A1, P4), 14/16 (DI-A2, P3), 8/10 (DI-B2, P4), and 9/10 (DI-B5, P5). Cells with abnormal karyotype were mostly considered a culture artifact.
Microsatellite analysis
To confirm that ciPSC lines derived from the original fibroblast line, we examined 5 canine microsatellites. All ciPSC lines displayed the same alleles as parental CTFs but differed from CSFs with different origins, indicating that ciPSCs and CTFs were equal but different from CSFs in identity (Supplementary Table S4). The probability that CTFs and derived cell lines were not from the same dog was <1.9×10−8.
In vitro differentiation
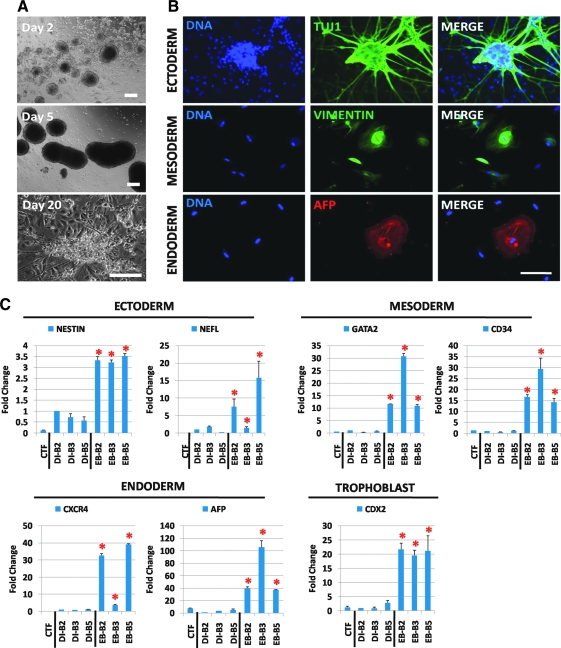
To evaluate the capability of differentiation in vitro, we induced ciPSC lines to differentiate using the EB formation assay (Fig. 4A). Cells derived from plated EBs on day 20 postdifferentiation were analyzed and found to be positive for the presence of cell derivatives from the 3 germ layers, including β-III neuron-specific tubulin (TUJ1) for the ectoderm, vimentin for the mesoderm, and alpha-fetoprotein (AFP) for the endoderm (Fig. 4B) [5,33]. Using qRT-PCR, we also found that differentiated ciPSCs silenced the canine OCT4 and NANOG (P<0.05; Fig. 3A). Differentiation-related genes in EB cells derived from DI-B2, DI-B3, and DI-B5 ciPSCs—that is, ectoderm (NESTIN and NEFL), mesoderm (CD34 and GATA2), and endoderm (CXCR4 and AFP)—were upregulated (P<0.05; Fig. 4C). Interestingly, we observed large multinuclear cells resembling giant cells from the trophectoderm in differentiated cells (Supplementary Fig. S9). We therefore evaluated the expression of trophoblast marker CDX2, which was highly expressed in EB cells but not in the original fibroblasts or undifferentiated ciPSCs (P<0.05; Fig. 4C). These results demonstrate that the vast majority of our ciPSC lines could differentiate into the 3 germ layers and express lineage-specific markers.
FIG. 4.
Differentiation of ciPSCs into EBs. (A) The morphology of floating and attached EBs. Pictures represent the EBs on days 2, 5, and 20 after isolation of ciPSC colonies for EB formation culture. (B) Ectoderm, mesoderm, and endoderm cell derivatives are marked by β-III neuron-specific tubulin (TUJ1), vimentin, and alpha-fetoprotein (AFP), respectively. (C) qRT-PCR analysis of relative transcript amounts of differentiation genes in CTF; the 3 ciPSC lines DI-B2, DI-B3, and DI-B5; and the EBs from these 3 ciPSC lines. Differentiation genes include NESTIN and NEFL (representing ectoderm and CD34), GATA2 (representing mesoderm and CXCR4), AFP (representing endoderm), and CDX2 (representing trophoblast cells). Values in the y-axis represent fold changes relative to canine RPL13 expression. Scale bar: 250 μm for A and B; (*P<0.05). Color images available online at www.liebertonline.com/scd
Significant efforts were made to obtain teratomas from these ciPSCs, to no avail. Under the same conditions, our laboratory has been able to derive teratomas from human iPSCs and ESCs [24,25]. Our observation is in agreement with that reported for canine ESCs [6,21,22,34]. It is possible that the models used for teratoma formation, that is, injection into immunodeficient mice, are not suitable for dog cells. Generation of chimeric dogs using ciPSCs will ultimately help elucidate the developmental potential of these cells.
