Abstract
The single gene encoding limit dextrinase (pullulan 6-glucanohydrolase; EC 3.2.1.41) in barley (Hordeum vulgare) has 26 introns that range in size from 93 to 822 base pairs. The mature polypeptide encoded by the gene has 884 amino acid residues and a calculated molecular mass of 97,417 D. Limit dextrinase mRNA is abundant in gibberellic acid-treated aleurone layers and in germinated grain. Gibberellic acid response elements were found in the promoter region of the gene. These observations suggest that the enzyme participates in starch hydrolysis during endosperm mobilization in germinated grain. The mRNA encoding the enzyme is present at lower levels in the developing endosperm of immature grain, a location consistent with a role for limit dextrinase in starch synthesis. Enzyme activity was also detected in developing grain. The limit dextrinase has a presequence typical of transit peptides that target nascent polypeptides to amyloplasts, but this would not be expected to direct secretion of the mature enzyme from aleurone cells in germinated grain. It remains to be discovered how the enzyme is released from the aleurone and whether another enzyme, possibly of the isoamylase group, might be equally important for starch hydrolysis in germinated grain.
Starch is the major carbohydrate reserve in cereal grains, where it is located in the nonliving cells of the starchy endosperm and constitutes up to 60% of total grain dry weight (Aman et al., 1985). Starch consists of the essentially linear (1→4)-α-glucan amylose, together with the branched (1→4,1→6)-α-glucan amylopectin. The two polysaccharides are organized in semicrystalline starch granules, which in barley (Hordeum vulgare) grain contain 70% to 75% amylopectin and 25% to 30% amylose (for review, see MacGregor and Fincher, 1993).
Following germination, the glucosyl residues of amylose and amylopectin are released to support seedling growth by the concerted action of α-amylases, β-amylases, debranching enzymes, and α-glucosidases. The debranching enzymes catalyze the hydrolysis of (1→6)-α-glucosidic linkages in amylopectin or in (1→4,1→6)-α-oligoglucosides released by α-amylases. Because the (1→6)-α-glucosyl linkages in these oligoglucosides, which are also referred to as limit dextrins, are not hydrolyzed by α- or β-amylases, and because the action of α-glucosidase on branched oligoglucosides is relatively slow, debranching enzymes are considered to play a central role in the complete depolymerization of starch to Glc. The debranched oligosaccharides are susceptible to further hydrolysis by amylases and α-glucosidases (Lee et al., 1971).
Starch-debranching enzymes have been divided into two groups based on differences in their substate specificities and action patterns (Lee et al., 1971). The first group includes the pullulanases (pullulan 6-glucanohydrolase; EC 3.2.1.41), endohydrolases capable of hydrolyzing (1→6)-α-linkages in pullulan, a polysaccharide consisting of maltotriosyl residues linked by (1→6)-α-linkages. The second group includes isoamylases (glycogen 6-glucanohydrolase; EC 3.2.1.68), endohydrolases that hydrolyze (1→6)-α-glucosyl linkages in glycogen and amylopectin but not in pullulan.
The debranching enzyme described here from germinated barley grain falls into the pullulanase group; however, the natural substrates of the enzyme are amylopectin or oligomeric limit dextrins rather than pullulan. For this reason, and because there is a marked preference for the hydrolysis of (1→6)-α-glucosyl linkages in oligosaccharides compared with polysaccharides, the enzyme is usually referred to as limit dextrinase (Lee et al., 1971). This nomenclature will be used here, although the enzyme is also referred to as the R-enzyme (Nakamura et al., 1996).
As a result of the proposed role for limit dextrinases in starch hydrolysis in germinated barley grains, and because of the associated importance of this process in the malting and brewing industries, considerable effort has been directed toward the purification and characterization of the barley enzyme (Sissons et al., 1992; Longstaff and Bryce, 1993; MacGregor et al., 1994a; Kristensen et al., 1998). The enzyme has been reported in both developing (Sissons et al., 1993) and germinated grain (Kristensen et al., 1998). Inactive “bound” forms and active “soluble” forms have been described (Longstaff and Bryce, 1993), and specific limit dextrinase inhibitors have also been detected in grain extracts (MacGregor et al., 1994b). These latter factors complicate the interpretation of limit dextrinase activity measurements and therefore limited our ability to fully describe the regulation of limit dextrinase gene expression in barley grain. The availability of cDNA probes for northern- blot analyses would clearly be advantageous for the provision of more direct information on the transcriptional activity of the limit dextrinase gene.
