Abstract
Anti-polysaccharide antibody responses in mice are often oligoclonal, and the mechanisms involved in Ag-specific clone production and selection remain poorly understood. We evaluated the relative contribution of DH germline content versus N nucleotide addition in a classic oligoclonal, T-independent antibody response [α1→3 Dextran (DEX)] by challenging adult TdT sufficient (TdT+/+) and TdT deficient (TdT−/−) gene-targeted mice limited to the use of a single DH gene segment (D-limited mice) with Enterobacter cloacae. D-limited mice achieved anti-DEX-specific antibodies at levels broadly comparable to that of wild type (WT) BALB/c mice. Sequence analysis of CDR-H3 intervals obtained by PCR amplification of VH domain DNA from DEX-specific plasmablasts revealed the near universal presence of an aspartic acid residue (D99) at the V-D junction irrespective of the composition of the DH locus. While WT mice were able to use germline DH (DQ52, DSP or DST) gene segment sequence, TdT activity or both to produce D99, all three of the D-limited mouse strains relied exclusively on N addition. And, in the absence of TdT, D-limited mice failed to produce a DEX response. Coupled with previous studies demonstrating a reduced response to DEX in TdT−/− mice with a WT DH locus, we conclude that in the case of the anti-DEX repertoire, which uses a short CDR-H3, the anti-DEX response relies more intensely on sequences created by postnatal N nucleotide addition than on the germline sequence of the DH.
Keywords: DH reading frame, CDR3, antibodies, repertoire development, J558 idiotype
Introduction
The ability of vaccines to elicit an effective defense against pathogens such as Haemophilus influenzae type b, Streptococcus pneumoniae and Neisseria meningitidis depends largely on the induction of protective antibodies to capsular polysaccharides or T-independent Ags associated with these microorganisms. (1–3). This protection can depend on the ontogenetically regulated production of antibodies with Ag-binding sites of specific sequence or structure that bind to critical epitopes on the eliciting Ag. For example, Streptococcus pneumoniae protection is best offered by antibodies produced by N-region deficient B-1a cells arising just after birth that bind to phosphorylcholine and bear the T15 idiotype (4). A better understanding of the mechanisms involved in the generation, ontogeny, production and selection of B cells bearing antibodies with antigen-binding sites specific to polysaccharides and T-independent Ags is thus needed in order to optimize the design and the ontogenetic timing of vaccines directed against pathogens bearing these types of immunodominant antigens.
Mice respond to many polysaccharides in a T-cell independent manner with the rapid production of an oligoclonal antibody response consisting primarily of IgM and IgG3 antibodies (5–7). The oligoclonal nature of this type of response facilitates the examination of factors that are important for the generation of polysaccharide-specific antibody diversity.
The Ag-binding site of an antibody, as classically defined, is created by the juxtaposition of three hypervariable complementarity determining regions (CDR) from the heavy chain and three CDRs from the light chain (8). In VH-restricted mice, the variability introduced by the third CDR of the heavy chain (CDR-H3) has been shown to be sufficient for the generation of the primary antibody specificities to proteins, but surprisingly not to selected polysaccharides (9). CDR-H3 is created by the combinatorial rearrangement of Variable (VH), Diversity (DH), and Joining (JH) segments in conjunction with the variable insertion of random N nucleotides and the variable loss or P nucleotide gain of terminal nucleotide sequence (10). The DH gene segment in its entirety is a major component of CDR-H3 and both ends of the DH can undergo the extensive loss or gain of sequence. Coupled with its enormous potential for sequence, and thus structural, variation, its central position at the core of the classic antigen binding site permits the amino acids contributed by CDR-H3 to often play a critical role in the recognition and binding of the antigen to the antibody (9, 11).
α 1→3 Dextran (DEX) is a branched polymer of α 1→3 glucose sugar moieties displaying epitopes that are expressed by a variety of organisms such as Enterobacter cloacae (12), Histoplasma capsulatum yeast cell wall (13) and Aspergillus fumigatus (Dizon B.L. and J.F. Kearney, unpublished observations). The antibody response of adult normal BALB/c mice to DEX is T-cell-independent, oligoclonal and consists entirely of antibodies bearing the λ1 light chain (14). The majority of anti-DEX antibodies express either J558 or M104E idiotypic determinants (14–16). Amino acid sequence analysis of DEX-binding hybridoma proteins has shown VH region homology, with diversity clustered in that portion of CDR-H3 contributed by the DH and N addition. Unsurprisingly, this region contributes heavily to the individual idiotype identity expressed by distinct B cell clones (17). The heavy chains of the prototypic J558 and M104E clones use identical J558.3 VH and JH1 gene segments, but differ by two amino acids located within CDR-H3 (R100 and Y101 for J558 and Y100 and D101 for M104E) (18). Ontogenetic studies of the BALB/c anti-DEX response show that while the M104E idiotype predominates in newborn mice, almost 70% of adult anti-DEX antibodies express the J558 idiotype, which requires N addition for its production (18, 19). Despite this dependence on TdT, the J558 clone has the same short length CDR-H3 as all other anti-DEX clones reported (18). This is not surprising as the length of CDR-H3 in antibodies to a number of polysaccharide antigens and other T-independent antigens is often strictly maintained (20, 21).
To assess the role of DH sequence and content on the antibody response to α 1→3 Dextran (DEX), we challenged mice limited to the usage of one DH gene segment (referred to as D-limited mice in this manuscript) with Enterobacter cloacae. One strain of D-limited mice uses only the DFL16.1 gene segment that preferentially produces antibodies enriched for tyrosine, serine and glycine in CDR-H3 (ΔD-DFL) (22). A second strain (ΔD-DµFS) (23) preferentially produces antibodies enriched for valine and threonine, which are encoded by DFL16.1 reading frame 2, but it can also use reading frame 1 to produce antibodies enriched with DH encoded tyrosine, serine and glycine, albeit with CDR-H3s that tend to be shorter than those from ΔD-DFL. The third strain (ΔD-DiD) (24) preferentially introduces arginine, serine, asparagine and histidine into CDR-H3. In these mice, the inclusion of a normal range of amino acids requires N addition or rearrangement by inversion, which is a rare event.
We found that the anti-DEX antibody response in the two strains of mice most capable of producing CDR-H3s of wild-type amino acid composition was robust. The loss of amino acids normally contributed by the DH gene segments that had been deleted in the D-limited mice appeared to be compensated by the activity of TdT, which was required for D-limited mice to encode an aspartic acid residue at position 99 (D99) that is conserved in the CDR-H3 regions of anti-DEX antibodies. However, when challenged with Enterobacter cloacae ΔD-DµFS mice can elicit a superior J558 Id expressing anti-DEX response when compared to other D-limited and WT mice. Taken together, these results show the extreme selectivity exerted by this classic T-independent Ag on the antibody repertoire, with specific amino acids in CDR-H3 proven to be not only sufficient, but necessary to permit a normal response.