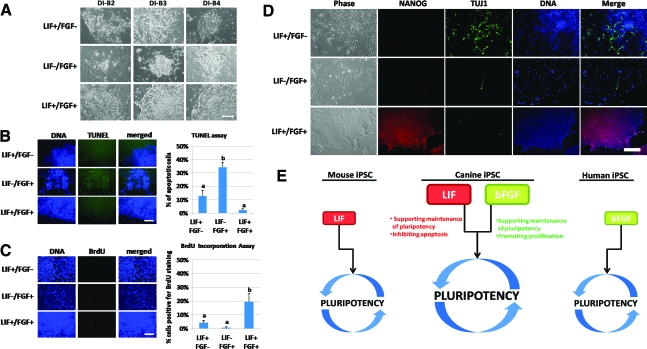
LIF and bFGF dependency
We examined the dependency of growth factors during ciPSC maintenance and found that, when LIF or bFGF were independently withdrawn from the culture medium, ciPSCs did not maintain their undifferentiated morphology (P<0.05; Fig. 5A and Supplementary Fig. S10). To investigate the role of LIF and bFGF in maintaining self-renewal, we cultured ciPSCs on Matrigel-coated plates (Invitrogen) with MEF-conditioned ciPSC media supplemented with only LIF (LIF+/FGF−) or bFGF (LIF−/FGF+) or both (LIF+/FGF+). TUNEL assays demonstrated that no difference existed in the percentage of apoptotic cells in the LIF+/FGF− and LIF+/FGF+ treatments, whereas the percentage in the LIF−/FGF+ cells was significantly higher (P<0.05; Fig. 5B). Using BrdU incorporation assay, we also determined that LIF+/FGF+ ciPSCs exhibited the highest proliferation rates (P<0.05; Fig. 5C). To test the effects of LIF and bFGF on pluripotency maintenance—measured by NANOG expression levels—we cultured ciPSCs for 7 days and immunostained them (Fig. 5D). Results indicated that removing either LIF or bFGF is sufficient to lose the pluripotency marker NANOG, suggesting that ciPSCs need both LIF and bFGF to maintain self-renewal. Our data indicate that withdrawing LIF also triggers signs of apoptosis, and bFGF is associated with proliferation of undifferentiated ciPSCs (Fig. 5E).
FIG. 5.
