Abstract
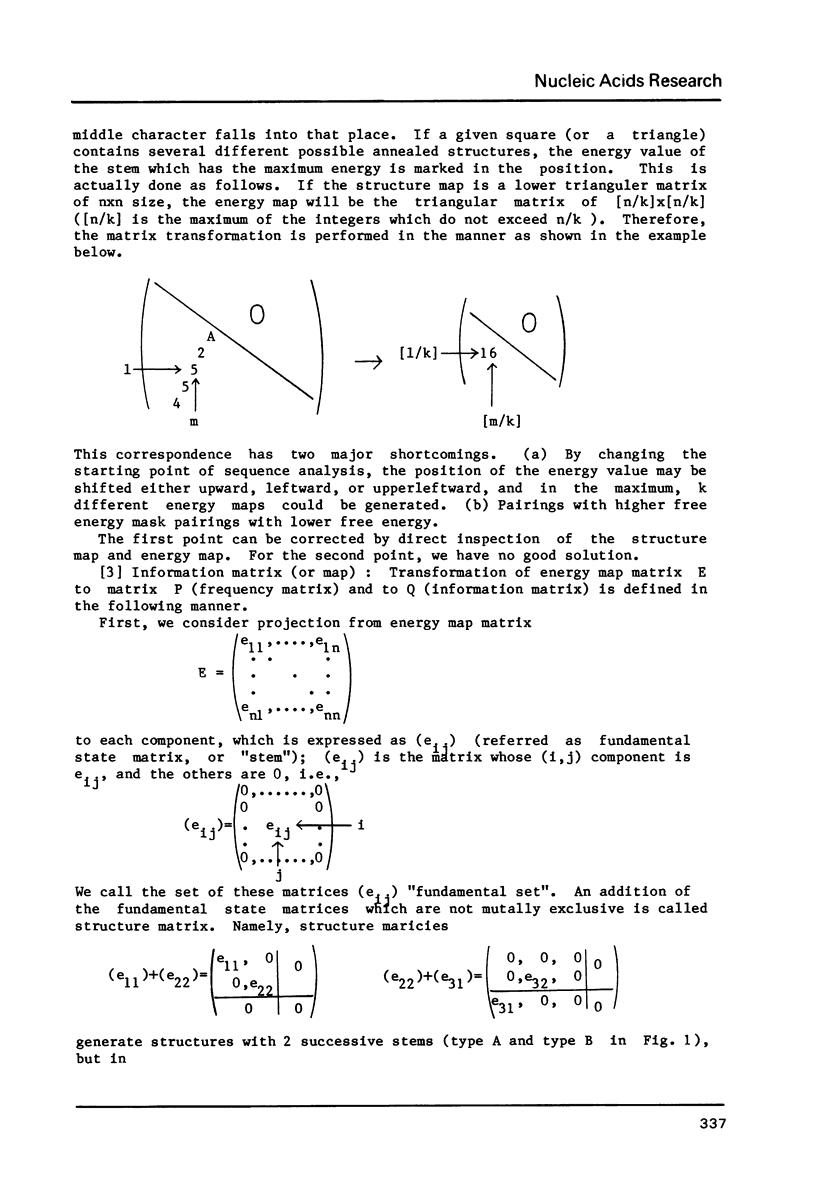
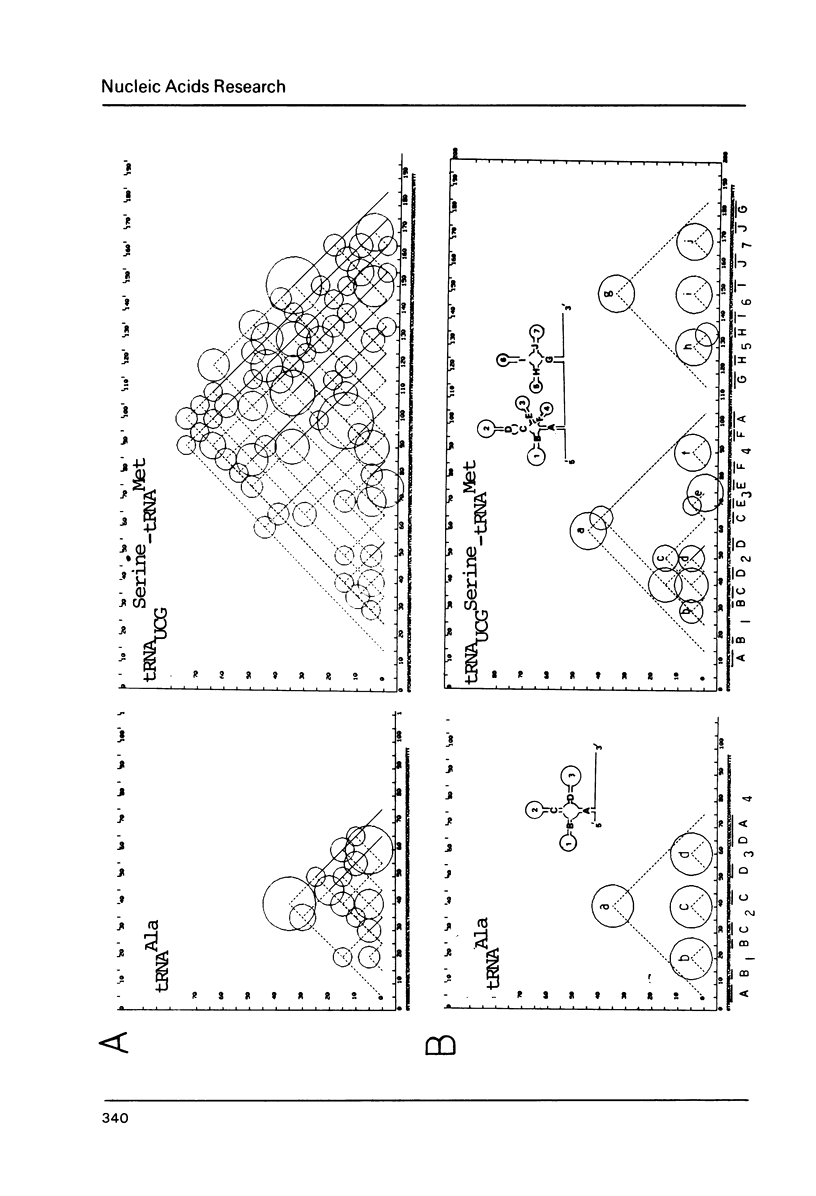
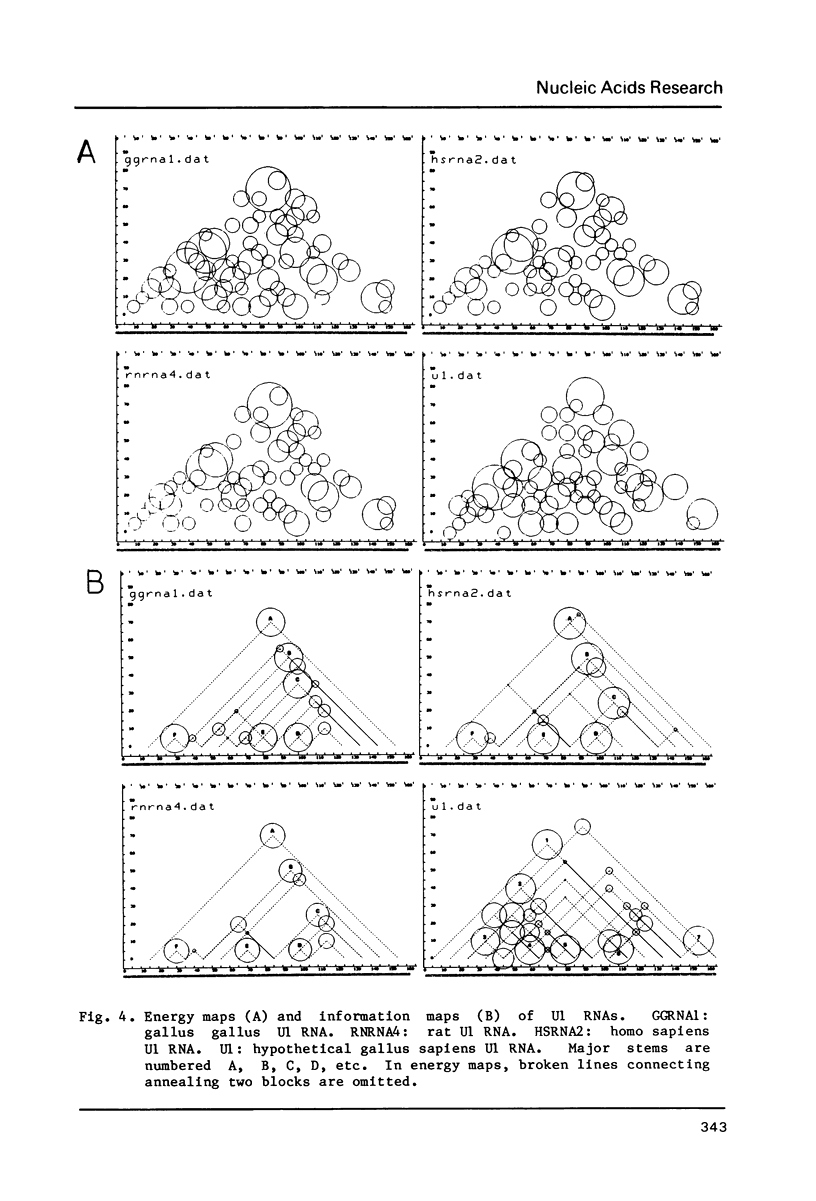
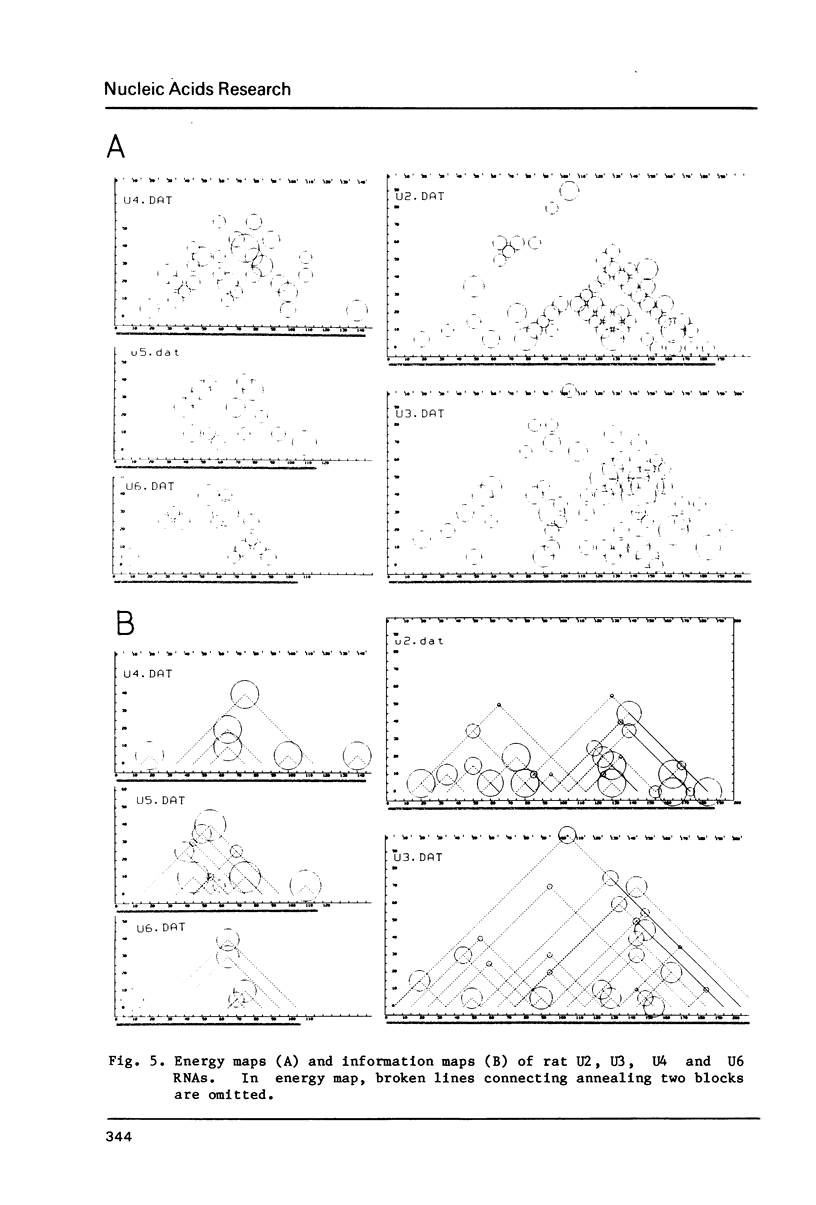
This paper presents a computer analysis of statistical secondary structure of nucleic acids. For a given single stranded nucleic acid, we generated "structure map" which included all the annealing structures in the sequence. The map was transformed into "energy map" by rough approximation; here, the energy level of every pairing structure consisting of more than 2 successive nucleic acid pairs was calculated. By using the "energy map", the probability of occurrence of each annealed structure was computed, i.e., the structure was computed statistically. The basis of computation was the 8-queen problem in the chess game. The