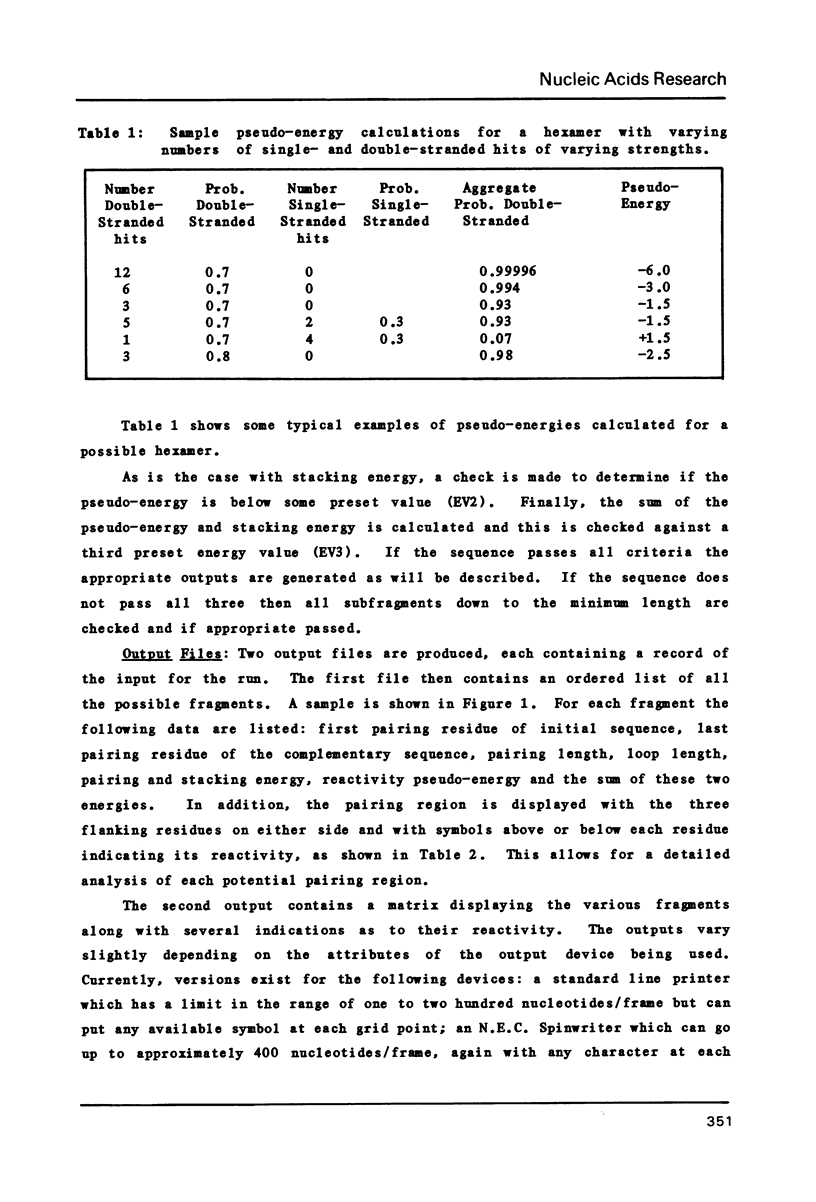
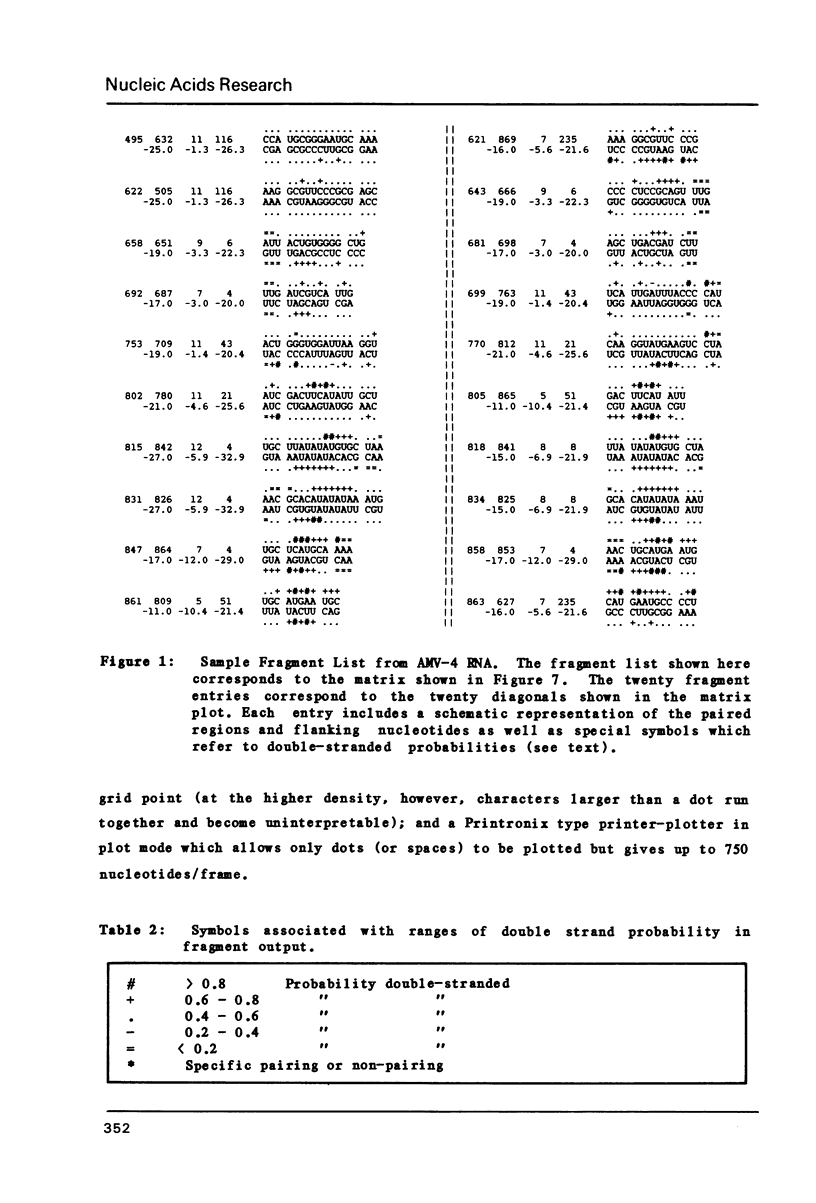
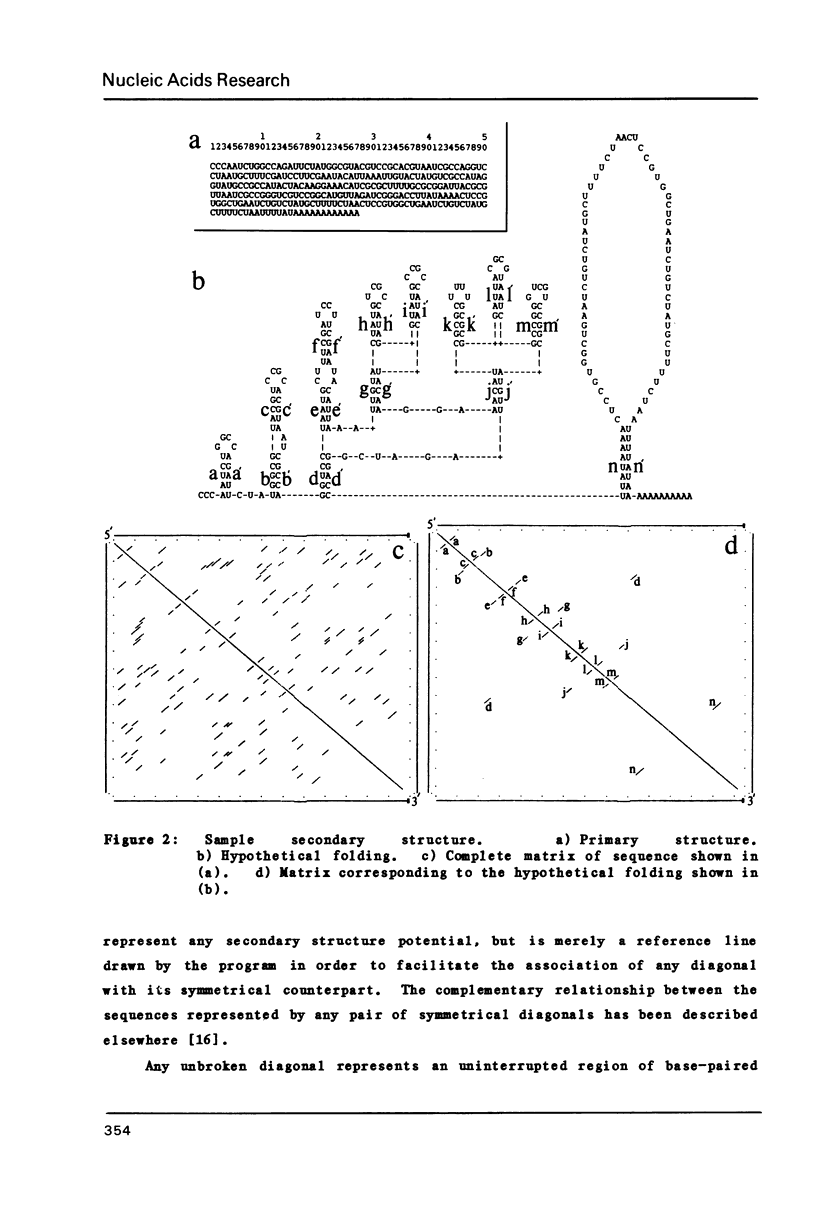
Abstract
We present a computer-aided method for determining nucleic acid secondary structure. The method utilizes a program which has the capability to filter matrix diagonal data on the basis of diagonal length, stabilization energy, and chemical and enzymatic data. The program also allows the user to assign selected regions of the structure as uniquely single-stranded or paired, and to filter out "trade-off" structures on the basis of such pairing. In order to demonstrate the utility of the program we present a preliminary secondary structure for the 3' end of alfalfa mosaic virus RNA 4 (AMV-4 RNA). This structure is based on an analysis which includes the use of in vitro partial enzymatic digestion of the RNA.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auron P. E., Rindone W. P., Vary C. P., Celentano J. J., Vournakis J. N. Computer-aided prediction of RNA secondary structures. Nucleic Acids Res. 1982 Jan 11;10(1):403–419. doi: 10.1093/nar/10.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Duerinck F., Haegmean G., Merregaert J., Jou W. M., Raeymakers A., Volckaert G., Ysebaert M., Van de Kerckhove J. A-protein gene of bacteriophage MS2. Nature. 1975 Jul 24;256(5515):273–278. doi: 10.1038/256273a0. [DOI] [PubMed] [Google Scholar]
- Gardner J. F. Regulation of the threonine operon: tandem threonine and isoleucine codons in the control region and translational control of transcription termination. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1706–1710. doi: 10.1073/pnas.76.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotz C., Zwieb C., Brimacombe R., Edwards K., Kössel H. Secondary structure of the large subunit ribosomal RNA from Escherichia coli, Zea mays chloroplast, and human and mouse mitochondrial ribosomes. Nucleic Acids Res. 1981 Jul 24;9(14):3287–3306. doi: 10.1093/nar/9.14.