Role of leukemia inhibitory factor (LIF) or basic fibroblast growth factor (bFGF) in survival, proliferation, and pluripotency maintenance of ciPSCs. (A) Morphology of ciPSCs from cell lines DI-B2, DI-B3, and DI-B4 on day 6 without passaging when cultured with human LIF only (LIF+/FGF−), bFGF only (LIF−/FGF+), and both human LIF and bFGF (LIF+/FGF+). (B) Terminal deoxynucleotidyl transferase dUTP nick-end labeling assay in ciPSCs when cultured with LIF+/FGF−, LIF−/FGF+, or LIF+/FGF+ for 4 days. Quantification results were analyzed by PROC GLM from SAS. Values in y-axis represent the percentage of apoptotic cells among the total cells. (C) 5-Bromo-2-deoxyuridine (BrdU) incorporation assay for ciPSCs cultured with supplement of LIF+/FGF−, LIF−/FGF+, or LIF+/FGF+ for 4 days. BrdU+ cells were counted as the cells with de novo synthesized DNA. The quantification results were analyzed by PROC GLM from SAS. Values in y-axis represent the percentage of BrdU+ cells among the total cells. (D) Immunofluorescent staining of pluripotency marker NANOG and differentiation marker TUJ1 in ciPSCs cultured for 7 days with LIF+/FGF−, LIF−/FGF+, or LIF+/FGF+. (E) The potential functions of LIF and bFGF during pluripotency maintenance of ciPSCs. Withdrawal of either LIF or bFGF, which resembles mouse or human ESC culture conditions, causes spontaneous differentiation and cell death or slowdown of proliferation. Pluripotency of ciPSCs can be maintained with both LIF and bFGF present in the culture medium. Scale bar: 250 μm. Different letters (a and b) indicate P<0.05. Color images available online at www.liebertonline.com/scd
Discussion
This study demonstrated that canine somatic cells isolated from an adult animal can be dedifferentiated into pluripotent cells. Following the strategy described for humans, we successfully induced fibroblasts to become pluripotent cells by transduction of 4 transcription factors—OCT4, KLF4, SOX2, and c-MYC (OKSIM) [24,25]. We successfully expanded and characterized 7 ciPSC lines: DI-A1, DI-A2, and DI-B1 to B5. Like human and mouse ESCs, the proliferation of ciPSCs required coculturing with MEFs [11,12]. Surprisingly, the generation of ciPSCs required the presence of both LIF and bFGF. We also found that ciPSCs, like their human counterparts, expressed many pluripotency-associated factors—including OCT4, SOX2, NANOG, TRA-1-60, TERT, FOXD3, and SSEA-4 [1,5,35]—while silencing the OKSIM transgene in most ciPSC lines.
The cell line used to derive our ciPSCs, CTF, was isolated from the testicle of an adult dog. Therefore, in an effort to rule out the possibility that the original cells were already pluripotent, we compared the gene expression profile of a set of pluripotency-associated genes with that of another canine cell line isolated from the skin of a different animal (CSF). At the time of these experiments, the CSF line was >10 passages old. Our qRT-PCR results showed that the expression of pluripotency genes in CTFs was negligible and as low as in CSFs. Further, the morphology of CTFs had all the characteristics of a typical fibroblast, consistent with the expression of the proteins fibronectin and vimentin. Although we cannot completely rule out the possible presence of a germline-derived cell within the culture of CTFs, our results indicate that, at the time of OKSIM infection, the cells were not pluripotent and were most likely stromal fibroblasts.
We found that the DI-A1 and DI-A2 ciPSC lines expressed lower levels of NANOG than the other ciPSC lines. This could be due to the OKSIM transgene remaining expressed, indicating incomplete reprogramming [36]. We also considered failure to derive EBs in these 2 lines as evidence of incomplete reprogramming [37].
At present, there is no report on the methylation status of canine pluripotency genes. Our bisulfite genome sequencing showed that the NANOG promoter was demethylated in the DI-A2 and DI-B5 cell lines. However, the methylation status of OCT4 was similar in CTFs and ciPSCs or even more methylated in ciPSCs. Interestingly, our results were similar to data recently published, suggesting that murine iPSCs maintained methylation signature characteristics similar to their differentiated donor cells in OCT4 and NANOG regulatory regions [38]. Although a more comprehensive epigenetic analysis for ciPSCs and CTFs is needed, our results suggest that the epigenetic status of ciPSCs may be similar but not identical to the donor fibroblasts and that, although the epigenetic memory of donor fibroblasts remains intact in some residues, it may not alter the overall characteristics of the ciPSCs derived from them. Additional regulatory factors enhancing epigenetic reprogramming might be necessary to help optimize the current reprogramming system, such as the use of microRNAs and small molecules [39–41].