validity of our computer programme was checked by computing tRNA structure which has been well established. Successful application of this programme to small nuclear RNAs of various origins is demonstrated.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Branlant C., Krol A., Ebel J. P., Gallinaro H., Lazar E., Jacob M. The conformation of chicken, rat and human U1A RNAs in solution. Nucleic Acids Res. 1981 Feb 25;9(4):841–858. doi: 10.1093/nar/9.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch H., Reddy R., Rothblum L., Choi Y. C. SnRNAs, SnRNPs, and RNA processing. Annu Rev Biochem. 1982;51:617–654. doi: 10.1146/annurev.bi.51.070182.003153. [DOI] [PubMed] [Google Scholar]
- Garber R. L., Gage L. P. Transcription of a cloned Bombyx mori tRNA2Ala gene: nucleotide sequence of the tRNA precursor and its processing in vitro. Cell. 1979 Nov;18(3):817–828. doi: 10.1016/0092-8674(79)90134-x. [DOI] [PubMed] [Google Scholar]
- Mao J., Schmidt O., Söll D. Dimeric transfer RNA precursors in S. pombe. Cell. 1980 Sep;21(2):509–516. doi: 10.1016/0092-8674(80)90488-2. [DOI] [PubMed] [Google Scholar]
- Rogers J., Clarke P., Salser W. Sequence analysis of cloned cDNA encoding part of an immunoglobulin heavy chain. Nucleic Acids Res. 1979 Jul 25;6(10):3305–3321. doi: 10.1093/nar/6.10.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Studnicka G. M., Rahn G. M., Cummings I. W., Salser W. A. Computer method for predicting the secondary structure of single-stranded RNA. Nucleic Acids Res. 1978 Sep;5(9):3365–3387. doi: 10.1093/nar/5.9.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]