Materials and Methods
Mice
BALB/c mice homozygous for the ΔD-DFL, ΔD-DiD and ΔD-DµFS DH loci were generated as previously described (22–24). Wild type BALB/c mice were purchased from Jackson Laboratories (Bar Harbor, ME). All mice were bred and housed within the specific pathogen-free facility at The University of Alabama at Birmingham and used at 8 to 12 weeks of age according to protocols approved by the University of Alabama at Birmingham Institutional Animal Care and Use Committee.
Immunization and serum analysis
Mice were immunized i.v. via the tail vein with 108 DEX-expressing paraformaldehyde-fixed Enterobacter cloacae (strain MK7). ELISA was used to quantify serum antibodies by methods previously described (12). Plates were coated with either DEX at 2 µg /mL (Dr. Slodki, USDA, Peoria, IL) or the anti-J558 idiotype EB3–7 at 1 µg /mL (25). Plates were then washed 3 times with PBS and blocked with 1% BSA solution in PBS for at least 1 hour. Serum samples diluted in blocking buffer were then added and incubated for 2 hours at 37° C. After washing, bound serum immunoglobulins were developed with alkaline-phosphatase conjugated anti-IgM for DEX-coated plates or anti-λ for EB3–7 coated plates (Southern Biotechnology Associates, Birmingham, AL). Anti-DEX MOPC 104E antibody (Sigma-Aldrich) was used to generate standard curves.
Flow Cytometry and Cell Sorting
Single cell suspensions of spleen cells or peritoneal cells were treated with Ab 93 (26) to block Fc receptors. DEX labeled with Alexa 488 and Alexa 647 anti-λ JC5-1 were used to label DEX+ λ+ plasmablasts (B220 +/− Syndecan-1+) which were sorted from mice 7 days post-immunization with DEX-expressing Enterobacter cloacae. Cells were sorted into RPMI 1640 media supplemented with 10% fetal calf serum and the collection tubes were maintained at 4°C throughout the sort. The sorted cells were then centrifuged and digestion buffer (50 mM Tris-HCl, 100 mM EDTA, 100mM NaCl, 1% SDS, pH 8.0) + Proteinase K (0.5mg/mL) was added to the cell pellet and incubated overnight at 50°C. DNA was precipitated using isopropanol and the DNA pellet was washed with 70% ethanol, air-dried and resuspended in molecular grade water. All samples were analyzed using FACSAria cell sorter (BD Biosciences, San Jose, CA). All antibodies were purchased from BD Biosciences except for Ab 93 and JC5-1 that were developed in our laboratory.
Polymerase Chain Reaction
V(D)J rearrangements were amplified by PCR from genomic DNA extracted from sorted plasmablasts using primers specific for gene segments J558.3 VH 5’-AGCTGCAACAATCTGGACCT-3’ and JH1 5’-CCCCAGACATCGAAGTACCA-3’. PCR conditions were 95° for 3 min then 95° for 1 min, 58° for 1 min and 72° for 1 min for 35 cycles then 72° for 7 min.
Cloning and Sequencing
PCR products were cloned into TOPO-TA vector and transformed into TOP10 competent bacteria (Invitrogen, Carlsbad, CA). Templiphi Amplification Kits were used to isolate plasmid DNA from colonies (Amersham Biosciences, Piscataway, NJ). Plasmid DNA was sequenced at the Center for AIDS Research sequencing facility at the University of Alabama at Birmingham.
Sequence analysis
Any nucleotides that could not have been derived from a coding sequence or P addition were considered as N nucleotides. To assign DH gene segments utilized by CDR-H3 at least five nucleotides had to match germline DH gene sequence. Sequences that did not meet this criterion were labeled as CDR-3Hs with “No DH” and their numbers was divided by the total number of sequences to obtain percent of total sequences with “No DH” CDR-3Hs. In the case of DH-JH homology joining, where nucleotides could be assigned to either DH or JH, nucleotides were assigned to DH. CDR-H3 was identified as the region between the 3' VH encoded conserved cysteine (TGT) at Kabat position 92 (IMGT 104) and the 5' JH-encoded conserved tryptophan (TGG) at Kabat position 103 (IMGT 118). Two types of repeated sequences were found. The first were completely identical duplicates (identical VH, DH, and JH segments as well as junctional sequences). These were counted only once. The second were near-identical, differing by one or more nucleotides in the VH. These were attributed to somatic mutation and were counted as unique. Somatic mutation occurs in T-independent responses albeit at lower levels than T-dependent responses (27). The numbers of both types of duplicates are reported.
Statistics
Data comparing three or more groups were analyzed by a one-way ANOVA test, followed by Tukey’s post test, for data with normal distribution and Kruskal-Wallis test, followed by Dunn’s post test, for data that did not distribute normally. Statistical significance was determined by a p value of <0.05.
Results
Limiting DH diversity does not impair the antibody response to DEX
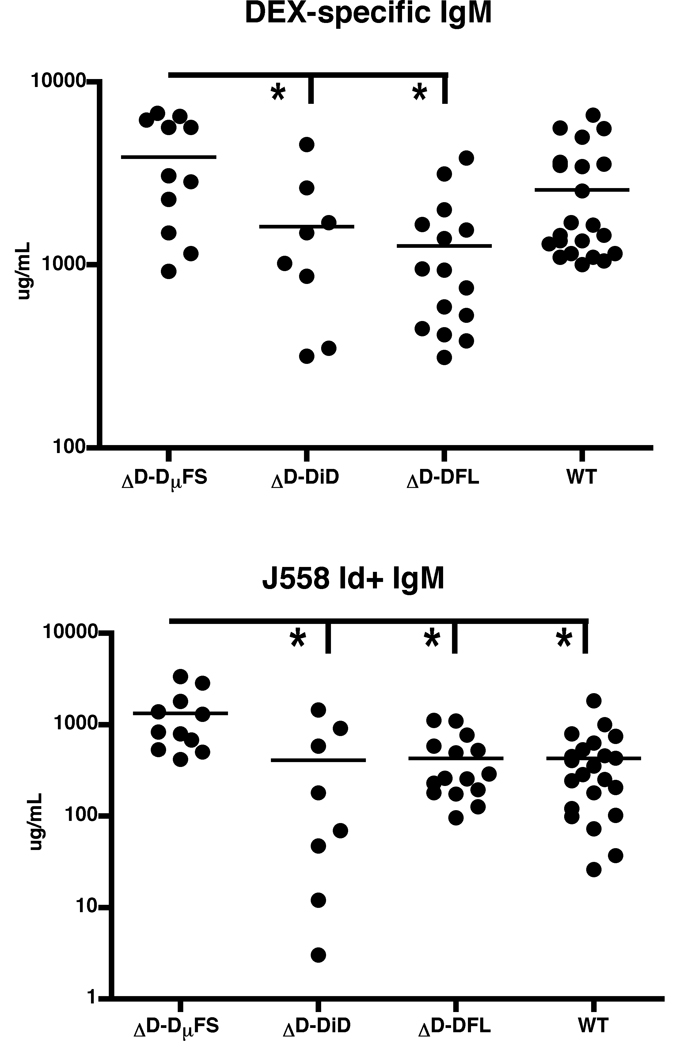
To test the role of limiting DH usage and hence CDR-H3 diversity in the antibody response to DEX, we used Enterobacter cloacae to immunize mice of three different D-limited strains as well as mice with WT DH loci. At 7 days, all three D-limited strains produced a response to DEX (Figure 1A). The highest level was observed in the ΔD-DµFS mice, which was slightly greater than WT and significantly greater than ΔD-iD and ΔD-DFL. We next determined whether the anti-DEX antibodies from D-limited mice express the J558 idiotype that normally predominates in the adult WT BALB/c response. The level of anti-DEX antibodies with the J558 idiotype was equivalent among the ΔD-iD, ΔD-DFL and WT BALB/c mice, but again higher in the ΔD-DµFS strain (Figure 1B). Thus, limiting CDR-H3 diversity by limiting DH segment availability still permitted robust production of anti-DEX antibodies upon bacterial challenge. However, restricting the repertoire to a single DH gene segment that enriches for the use of hydrophobic reading frame 2 in place of hydrophilic reading frame 1 paradoxically generated antibody responses with enhanced levels of expression of the hydrophilic J558 idiotype.