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla J., Crothers D. M. Free energy of imperfect nucleic acid helices. II. Small hairpin loops. J Mol Biol. 1973 Feb 5;73(4):497–511. doi: 10.1016/0022-2836(73)90096-x. [DOI] [PubMed] [Google Scholar]
- Kozak M. Influence of mRNA secondary structure on binding and migration of 40S ribosomal subunits. Cell. 1980 Jan;19(1):79–90. doi: 10.1016/0092-8674(80)90390-6. [DOI] [PubMed] [Google Scholar]
- Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982 Nov;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Lee F., Yanofsky C. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4365–4369. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M. R., Boyle J. A., Mount S. M., Wolin S. L., Steitz J. A. Are snRNPs involved in splicing? Nature. 1980 Jan 10;283(5743):220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- Maizel J. V., Jr, Lenk R. P. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7665–7669. doi: 10.1073/pnas.78.12.7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M. A., Shatkin A. J. Initiation of reovirus transcription by inosine 5'-triphosphate and properties of 7-methylinosine-capped, inosine-substituted messenger ribonucleic acids. Biochemistry. 1980 Dec 23;19(26):5960–5966. doi: 10.1021/bi00567a003. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Kop J., Wheaton V., Brosius J., Gutell R. R., Kopylov A. M., Dohme F., Herr W., Stahl D. A., Gupta R. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 1981 Nov 25;9(22):6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F., Woese C. R. Secondary structure of 16S ribosomal RNA. Science. 1981 Apr 24;212(4493):403–411. doi: 10.1126/science.6163215. [DOI] [PubMed] [Google Scholar]