In addition to their role in starch hydrolysis in germinated grain, debranching enzymes have also been implicated in starch synthesis, where it is proposed that an appropriate balance between branching and debranching enzymes is required to achieve the final degree of branching in amylopectin (Pan and Nelson, 1984; James et al., 1995; Martin and Smith, 1995; Ball et al., 1996; Nakamura et al., 1997; Rahman et al., 1998). A debranching enzyme would also be capable of producing primers for the starch synthase reaction if required (Duffus and Cochrane, 1993), a function that would explain the presence of limit dextrinase in developing rice grain (Nakamura et al., 1996).
We have isolated cDNAs encoding a barley limit dextrinase and deduced the complete primary structure of the enzyme, together with a number of its physical and chemical properties. cDNA and PCR techniques have been used to monitor levels of limit dextrinase mRNA transcripts in various tissues, including developing and germinated grain, and to examine the influence of the phytohormone GA3 on transcriptional activity of the gene in isolated aleurone layers. Enzyme activity was detected in both developing and germinated grain. A limit dextrinase gene containing 26 introns ranging in size from 93 to 822 bp has been sequenced, and cis-acting elements that may mediate the GA3 response have been identified in the promoter region of the gene. Finally, a presequence with structural features similar to those of transit peptides has been detected. Although this can be reconciled with a function for the enzyme in starch synthesis in amyloplasts in the developing grain, it is more difficult to envisage how a transit peptide would direct secretion of the enzyme from the aleurone layer in germinated grain and, therefore, how limit dextrinase could participate in starch hydrolysis.
MATERIALS AND METHODS
Plant Material
Embryoless half-grains of barley (Hordeum vulgare L. cv Haruna Nijo) were surface-sterilized for 30 min with 1% (v/v) NaOCl, washed thoroughly with 10 mm HCl and water, and soaked in the dark for 3 d in sterile Petri dishes at 25°C. The starchy endosperm was scraped away and the isolated aleurone layers were incubated in sterile 10 mm CaCl2, 2 μm GA3, 10 μg/mL chloramphenicol, 100 μg/mL neomycin sulfate, and 100 units/mL nystatin at 25°C in the dark with continuous shaking for 48 h (Chrispeels and Varner, 1967).
Intact grains were also surface-sterilized and were then germinated in the dark at 20°C for up to 7 d. Moisture content was maintained at about 40% (v/v) using the antibiotic mixture described above.
Samples of developing grains at various stages of endosperm development were harvested from 6 to 36 DPA, when grain fresh weights ranged from approximately 1 to 72 mg. Because of differences in flowering times within and between individual heads, developing grains were pooled for RNA isolation according to their weight. Harvested grains were immediately frozen in liquid N2 and stored at −80°C until RNA extraction.
Amino Acid Sequence Analysis
Barley limit dextrinase preparations were generously provided by Dr. Michael Symons (Department of Primary Industries, Toowoomba, Queensland, Australia) and Dr. A.W. MacGregor (Canadian Grain Commission, Grain Research Laboratory, Winnipeg, Manitoba, Canada). Endoproteinase Asp-N fragments were generated essentially as described by Hagmann et al. (1995) and purified by microbore reversed-phase HPLC on a C18 column (Chen et al., 1993). Selected peptides and enzyme fractions were sequenced in a protein sequencer (model G1005A, Hewlett-Packard) using the version 3.0 sequencing routine, which is based on Edman degradation chemistry.
Detection of Limit Dextrinase Activity
Mature and developing grains and isolated aleurone layers were homogenized with a Teflon pestle in Eppendorf tubes in 100 mm sodium acetate buffer, pH 5.5, containing 25 mm DTT, 50 mm CaCl2, and 0.05% (w/v) BSA. To ensure that limit dextrinase inhibitors were released from the enzyme so that total activity could be more accurately measured, homogenates were held for 5 h at 40°C (Schroeder and MacGregor, 1998). Homogenates were subsequently centrifuged for 30 min at 11,000g and supernatants were precipitated with 85% saturated (NH4)2SO4 at 4°C for 2 h. Precipitates were redissolved in extraction buffer and desalted by dialysis before activity assays.