Differentiation potential is one feature critical to determine the utility of pluripotent stem cells for regenerative medicine. Immunocytochemical and qRT-PCR analyses of EBs from the DI-B1 to B5 ciPSCs found significantly increased expressions of markers for cell derivatives of the 3 germ layers and significantly downregulated pluripotency gene expression. Also noteworthy, cells appeared, resembling trophectoderm cells, with upregulated expression of trophoblast marker CDX2, a feature similar to that reported in pig iPSCs [13]. Why porcine and canine pluripotent cells produce cells with features of extra-embryonic tissues, whereas human and mouse cells do not, remains unresolved.
To understand the requirement of growth factors, we attempted to culture ciPSCs with media used for mouse or human ESCs or iPSCs [11,12]. Unlike mouse or human ESCs, which required LIF or bFGF, respectively, for survival, removing LIF or bFGF caused, respectively, the loss of pluripotency markers and apoptosis or the loss of pluripotency markers and the slowdown of proliferation (Fig. 5E). The role of LIF in self-renewal maintenance was widely reported in the mouse ESCs [42]. In the presence of LIF receptors, LIF supports pluripotency by activating the Janus kinase/signal transducer and activator of transcription 3 pathway [42]. In dogs, LIF receptor was reportedly expressed in kidney cells; these canine cells responded to human LIF by further activating the Janus kinase/signal transducer and activator of transcription 3 pathway [5,42,43]. The requirement of LIF for ciPSC culture also agrees with the culture conditions reported for canine ESCs [5,21]. Interestingly, we noticed that absence of LIF triggers severe apoptosis. Previous reports have indicated an antiapoptotic role for LIF when culturing primordial germ cells, oligodendrocytes, and cardiomyocytes, but the mechanism governing this was not yet understood [44–46]. Human ESCs, recognized as pluripotent cells in the epiblast stage, and mouse epiblast stem cells reportedly depend on bFGF but do not react with LIF [47,48]. We speculate that bFGF may act in ciPSCs through similar signaling pathways, that is, stimulating MEFs to synthesize activin A—which, in turn, activates Smad2/3 and promotes NANOG expression—and activating the FGF/ERK pathway, thus promoting proliferation [42,47]. Naive mouse ESCs are described as comparable to cells from the blastocyst inner cell mass (ICM) [42] and are LIF/STAT3 pathway dependent. As ciPSCs present dual-factor dependency, it will be necessary to determine the position of ciPSCs in the “pluripotency map” and to clarify their apparent ICM/epiblast concomitant state. A better understanding of ciPSC pluripotency regulation may enhance our understanding of the molecular mechanisms responsible for the transition from ICM to epiblast cells.
The physiologies, anatomies, disease presentations, and clinical responses of dogs and humans are very similar, making the dog a very promising model for human disease research [6]. Among ∼400 known hereditary canine diseases, over half have equivalent human diseases, including retinal diseases, epilepsy, narcolepsy, cardiomyopathies, muscular dystrophy, and malignant tumors such as prostate cancer [6,49]. In terms of stem cell kinetics—for example, hematopoietic stem cells—and responsiveness to cytokines, the dogs are more biologically comparable with humans than mice, making it the most commonly used species for early transplantation research in human regenerative medicine [6]. However, until now, approaches that involve deriving natural canine pluripotent stem cells have been poorly explored. The successful establishment of a robust ciPSC derivation and culture system offers a novel template for human regenerative medicine studies. It will help us to understand and treat human diseases, including those of genetic origin. In addition, provided that these ciPSCs are germline competent, chimeric animals carrying specific gene mutations will help understand complex diseases and enable the development of new gene and cell therapy strategies [22]. The unexpected finding of dual-growth-factor dependency in ciPSCs provides an opportunity to understand mechanisms of stem self-renewal and maintenance of pluripotency.