Figure 1. Anti-DEX antibody responses elicited by D-limited mice.
Adult WT, ΔD-DµFS, ΔD-DiD and ΔD-DFL mice were immunized i.v. with DEX-expressing Enterobacter cloacae and serum collected 7 days post-immunization. ELISA was used to determine (A) DEX-specific IgM antibodies and (B) J558 Id expressing antibodies. * p < 0.05
Extensive nucleotide modifications at both ends of the DH gene segment of the anti-DEX repertoire of D-limited mice
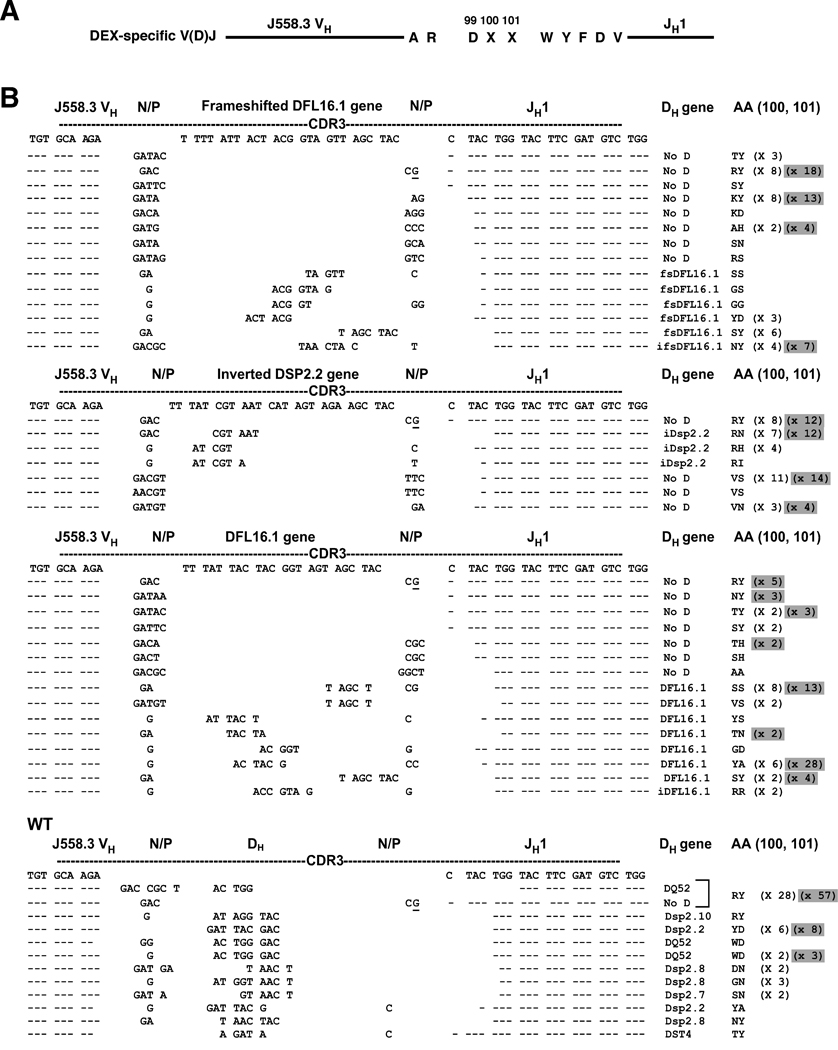
To determine the molecular characteristics of the anti-DEX response elicited by D-limited mice, we PCR-amplified and sequenced heavy chain V(D)J rearrangements from genomic DNA of DEX+ λ+ plasmablasts sorted from the spleens of E. cloacae-immunized mice (Figure 2). Among the WT sequences, 28 (58%) used an identical V(D)J join which could either be attributed to DQ52 with seven nucleotides of N addition, or to no DH with four nucleotides of N addition and one P nucleotide. The remaining 42% of sequences could be attributed to either DQ52, DSP or DST DH gene segments in conjunction with 2–8 nucleotides of N addition. Thus, a DH gene could potentially be assigned to all of the WT sequences (Figure 2 and Figure S1).
Figure 2. CDR-H3 sequences from DEX+ plasmablasts.
DEX+ λ+ plasmablasts were sorted 7 days post-immunization with Enterobacter cloacae and V(D)J rearrangements were PCR-amplified from genomic DNA. (A) The amino acid sequence of a typical DEX-specific heavy chain V(D)J rearrangement. The amino acids shown are canonical in all rearrangements reported in this study with the exception of the two amino acids (labeled XX) at positions 100 and 101. (B) CDR-H3 sequences are compared to those from 3’ end of VH J558.3, the frameshifted DFL16.1 gene (for ΔD-DµFS), the inverted DSP2.2 gene (for ΔD-DiD), the DFL16.1 gene (for ΔD-DFL) and the 5’ end of JH1. In the case of sequence overlap nucleotides were arbitrarily assigned to the DH gene segment. P-nucleotides are underlined. AA: the two amino acids at positions 100 and 101 that differentiate each DEX-specific heavy chain sequence from the others. Sequences from D-limited mice are compared to our database of WT sequences. In WT sequences RY amino acids can have two different assignments and are indicated by a bracket. To assign DH gene segments utilized by CDR-H3 at least five nucleotides had to match germline DH gene sequence. Sequences that did not meet this criterion were labeled as CDR-3Hs with “No DH”. PCR amplification was done from at least three independent FACS sorts for each mouse strain. Two types of repeated sequences were found. The first were completely identical duplicates (identical VH, DH, and JH segments as well as junctional sequences). These were counted only once. The second were near-identical, differing by one or more nucleotides in the VH. These were attributed to somatic mutation and were counted as unique. The numbers of both types of duplicates are reported in parenthesis next to each sequence. The number of identical duplicates is highlighted in grey.