Limit dextrinase activity was measured with Red Pullulan (Megazyme International, Bray, County Wicklow, Ireland) or DextriZyme tablets (Deltagen, Boronia, Victoria, Australia) according to the method of McCleary (1992) as modified by MacGregor et al. (1994a). Activity remained linear with time for at least 2 h and is expressed as milliunits per milligram of protein per hour. Protein was measured with Coomassie brilliant blue (Bradford, 1976) using BSA as a standard.
Activity was also observed on IEF gels. (NH4)2SO4-precipitated proteins were separated on a flatbed IEF apparatus (Pharmacia Biotech) in 1-mm polyacrylamide gels using a pH gradient of 3.5 to 9.5 (Hrmova and Fincher, 1993). Prefocused gels were run at 600 V for 40 min, followed by 800 V for another 10 min. Proteins were detected with Coomassie brilliant blue after gels were fixed in 20% (w/v) TCA. Apparent pI values were estimated by reference to marker proteins with pI values in the range of 4.45 to 9.6 (Bio-Rad). Activity was detected by immediately overlaying the IEF gels with a 1-mm, 2% (w/v) agar gel containing 0.5% (w/v) Red Pullulan, and heated to 37°C. After the active enzyme caused gel clearing, the agar overlay was fixed in 85% (v/v) ethanol for 16 h.
RNA Extraction and Northern-Blot Analysis
Total RNA was extracted from tissue ground to a fine powder under liquid N2 using sodium glycinate buffer (Chandler and Jacobsen, 1991) for isolated aleurone layers or a commercially available phenol/guanidine isothiocyanate procedure (Trizol, GIBCO-BRL) for developing grain and other tissues, including leaves, roots, and coleoptiles from 5- and 10-d-old seedlings and scutella from 1-d-germinated grain.
For northern-blot analyses RNA samples (15 μg) were separated in 1% (w/v) agarose gels containing 2.2 m formaldehyde and transferred to nylon membranes (Hybond N+, Amersham). Membranes were baked for 1 h at 80°C, RNA was fixed under short-wavelength UV light for 7 min, and the membranes were probed with 32P-labeled limit dextrinase cDNA under conditions described previously (Banik et al., 1997). To ensure approximately equal loading of RNA in individual lanes, gels were stained with ethidium bromide for comparison of the intensities of rRNA bands.
RT-PCR Amplification of mRNAs
To detect low levels of mRNA encoding limit dextrinase in the starchy endosperm of developing grain and in young vegetative tissues, cDNA was prepared from 3 μg of total RNA following the procedure of Frohman et al. (1988) using RT (Superscript II, GIBCO-BRL) and a RACE primer (TRACE, 5′-GACTCGAGTCGACATCGAT17-3′; Frohman et al., 1988) at 50°C. The final reaction volume was 50 μL. Aliquots of 2 μL were subsequently used in PCR reactions with the 5′ and 3′ limit dextrinase primers (5′-GTGCATTTGCATATCAGG-3′ and 5′-TAAGGCTTTGAAGAGCAGA-3′, respectively). The PCR program was 35 cycles of 94°C for 40 s, 50°C for 40 s, and 72°C for 60 s. The single, 641-bp product was isolated from gels using Geneclean (Bresatec, Adelaide, Australia), digested with diagnostic restriction enzymes to check fragmentation patterns, and, following transfer to Hybond N+ membranes, probed with a fragment of the limit dextrinase cDNA at high-stringency conditions (0.1× SSC and 0.1% [w/v] SDS at 65°C). Selected PCR products were also sequenced to confirm that they encoded the barley limit dextrinase.
To ensure that approximately equal amounts of RNA were used for amplification, control PCR reactions in which mRNA encoding the constitutive glycolytic pathway enzyme GAPDH from barley (accession no. U36650) were also performed. The 5′ and 3′ GAPDH primers were 5′-CCACCGGTGTCTTCACTGACAAGG-3′ and 5′-GCCTTAGCATCAAAGATGCTGG-3′, respectively, and the 550-bp product was observed after either 28 or 35 cycles (94°C for 40 s, 65°C for 40 s, and 72°C for 60 s).