Supplementary Material
Acknowledgments
The authors appreciate the support from the MSU Foundation, MSU's vice president of research, and the Michigan Agricultural Experiment Station as well as the Naylor Family Foundation. This work was supported in part by grant R01GM095347 from the National Institute of General Medical Sciences to J.G.K.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Thomson JA. Itskovitz-Eldor J. Shapiro SS. Waknitz MA. Swiergiel JJ. Marshall VS. Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomson JA. Kalishman J. Golos TG. Durning M. Harris CP. Becker RA. Hearn JP. Isolation of a primate embryonic stem cell line. Proc Natl Acad Sci U S A. 1995;92:7844–7848. doi: 10.1073/pnas.92.17.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iannaccone PM. Taborn GU. Garton RL. Caplice MD. Brenin DR. Pluripotent embryonic stem cells from the rat are capable of producing chimeras. Dev Biol. 1994;163:288–292. doi: 10.1006/dbio.1994.1146. [DOI] [PubMed] [Google Scholar]
- 5.Vaags AK. Rosic-Kablar S. Gartley CJ. Zheng YZ. Chesney A. Villagomez DA. Kruth SA. Hough MR. Derivation and characterization of canine embryonic stem cell lines with in vitro and in vivo differentiation potential. Stem Cells. 2009;27:329–340. doi: 10.1634/stemcells.2008-0433. [DOI] [PubMed] [Google Scholar]
- 6.Schneider MR. Wolf E. Braun J. Kolb HJ. Adler H. Canine embryo-derived stem cells and models for human diseases. Hum Mol Genet. 2008;17:R42–R47. doi: 10.1093/hmg/ddn078. [DOI] [PubMed] [Google Scholar]
- 7.Evans MJ. Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 8.Tada M. Takahama Y. Abe K. Nakatsuji N. Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol. 2001;11:1553–1558. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- 9.Wilmut I. Schnieke AE. McWhir J. Kind AJ. Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 10.Hall VJ. Stojkovic P. Stojkovic M. Using therapeutic cloning to fight human disease: a conundrum or reality? Stem Cells. 2006;24:1628–1637. doi: 10.1634/stemcells.2005-0592. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi K. Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 12.Yu J. Vodyanik MA. Smuga-Otto K. Antosiewicz-Bourget J. Frane JL. Tian S. Nie J. Jonsdottir GA. Ruotti V, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 13.Ezashi T. Telugu BP. Alexenko AP. Sachdev S. Sinha S. Roberts RM. Derivation of induced pluripotent stem cells from pig somatic cells. Proc Natl Acad Sci U S A. 2009;106:10993–10998. doi: 10.1073/pnas.0905284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao J. Cui C. Chen S. Ren J. Chen J. Gao Y. Li H. Jia N. Cheng L. Xiao H. Xiao L. Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell. 2009;4:11–15. doi: 10.1016/j.stem.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 15.Liu H. Zhu F. Yong J. Zhang P. Hou P. Li H. Jiang W. Cai J. Liu M, et al. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3:587–590. doi: 10.1016/j.stem.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 16.Lindblad-Toh K. Wade CM. Mikkelsen TS. Karlsson EK. Jaffe DB. Kamal M. Clamp M. Chang JL. Kulbokas EJ, 3rd, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 17.Garibal J. Hollville E. Bell AI. Kelly GL. Renouf B. Kawaguchi Y. Rickinson AB. Wiels J. Truncated form of the Epstein-Barr virus protein EBNA-LP protects against caspase-dependent apoptosis by inhibiting protein phosphatase 2A. J Virol. 2007;81:7598–7607. doi: 10.1128/JVI.02435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ostrander EA. Galibert F. Patterson DF. Canine genetics comes of age. Trends Genet. 2000;16:117–124. doi: 10.1016/s0168-9525(99)01958-7. [DOI] [PubMed] [Google Scholar]