All of the members of the DQ52, DSP and DST families were deleted in the D-limited mice. In those mice limited to use of DFL16.1 (ΔD-DFL) approximately three in four of the sequences could be attributed to this gene segment. In contrast, among the mice limited to either the ΔD-DµFS frameshifted DFL16.1 gene segment or the inverted DSP2.2 ΔD-iD gene segment, only one in three of the anti-DEX sequences could be assigned a progenitor DH (Figures 2 and Figure S1). All of the DH gene segments in joins assigned to DSP or DFL DH gene segments were in reading frame 1 (RF1), including those that by our criterion could be assigned to the DµFS gene segment.
The length of CDR-H3 among all the sequences was the same. And, among those sequences without an identifiable DH gene segment, the needed sequence was derived from either N addition or from an increased contribution from JH1 (Figure S1). Collectively, therefore, the single DH gene available for each D-limited mouse appeared to require more extensive nucleotide deletions with compensatory terminal modifications to generate an anti-DEX response than WT.
The amino acid composition of CDR-H3 regions of anti-DEX antibodies from D-limited mice is heterogeneous yet shows conservation for an aspartic acid residue
The CDR-H3 sequence of WT anti-DEX V(D)J rearrangements exhibited variation in sequence only at positions 100 and 101 (shown on the far right column of Figure 2). These differences reflected both use of alternative DH gene segments in the WT and/or N/P addition in all four strains. This was not surprising since the anti-DEX rearrangements used a similar VH sequence and the same JH gene. Although positions 100 and 101 varied, position 99 contained an aspartic acid (D99) in all of the WT CDR-H3s. In some cases, D99 was entirely encoded by DH sequence, N addition by others and by both for the rest of the sequences.
Neither of the DFL16.1, Dµ FS or iD gene segments encode aspartic acid in their deletional reading frames. To test the extent to which N region addition could contribute D99 to the DEX antibodies produced by the D-limited mice, we examined the deduced amino acid in this position and also looked at amino acids 100 and 101 to ascertain the extended level of junctional diversity created in the D-limited mice.
Despite the inability of D-limited mice to contribute a germline-encoded D99 to the anti-DEX antibodies, all but one of the sequences obtained from these mice included this one particular amino acid. The single exception was an AAC encoded asparagine (N99) that was identified in only one of 35 unique sequences cloned from the ΔD-iD strain. All of the sequences obtained from the ΔD-DFL and ΔD-D° FS mice contained D99.
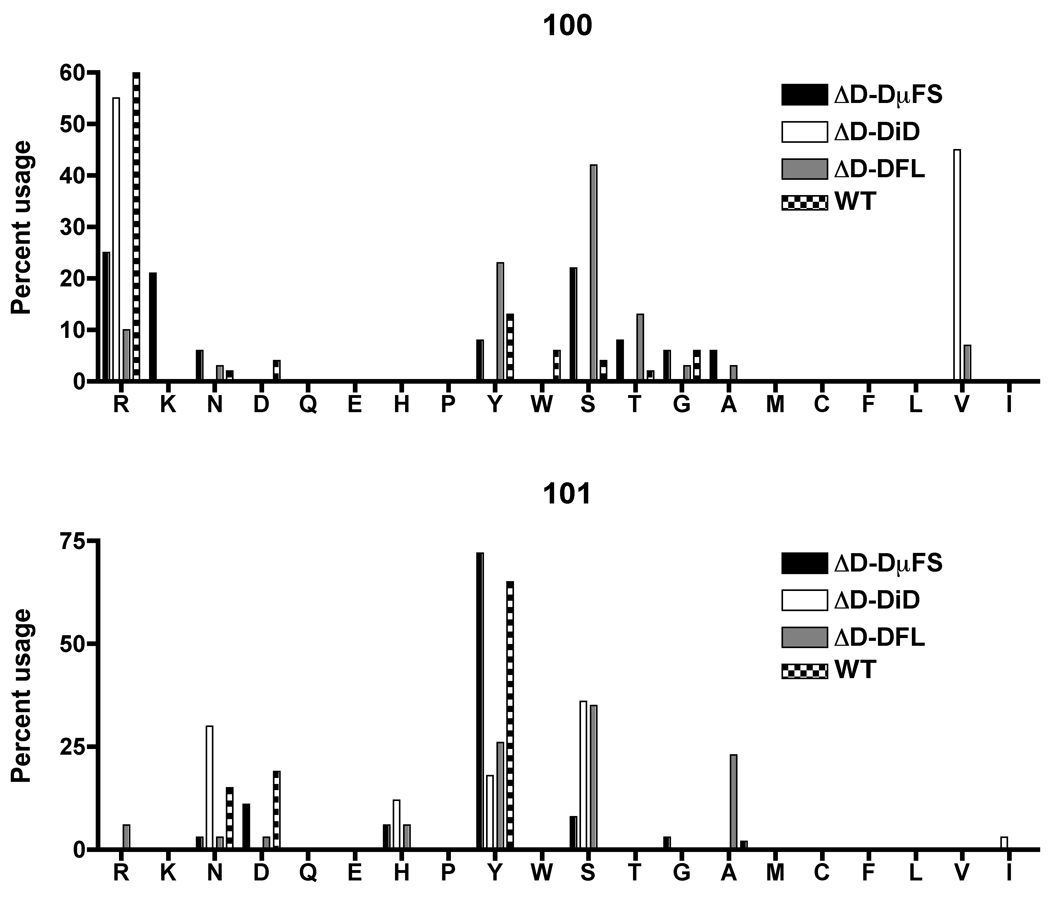
At position 100 and 101, amino acid usage proved more variable with each mouse strain displaying its own unique signature (Figure 3). ΔD-DFL CDR-H3 sequences showed increased usage of serine and tyrosine at amino acid positions 100 and 101, which closely followed the amino acid signature of DFL16.1 reading frame one. ΔD-DµFS CDR-H3 sequences were unique in their acquisition of an N nucleotide generated lysine residue at position 100 and an increased dependence on JH1 to introduce a germline-derived tyrosine at amino acid position 101. Anti-DEX sequences from the ΔD-iD mice were marked by the use of iD germline-encoded arginine at position 100 (R100) as well as by an N nucleotide generated valine at amino acid position 100 (V100) (Figure 3 and Figure 4).
Figure 3. Amino acid usage at Kabat positions 100 and 101 in CDR-H3 sequences from ΔD-DµFS, ΔD-DiD, ΔD-DFL and WT mice.
Distribution of individual amino acids in the CDR-H3 loop of sequences from homozygous D-limited and WT mice is shown.