- Nussinov R., Tinoco I., Jr, Jacobson A. B. Secondary structure model for the complete simian virus 50 late precursor mRNA. Nucleic Acids Res. 1982 Jan 11;10(1):351–363. doi: 10.1093/nar/10.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlakis G. N., Lockard R. E., Vamvakopoulos N., Rieser L., RajBhandary U. L., Vournakis J. N. Secondary structure of mouse and rabbit alpha- and beta-globin mRNAs: differential accessibility of alpha and beta initiator AUG codons towards nucleases. Cell. 1980 Jan;19(1):91–102. doi: 10.1016/0092-8674(80)90391-8. [DOI] [PubMed] [Google Scholar]
- Pustell J., Kafatos F. C. A high speed, high capacity homology matrix: zooming through SV40 and polyoma. Nucleic Acids Res. 1982 Aug 11;10(15):4765–4782. doi: 10.1093/nar/10.15.4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley G. J., Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Stiege W., Glotz C., Brimacombe R. Localisation of a series of intra-RNA cross-links in the secondary and tertiary structure of 23S RNA, induced by ultraviolet irradiation of Escherichia coli 50S ribosomal subunits. Nucleic Acids Res. 1983 Mar 25;11(6):1687–1706. doi: 10.1093/nar/11.6.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas N. S., Butcher P. D., Arnstein H. R. Polyribosome binding of rabbit globin messenger RNA and messenger ribonucleoprotein labelled with bacteriophage-T4 RNA ligase and 5'-[32P] phosphocytidine 3'-phosphate. Nucleic Acids Res. 1983 Jan 11;11(1):1–10. [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Vournakis J. N., Celantano J., Finn M., Lockard R. E., Mitra T., Pavlakis G., Troutt A., van den Berg M., Wurst R. M. Sequence and structure analysis of end-labeled RNA with nucleases. Gene Amplif Anal. 1981;2:267–298. [PubMed] [Google Scholar]
- Wurst R. M., Vournakis J. N., Maxam A. M. Structure mapping of 5'-32P-labeled RNA with S1 nuclease. Biochemistry. 1978 Oct 17;17(21):4493–4499. doi: 10.1021/bi00614a021. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]