- 19.Yamada S. Shimazu Y. Kawano Y. Nakazawa M. Naito K. Toyoda Y. In vitro maturation and fertilization of preovulatory dog oocytes. J Reprod Fertil Suppl. 1993;47:227–229. [PubMed] [Google Scholar]
- 20.Yamada S. Shimazu Y. Kawaji H. Nakazawa M. Naito K. Toyoda Y. Maturation, fertilization, and development of dog oocytes in vitro. Biol Reprod. 1992;46:853–858. doi: 10.1095/biolreprod46.5.853. [DOI] [PubMed] [Google Scholar]
- 21.Hayes B. Fagerlie SR. Ramakrishnan A. Baran S. Harkey M. Graf L. Bar M. Bendoraite A. Tewari M. Torok-Storb B. Derivation, characterization, and in vitro differentiation of canine embryonic stem cells. Stem Cells. 2008;26:465–473. doi: 10.1634/stemcells.2007-0640. [DOI] [PubMed] [Google Scholar]
- 22.Schneider MR. Wolf E. Braun J. Kolb HJ. Adler H. Canine embryonic stem cells: state of the art. Theriogenology. 2010;74:492–497. doi: 10.1016/j.theriogenology.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 23.Shimada H. Nakada A. Hashimoto Y. Shigeno K. Shionoya Y. Nakamura T. Generation of canine induced pluripotent stem cells by retroviral transduction and chemical inhibitors. Mol Reprod Dev. 77:2. doi: 10.1002/mrd.21117. [DOI] [PubMed] [Google Scholar]
- 24.Ross PJ. Suhr S. Rodriguez RM. Chang EA. Wang K. Siripattarapravat K. Ko T. Cibelli JB. Human induced pluripotent stem cells produced under xeno-free conditions. Stem Cells Dev. 2010;19:1221–1229. doi: 10.1089/scd.2009.0459. [DOI] [PubMed] [Google Scholar]
- 25.Suhr ST. Chang EA. Rodriguez RM. Wang K. Ross PJ. Beyhan Z. Murthy S. Cibelli JB. Telomere dynamics in human cells reprogrammed to pluripotency. PLoS ONE. 2009;4:e8124. doi: 10.1371/journal.pone.0008124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang EA. Beyhan Z. Yoo MS. Siripattarapravat K. Ko T. Lookingland KJ. Madhukar BV. Cibelli JB. Increased cellular turnover in response to fluoxetine in neuronal precursors derived from human embryonic stem cells. Int J Dev Biol. 2010;54:707–715. doi: 10.1387/ijdb.092851ec. [DOI] [PubMed] [Google Scholar]
- 27.Wang K. Chen Y. Chang EA. Knott JG. Cibelli JB. Dynamic epigenetic regulation of the Oct4 and Nanog regulatory regions during neural differentiation in rhesus nuclear transfer embryonic stem cells. Cloning Stem Cells. 2009;11:483–496. doi: 10.1089/clo.2009.0019. [DOI] [PubMed] [Google Scholar]
- 28.Shiota K. DNA methylation profiles of CpG islands for cellular differentiation and development in mammals. Cytogenet Genome Res. 2004;105:325–334. doi: 10.1159/000078205. [DOI] [PubMed] [Google Scholar]
- 29.Jaenisch R. Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 30.Mellersh CS. Langston AA. Acland GM. Fleming MA. Ray K. Wiegand NA. Francisco LV. Gibbs M. Aguirre GD. Ostrander EA. A linkage map of the canine genome. Genomics. 1997;46:326–336. doi: 10.1006/geno.1997.5098. [DOI] [PubMed] [Google Scholar]
- 31.Irion DN. Schaffer AL. Famula TR. Eggleston ML. Hughes SS. Pedersen NC. Analysis of genetic variation in 28 dog breed populations with 100 microsatellite markers. J Hered. 2003;94:81–87. doi: 10.1093/jhered/esg004. [DOI] [PubMed] [Google Scholar]
- 32.Neilan BA. Wilton AN. Jacobs D. A universal procedure for primer labelling of amplicons. Nucleic Acids Res. 1997;25:2938–2939. doi: 10.1093/nar/25.14.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi K. Tanabe K. Ohnuki M. Narita M. Ichisaka T. Tomoda K. Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 34.Hatoya S. Torii R. Kondo Y. Okuno T. Kobayashi K. Wijewardana V. Kawate N. Tamada H. Sawada T, et al. Isolation and characterization of embryonic stem-like cells from canine blastocysts. Mol Reprod Dev. 2006;73:298–305. doi: 10.1002/mrd.20392. [DOI] [PubMed] [Google Scholar]