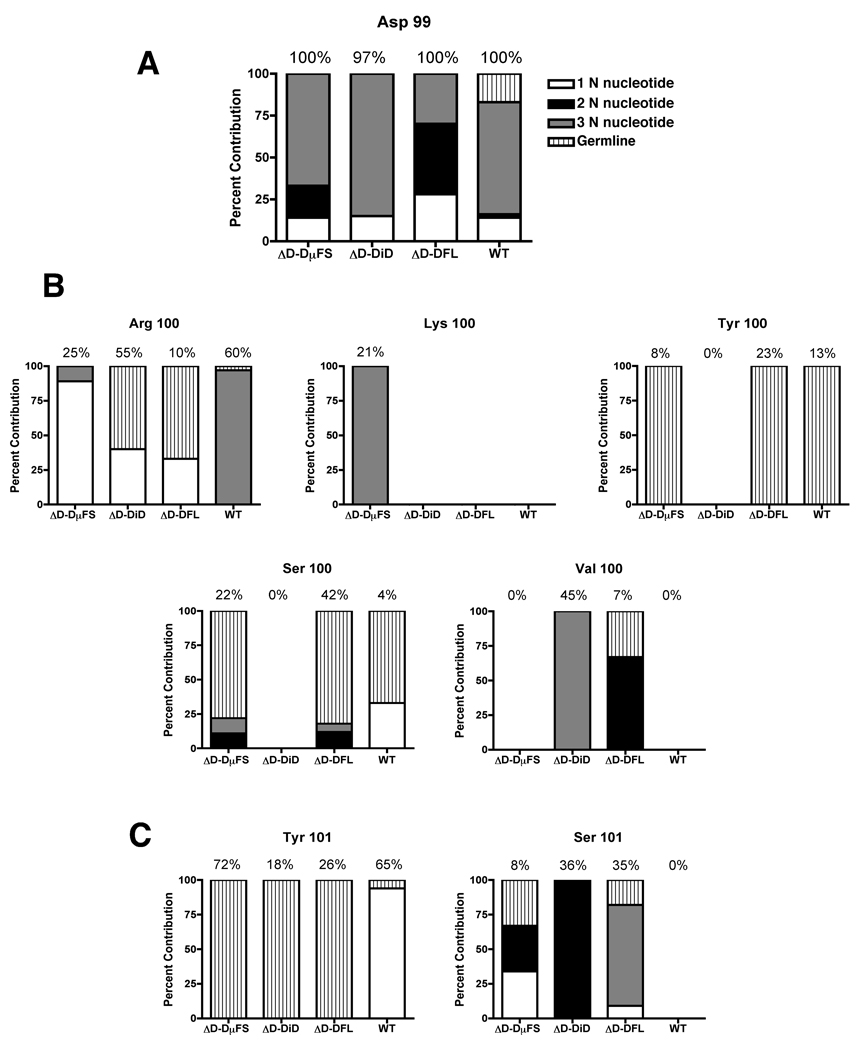
Figure 4. Contribution of N-addition in generating crucial amino acids in CDR-H3 sequences from ΔD-DµFS, ΔD-DiD, ΔD-DFL and WT mice.
The numbers indicate the number of nucleotides that are non-germline (N-nucleotides) that encode for (A) aspartic acid at position 99, (B) arginine, lysine, valine, tyrosine and serine at position 100, (C) tyrosine and serine at position 101 as well as the nucleotides that are germline DH or JH. All are shown as percent of total sequences. Percent usage of each amino acids for each mouse strain is shown on top of each bar.
In summary, as a population the DEX-specific CDR-H3 sequences from the D-limited mouse strains conserved D99 usage and each exhibited signature sequence characteristics that reflected the individual nature of the DH gene segment from which the sequences were derived. Somatic selection for anti-DEX binding capabilities at positions 100 and 101 was thus predicated by the sequence of the DH.
N-addition likely contributes to D99 generation in D-limited mice
In contrast to positions 100 and 101, the near universal presence of aspartic acid at position 99 in all three strains of D-limited mice, as well as WT, suggest that at this one particular position somatic selection played a dominant and necessary role. Unlike WT, where one-fifth of the D99s could be attributed to either the DSP2.2 or DST4 gene segments, all of the D-limited mice appeared to rely on N addition to generate D99 (Figure 4A). N-addition also played a role in the introduction of arginine, lysine and valine amino acids at position 100 in all four mouse strains, including WT (Figure 4B). In contrast, where present, the tyrosines at position 100 and 101 were universally encoded by either germline DH or JH sequence in the D-limited mice (Figure 4). Among the WT mice, at least two of the three nucleotides encoding tyrosine were similarly encoded by germline DH or JH sequence.
D-limited × TdT−/− fail to respond to DEX
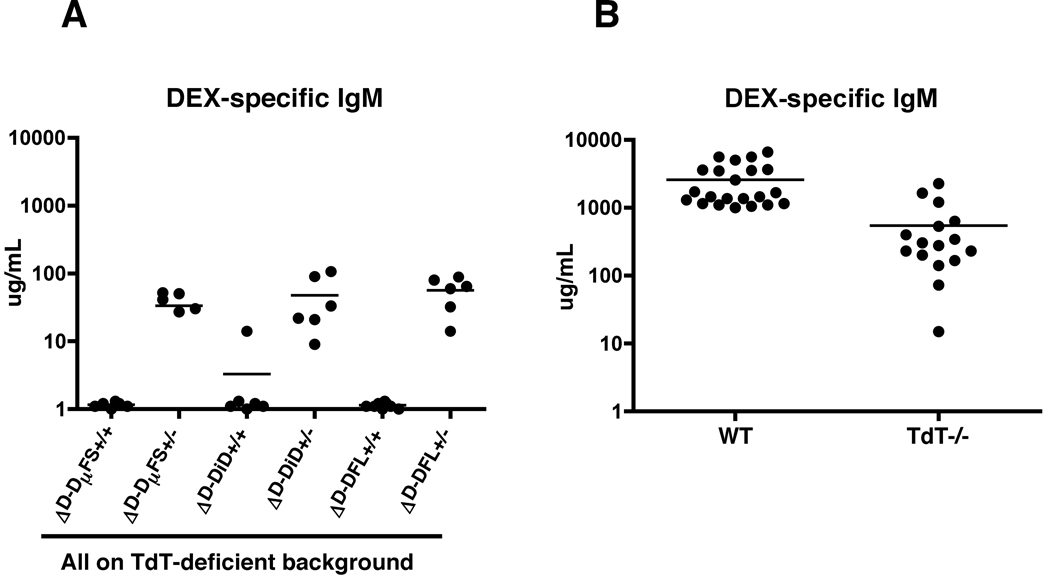
To test the extent of reliance on TdT activity to provide D99 and, by extension, to create the anti-DEX response, we bred the TdT knock-out allele onto each of the D-limited mouse strains and then performed similar E. cloacae immunizations. A response to DEX 7 days after immunization could not be detected in any of the D-limited mice homozygous for the knocked-in DH gene (Figure 5A). Moreover, the response elicited by the heterozygous D-limited mice was even lower than that of TdT-deficient animals (18). These results led us to conclude that the ability to include an aspartic acid in position 99 is essential for the production of Enterobacter cloacae induced anti-DEX antibodies in BALB/c mice and that in the presence or absence of DH encoded aspartic acid, TdT expression either facilitates or is absolutely required, respectively, to place this D99 in CDR-H3. By extension, these findings support the view that the ontogenetic differences in the anti-DEX response observed in BALB/c mice are directly related to physiologic control of TdT activity.
Figure 5. Anti-DEX antibody responses elicited by D-limited TdT-deficient mice.
(A) Adult homozygous ΔD-DµFS, ΔD-DiD, ΔD-DFL and heterozygous controls mice were immunized i.v. with DEX-expressing Enterobacter cloacae and serum collected 7 days post-immunization. ELISA was used to determine DEX-specific IgM antibodies. All mice in this study are on a homozygous TdT-deficient background. (B) DEX-specific IgM ELISA values for WT and TdT−/− (obtained from (18)) are shown for comparison.
Discussion
Antibodies to a number of polysaccharide antigens have several common attributes: CDR-H3 is implicated in binding to antigen (28, 29); its length is strictly maintained (20, 30) and its sequence is almost invariably characterized by the presence of hydrophilic tyrosine residues (21, 31, 32). Xu et al. (9) showed that in VH-restricted mice CDR-H3 diversity was sufficient for development of specific antibody responses to a variety of hapten and protein antigens, but not for two bacterial polysaccharide antigens. The latter finding was attributed to the failure of the single VH gene in their mouse model to accommodate the polysaccharide-specific response, which suggested VH dependency. Work by Casadevall and colleagues (33) showed that in addition to CDR-H3, CDR-H2 encoded amino acids critical for the generation of antibodies specific for the polysaccharide GXM. These studies highlighted the significant roles that both the VH as a whole and the CDR component of the VH as a part can play in the generation of polysaccharide-specific antibodies.
In this study we addressed the relative roles of DH gene segment content and N addition, both key components of CDR-H3 diversity, on the nature of the antibody response to the bacterial polysaccharide DEX following bacterial challenge. We have previously shown that restriction of TdT alone leads to a lower total anti-DEX response and drastically diminished expression of the normally dominant J558 idiotype (18). We now show that limiting the repertoire to the use of a DH segment that preferentially encodes for hydrophilic (ΔD-DFL), charged (ΔD-DiD) or hydrophobic (ΔD-DµFS) amino acids does not dramatically impair the magnitude of the DEX-specific antibody response, per se. We further show that in the absence of a critical DH germline encoded amino acid (D99), D-limited mice wholly depend on the activity of TdT to generate this residue, which our data would suggest is absolutely required for anti-DEX antibody responses. Thus, in the presence of genetic constraints on DH usage, the compensatory diversifying mechanisms of N addition and exonucleolytic nibbling permitted and influenced the magnitude of the anti-DEX response with the short CDR-H3 sequences that are typical of this response in WT BALB/c mice.
In response to Enterobacter cloacae, our D-limited mice, especially ΔD-DµFS, produced an anti-DEX antibody response that was similar, although not identical, to WT mice. The drive to generate DEX-specific antibodies that contain a highly focused CDR-H3 amino acid content suggests that the physicochemical properties of the antigen are playing a role in selecting clones within a specific range of hydrophobicity. Polymers of glucose such as α 1→3 and α 1→6 Dextran (DEX B512) are generally neutral (34). Therefore, we propose that the interaction between DEX-specific B cells and their cognate antigen may require a preponderance of neutral to hydrophilic moieties at their binding site.
ΔD-DµFS mice were capable of generating higher levels of J558-expressing antibodies compared to the other mouse groups. This is likely due to the preponderance of tyrosine residues in amino acid position 101 (Y101) in CDR-H3 sequences isolated from plasmablasts of DEX-immunized ΔD-DµFS mice. Previous reports have shown that the capacity of EB3–7 antibody (anti-J558 Id antibody) to bind J558 was diminished if this tyrosine residue was chemically modified (35). This specific affinity of EB3–7 to bind Y101 may explain the higher levels of J558 Id expression in the serum of ΔD-DµFS DEX-immunized mice. In WT, Y101 was variably the product of JH1, N addition or DQ52. In all of the sequences from the D-limited mice Y101 was only the product of germline DH or JH1. And, of all the D-limited mice, ΔD-DµFS used this residue most frequently. This preference for JH1 Y101 in ΔD-DµFS may reflect the differences in the way the DµFS gene segment is handled by the recombination and selection system in the bone marrow. Evidence for this was reported earlier by examining sequences of VH 7183DJCµ transcripts from different stages of B cell development where ΔD-DµFS rearrangements showed increased nucleolytic loss at the 3’ end of the DH gene segment compared to controls (23). This was true for the minor population of B cells in ΔD-DµFS mice that were selected to use DH reading frame 1 instead of the genetically enforced DH reading frame 2. A tendency to compensate for a shorter DH reading frame 1 sequence with a JH terminal residue could provide a partial explanation for the enhanced response.
A large proportion of the DEX-specific CDR-H3 sequences that were PCR-amplified from immunized D-limited mice contained no identifiable DH gene segments. The mechanisms that can reduce or limit DH content in CDR-H3 have been previously thoroughly discussed by Rohatgi et al (36). Briefly, CDR-H3s without identifiable DH content can be generated by 1) Recombination Signal Sequence replacement, hence permitting direct VH to JH joining (37) 2) violation of the 12/23 bp rule as demonstrated in the DH gene deficient mouse (38) or 3) excessive nucleolytic activity at the DH gene segment. The DEX-specific CDR-H3 sequences that we obtained from plasmablasts contained many of these types of extensive nucleotide modifications, including nucleotide deletions or additions. In D-limited mice, with the exception of ΔD-DFL mice, these junctional alterations drastically modified the genetically enforced DH gene segment and generated a CDR-H3 region hydrophilic in nature and of the appropriate length to allow the host B cell to respond robustly to DEX. It is important to note that V(D)J joins that had assignable DH gene segments were mostly in reading frame 1 (RF1). Taken together, several different mechanisms may be in play to ensure that the heavy chain antibody repertoire conforms to a certain architecture despite genetic manipulations.
Alternative V(D)J recombination events, other than generation of CDR-H3 regions with “No DH”, were available for D-limited mice to generate a WT DEX-specific repertoire. Examination of the WT anti-DEX response shows a propensity for use of the DSP gene family. ΔD-DiD mice had the capacity to invert its iDSP2.2 gene to increase the likelihood of generating a DEX-specific repertoire that displays increased usage of DSP2.2 gene with its component aspartic acid. Despite this fact, V(D)J recombination by inversion was not detected in DEX-specific sequences isolated from these mice. This confirms our earlier observation that V(D)J recombination by deletion is favored over inversion despite the more WT-like antibody repertoire that can develop if inversion was the predominant recombination event (24).
The most striking finding is the apparent absolute requirement for an aspartic acid at position 99 of the CDR-H3 sequence (D99) in order to generate an anti-DEX IgM response. We have previously shown that under WT conditions D99 can be contributed by either DSP2.2 or DST4, or by TdT activity. Both DSP2.2 and DST4 are deleted in the D-limited mice, and we have shown that in these mice D99 was absolutely dependent on TdT activity. This absolute dependence on D99 in general, and on the need for N addition in the D-limited mice in particular, was underscored by the observation that in the absence of TdT the response to DEX was completely abrogated. The singular importance of particular amino acids in specific locations in T-independent responses is not unique to the DEX response. An aspartic acid at the VH-DH junction has been previously shown to be essential for the binding of anti-phosphorylcholine (PC) antibodies such as the M603 clone (39).