- 35.Ginis I. Luo Y. Miura T. Thies S. Brandenberger R. Gerecht-Nir S. Amit M. Hoke A. Carpenter MK. Itskovitz-Eldor J. Rao MS. Differences between human and mouse embryonic stem cells. Dev Biol. 2004;269:360–380. doi: 10.1016/j.ydbio.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 36.Chan EM. Ratanasirintrawoot S. Park IH. Manos PD. Loh YH. Huo H. Miller JD. Hartung O. Rho J. Ince TA. Daley GQ. Schlaeger TM. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 37.Sommer CA. Sommer AG. Longmire TA. Christodoulou C. Thomas DD. Gostissa M. Alt FW. Murphy GJ. Kotton DN. Mostoslavsky G. Excision of reprogramming transgenes improves the differentiation potential of iPS cells generated with a single excisable vector. Stem Cells. 28:64–74. doi: 10.1002/stem.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim K. Doi A. Wen B. Ng K. Zhao R. Cahan P. Kim J. Aryee MJ. Ji H, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ieda M. Fu JD. Delgado-Olguin P. Vedantham V. Hayashi Y. Bruneau BG. Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huangfu D. Osafune K. Maehr R. Guo W. Eijkelenboom A. Chen S. Muhlestein W. Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 41.Huangfu D. Maehr R. Guo W. Eijkelenboom A. Snitow M. Chen AE. Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okita K. Yamanaka S. Intracellular signaling pathways regulating pluripotency of embryonic stem cells. Curr Stem Cell Res Ther. 2006;1:103–111. doi: 10.2174/157488806775269061. [DOI] [PubMed] [Google Scholar]
- 43.Buk DM. Waibel M. Braig C. Martens AS. Heinrich PC. Graeve L. Polarity and lipid raft association of the components of the ciliary neurotrophic factor receptor complex in Madin-Darby canine kidney cells. J Cell Sci. 2004;117:2063–2075. doi: 10.1242/jcs.01049. [DOI] [PubMed] [Google Scholar]
- 44.Zou Y. Takano H. Mizukami M. Akazawa H. Qin Y. Toko H. Sakamoto M. Minamino T. Nagai T. Komuro I. Leukemia inhibitory factor enhances survival of cardiomyocytes and induces regeneration of myocardium after myocardial infarction. Circulation. 2003;108:748–753. doi: 10.1161/01.CIR.0000081773.76337.44. [DOI] [PubMed] [Google Scholar]
- 45.Kerr BJ. Patterson PH. Leukemia inhibitory factor promotes oligodendrocyte survival after spinal cord injury. Glia. 2005;51:73–79. doi: 10.1002/glia.20177. [DOI] [PubMed] [Google Scholar]
- 46.Pesce M. Farrace MG. Piacentini M. Dolci S. De Felici M. Stem cell factor and leukemia inhibitory factor promote primordial germ cell survival by suppressing programmed cell death (apoptosis) Development. 1993;118:1089–1094. doi: 10.1242/dev.118.4.1089. [DOI] [PubMed] [Google Scholar]
- 47.Greber B. Wu G. Bernemann C. Joo JY. Han DW. Ko K. Tapia N. Sabour D. Sterneckert J. Tesar P. Scholer HR. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell. 6:215–226. doi: 10.1016/j.stem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 48.Hanna J. Cheng AW. Saha K. Kim J. Lengner CJ. Soldner F. Cassady JP. Muffat J. Carey BW. Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci U S A. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Starkey MP. Scase TJ. Mellersh CS. Murphy S. Dogs really are man's best friend—canine genomics has applications in veterinary and human medicine! Brief Funct Genomic Proteomic. 2005;4:112–128. doi: 10.1093/bfgp/4.2.112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.