DEX-specific sequences from WT mice show a superior capacity to generate R100 and Y101 in the CDR-H3, which are the amino acid residues that characterize the J558 idiotype. If the generation of these two residues was absolutely dependent on TdT activity then we should expect all D-limited mice to generate an equivalent number of sequences to the WT. However, our analysis of the WT sequences revealed that there are two ways to generate R100 and Y101 in WT mice (Figure 2), one of which involves the DQ52 DH gene segment (which is only available in the WT strain). It is possible that DQ52 usage has thus provided WT mice with an advantage in generating the preferred D99, R100 and Y101 CDR-H3 anti-DEX sequence.
Past studies have shown that D-limited mice elicit a lower antibody response to purified DEX (22–24). In this study, we analyzed the DEX-response elicited by the gram-negative commensal Enterobacter cloacae and it is very likely that pathogen-associated molecular patterns (PAMPs), such as LPS, engaged Toll-like receptors and other receptors involved in the innate arm of the immune response, possibly boosting the antibody response. Conversely, it is possible that the antibody response to the bacteria as a whole yields a more diverse antibody response to other bacterial antigens and epitopes that affects the relative level of response to DEX differently depending on the composition of the DH locus and, by extension, the antibody repertoire as a whole.
This report shows that mechanisms controlling the nature of the antibody response to DEX operate to ensure that antibodies generated exhibit certain characteristics that were independent of the sequence composition of the DH. These restrictive mechanisms are not well understood and have received little attention in immunological research (33). Possible mechanisms include strict structural constraints for the antigen–antibody interaction such that only some V regions, or other HCDR component can be selected, idiotypic regulation, deletion of self reactive clones resulting in the emergence of a dominant clone or selection of a protective clone by self or commensal antigen (40–42).
Supplementary Material
Acknowledgements
The authors are very grateful to Jeremy Foote for very helpful discussions and Yao Chen and Yingxin Zhuang for animal husbandry.
Footnotes
This work was supported by research funds from the National Institutes of Health (NIH) Grants AI045794-33 to J.F.K. and AI48115, AI088498, AI090742, AI090902 to H.W.S.J.
References
- 1.Mazmanian SK, Kasper DL. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat Rev Immunol. 2006;6:849–858. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]
- 2.Lesinski GB, Westerink MA. Vaccines against polysaccharide antigens. Curr Drug Targets Infect Disord. 2001;1:325–334. doi: 10.2174/1568005014605964. [DOI] [PubMed] [Google Scholar]
- 3.Weintraub A. Immunology of bacterial polysaccharide antigens. Carbohydr Res. 2003;338:2539–2547. doi: 10.1016/j.carres.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Benedict CL, Kearney JF. Increased junctional diversity in fetal B cells results in a loss of protective anti-phosphorylcholine antibodies in adult mice. Immunity. 1999;10:607–617. doi: 10.1016/s1074-7613(00)80060-6. [DOI] [PubMed] [Google Scholar]
- 5.Insel RA, Adderson EE, Carroll WL. The repertoire of human antibody to the Haemophilus influenzae type b capsular polysaccharide. Int Rev Immunol. 1992;9:25–43. doi: 10.3109/08830189209061781. [DOI] [PubMed] [Google Scholar]
- 6.Zhou J, Lottenbach KR, Barenkamp SJ, Lucas AH, Reason DC. Recurrent variable region gene usage and somatic mutation in the human antibody response to the capsular polysaccharide of Streptococcus pneumoniae type 23F. Infect Immun. 2002;70:4083–4091. doi: 10.1128/IAI.70.8.4083-4091.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez-Fernandez A, Faro J, Fernandez C. Immune responses to polysaccharides: lessons from humans and mice. Vaccine. 2008;26:292–300. doi: 10.1016/j.vaccine.2007.11.042. [DOI] [PubMed] [Google Scholar]
- 8.Padlan EA. Anatomy of the antibody molecule. Mol Immunol. 1994;31:169–217. doi: 10.1016/0161-5890(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 9.Xu JL, Davis MM. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity. 2000;13:37–45. doi: 10.1016/s1074-7613(00)00006-6. [DOI] [PubMed] [Google Scholar]
- 10.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 11.Collins AM, Sewell WA, Edwards MR. Immunoglobulin gene rearrangement, repertoire diversity, and the allergic response. Pharmacol Ther. 2003;100:157–170. doi: 10.1016/j.pharmthera.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Kearney JF, McCarthy MT, Stohrer R, Benjamin WH, Jr, Briles DE. Induction of germ-line anti-alpha 1–3 dextran antibody responses in mice by members of the Enterobacteriaceae family. J Immunol. 1985;135:3468–3472. [PubMed] [Google Scholar]
- 13.Rappleye CA, Eissenberg LG, Goldman WE. Histoplasma capsulatum alpha-(1,3)-glucan blocks innate immune recognition by the beta-glucan receptor. Proc Natl Acad Sci U S A. 2007;104:1366–1370. doi: 10.1073/pnas.0609848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blomberg B, Geckeler WR, Weigert M. Genetics of the antibody response to dextran in mice. Science. 1972;177:178–180. doi: 10.1126/science.177.4044.178. [DOI] [PubMed] [Google Scholar]
- 15.Hansburg D, Perlmutter RM, Briles DE, Davie JM. Analysis of the diversity of murine antibodies to dextran B1355. III. Idiotypic and spectrotypic correlations. Eur J Immunol. 1978;8:352–359. doi: 10.1002/eji.1830080512. [DOI] [PubMed] [Google Scholar]
- 16.Carson D, Weigert M. Immunochemical analysis of the cross-reacting idiotypes of mouse myeloma proteins with anti-dextran activity and normal anti-dextran antibody. Proc Natl Acad Sci U S A. 1973;70:235–239. doi: 10.1073/pnas.70.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clevinger B, Schilling J, Hood L, Davie JM. Structural correlates of cross-reactive and individual idiotypic determinants on murine antibodies to alpha-(1 leads to 3) dextran. J Exp Med. 1980;151:1059–1070. doi: 10.1084/jem.151.5.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahmoud TI, Kearney JF. Terminal deoxynucleotidyl transferase is required for an optimal response to the polysaccharide alpha-1,3 dextran. J Immunol. 2010;184:851–858. doi: 10.4049/jimmunol.0902791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stohrer R, Kearney J. Ontogeny of B cell precursors responding to alpha 1- greater than 3 dextran in BALB/c mice. J Immunol. 1984;133:2323–2326. [PubMed] [Google Scholar]
- 20.Rudikoff S. Antibodies to beta(1,6)-D-galactan: proteins, idiotypes and genes. Immunol Rev. 1988;105:97–111. doi: 10.1111/j.1600-065x.1988.tb00768.x. [DOI] [PubMed] [Google Scholar]
- 21.Hutchins WA, Adkins AR, Kieber-Emmons T, Westerink MA. Molecular characterization of a monoclonal antibody produced in response to a group C meningococcal polysaccharide peptide mimic. Mol Immunol. 1996;33:503–510. doi: 10.1016/0161-5890(96)00012-0. [DOI] [PubMed] [Google Scholar]
- 22.Schelonka RL, Ivanov, Jung DH, Ippolito GC, Nitschke L, Zhuang Y, Gartland GL, Pelkonen J, Alt FW, Rajewsky K, Schroeder HW., Jr A single DH gene segment creates its own unique CDR-H3 repertoire and is sufficient for B cell development and immune function. J Immunol. 2005;175:6624–6632. doi: 10.4049/jimmunol.175.10.6624. [DOI] [PubMed] [Google Scholar]
- 23.Zemlin M, Schelonka RL, Ippolito GC, Zemlin C, Zhuang Y, Gartland GL, Nitschke L, Pelkonen J, Rajewsky K, Schroeder HW., Jr Regulation of repertoire development through genetic control of DH reading frame preference. J Immunol. 2008;181:8416–8424. doi: 10.4049/jimmunol.181.12.8416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ippolito GC, Schelonka RL, Zemlin M, Ivanov, Kobayashi R, Zemlin C, Gartland GL, Nitschke L, Pelkonen J, Fujihashi K, Rajewsky K, Schroeder HW., Jr Forced usage of positively charged amino acids in immunoglobulin CDR-H3 impairs B cell development and antibody production. J Exp Med. 2006;203:1567–1578. doi: 10.1084/jem.20052217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stohrer R, Lee MC, Kearney JF. Analysis of the anti-alpha 1 leads to 3 dextran response with monoclonal anti-idiotype antibodies. J Immunol. 1983;131:1375–1379. [PubMed] [Google Scholar]
- 26.Oliver AM, Grimaldi JC, Howard MC, Kearney JF. Independently ligating CD38 and Fc gammaRIIB relays a dominant negative signal to B cells. Hybridoma. 1999;18:113–119. doi: 10.1089/hyb.1999.18.113. [DOI] [PubMed] [Google Scholar]
- 27.Toellner KM, Jenkinson WE, Taylor DR, Khan M, Sze DM, Sansom DM, Vinuesa CG, MacLennan IC. Low-level hypermutation in T cell-independent germinal centers compared with high mutation rates associated with T cell-dependent germinal centers. J Exp Med. 2002;195:383–389. doi: 10.1084/jem.20011112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casadevall A, Scharff MD. The mouse antibody response to infection with Cryptococcus neoformans: VH and VL usage in polysaccharide binding antibodies. J Exp Med. 1991;174:151–160. doi: 10.1084/jem.174.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura H, Cook R, Meek K, Umeda M, Ball E, Capra JD, Marcus DM. Sequences of the VH and VL regions of murine monoclonal antibodies against 3-fucosyllactosamine. J Immunol. 1988;140:1212–1217. [PubMed] [Google Scholar]
- 30.Schilling J, Clevinger B, Davie JM, Hood L. Amino acid sequence of homogeneous antibodies to dextran and DNA rearrangements in heavy chain V-region gene segments. Nature. 1980;283:35–40. doi: 10.1038/283035a0. [DOI] [PubMed] [Google Scholar]
- 31.Otteson EW, Welch WH, Kozel TR. Protein-polysaccharide interactions. A monoclonal antibody specific for the capsular polysaccharide of Cryptococcus neoformans. J Biol Chem. 1994;269:1858–1864. [PubMed] [Google Scholar]
- 32.Evans SV, Rose DR, To R, Young NM, Bundle DR. Exploring the mimicry of polysaccharide antigens by anti-idiotypic antibodies. The crystallization, molecular replacement, and refinement to 2.8 A resolution of an idiotope-anti-idiotope Fab complex and of the unliganded anti-idiotope Fab. J Mol Biol. 1994;241:691–705. doi: 10.1006/jmbi.1994.1544. [DOI] [PubMed] [Google Scholar]
- 33.Nakouzi A, Casadevall A. The function of conserved amino acids in or near the complementarity determining regions for related antibodies to Cryptococcus neoformans glucuronoxylomannan. Mol Immunol. 2003;40:351–361. doi: 10.1016/s0161-5890(03)00149-4. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez-Fernandez M, Carrasco-Marin E, Alvarez-Dominguez C, Outschoorn IM, Leyva-Cobian F. Inhibitory effects of thymus-independent type 2 antigens on MHC class II-restricted antigen presentation: comparative analysis of carbohydrate structures and the antigen presenting cell. Cell Immunol. 1997;176:1–13. doi: 10.1006/cimm.1996.1078. [DOI] [PubMed] [Google Scholar]
- 35.Dickerman J, Clevinger B, Friedenson B. Loss of an individual idiotype on chemical modification. A strategy for assigning idiotypic determinants. J Exp Med. 1981;153:1275–1285. doi: 10.1084/jem.153.5.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohatgi S, Dutta D, Tahir S, Sehgal D. Molecular dissection of antibody responses against pneumococcal surface protein A: evidence for diverse DH-less heavy chain gene usage and avidity maturation. J Immunol. 2009;182:5570–5585. doi: 10.4049/jimmunol.0803254. [DOI] [PubMed] [Google Scholar]
- 37.Maizels N, Bothwell A. The T-cell-independent immune response to the hapten NP uses a large repertoire of heavy chain genes. Cell. 1985;43:715–720. doi: 10.1016/0092-8674(85)90244-2. [DOI] [PubMed] [Google Scholar]
- 38.Koralov SB, Novobrantseva TI, Hochedlinger K, Jaenisch R, Rajewsky K. Direct in vivo VH to JH rearrangement violating the 12/23 rule. J Exp Med. 2005;201:341–348. doi: 10.1084/jem.20041577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mi QS, Rezanka LJ, Lustig A, Zhou L, Longo DL, Kenny JJ. The M603 idiotype is lost in the response to phosphocholine in terminal deoxynucleotidyl transferase-deficient mice. Eur J Immunol. 2002;32:1139–1146. doi: 10.1002/1521-4141(200204)32:4<1139::AID-IMMU1139>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 40.Bona C. Molecular characteristics of anti-polysaccharide antibodies. Springer Semin Immunopathol. 1993;15:103–118. doi: 10.1007/BF00201095. [DOI] [PubMed] [Google Scholar]
- 41.Victor-Kobrin C, Manser T, Moran TM, Imanishi-Kari T, Gefter M, Bona CA. Shared idiotopes among antibodies encoded by heavy-chain variable region (VH) gene members of the J558 VH family as basis for cross-reactive regulation of clones with different antigen specificity. Proc Natl Acad Sci U S A. 1985;82:7696–7700. doi: 10.1073/pnas.82.22.7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nussbaum G, Anandasabapathy S, Mukherjee J, Fan M, Casadevall A, Scharff MD. Molecular and idiotypic analyses of the antibody response to Cryptococcus neoformans glucuronoxylomannan-protein conjugate vaccine in autoimmune and nonautoimmune mice. Infect Immun. 1999;67:4469–4476. doi: 10.1128/iai.67.9.4469-4476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.