Isolation of cDNAs
A cDNA library prepared from poly(A+) RNA from 12-d-old barley seedlings was purchased from Clontech Laboratories (Palo Alto, CA) and screened with a cDNA encoding the rice endosperm R-enzyme (clone D50602; Nakamura et al., 1996). A single positive clone containing a cDNA insert of 654 bp was isolated from 9 × 105 plaque-forming units using procedures described by Banik et al. (1996). The cDNA was subsequently used to screen a cDNA library from GA3-treated barley aleurone layers, which was generously provided by Dr. Mitali Banik. The aleurone library was prepared in λZAP (Uni-ZAP XR, Stratagene). In both cases, libraries were plated out at high density on Escherichia coli lawns. Duplicate plaque lifts on Hybond-C nitrocellulose membranes (Amersham) were prehybridized, hybridized, and washed under previously described conditions (Banik et al., 1996). Following plaque purification of positive clones, cDNA inserts were rescued into the phagemid pBluescript SK(+), and both strands were sequenced using the dideoxynucleotide chain-termination procedure (Sanger et al., 1977). Computer analyses of nucleotide sequences were performed with Genetics Computer Group (Madison, WI) software (Devereux et al., 1984) in the ANGIS suite of programs at the University of Sydney.
Although the screening procedure yielded a cDNA of 2.6 kb from the barley aleurone cDNA library, this was not a full-length cDNA. To obtain longer cDNAs, 1 μL of the aleurone cDNA library consisting of approximately 3 × 106 plaque-forming units was used for a mass excision of cDNA inserts, and the resulting phagemids were subjected to PCR amplification using the T3 primer as the 5′ PCR primer and an oligonucleotide corresponding to a 5′ sequence of the 2.6-kb cDNA (5′-TGGATGATACACGTCGAC-3′) as the 3′ PCR primer. This yielded a 316-bp PCR product, but its sequence still did not extend to the 5′ end of the mRNA, and it was concluded that the aleurone cDNA library contained no full-length clones. Finally, 5′-RACE PCR (Frohman et al., 1988) from a single-stranded cDNA population generated from total RNA extracted from GA3-treated aleurone layers using a 3′ oligonucleotide primer (5′-CAAGGCTGGCGACGTCGACAG-3′) based on the sequence of the 316-bp PCR product resulted in the isolation of a 507-bp fragment that enabled the sequence of the 5′ end of the mRNA to be determined. The PCR products were purified from agarose gels using Bresaclean (Bresatec), and ligated into pBluescript SK(+) for nucleotide sequence analysis.
Isolation of Genomic Clones
The barley genomic library was prepared by Mr. Ron Osmond (Department of Plant Science, University of Adelaide) from partially digested genomic DNA extracted from 7-d-old barley (cv Galleon) seedlings. The DNA fragments were ligated into the EcoRI site of the bacteriophage λDASH II vector (Stratagene). The library was plated out on lawns of E. coli XL1-Blue (P2) cells and screened by hybridization of membrane filter plaque replicas using the 654-bp cDNA fragment as a probe (Sambrook et al., 1989). Phage DNA was isolated from positive clones, digested with restriction endonucleases, and subjected to Southern-blot analysis to identify limit dextrinase gene fragments. The fragments were subcloned into the plasmid pBluescript SK(+) for sequencing. Deletion series were generated with the Erase-a-Base system (Promega).
Primer Extension
To determine the transcription start point of the gene, total RNA preparations (12–24 μg) from GA3-treated aleurone layers were annealed with 25 ng of end-labeled oligonucleotides (complementary to sequences in the 5′ region of the cDNA) in deionized 50% (v/v) formamide at 34°C for 16 h after heating the incubation mixture for 10 min at 85°C (Sambrook et al., 1989; Slakeski et al., 1990). The RNA-oligonucleotide complex was recovered by ethanol precipitation, and reverse transcription was performed as described by Sambrook et al. (1989). The primer-extension product was examined on a DNA-sequencing gel on which sequence reactions of the gene itself, primed with the same 5′ oligonucleotides, were also subjected to electrophoresis.
RESULTS
Isolation and Characterization of cDNAs
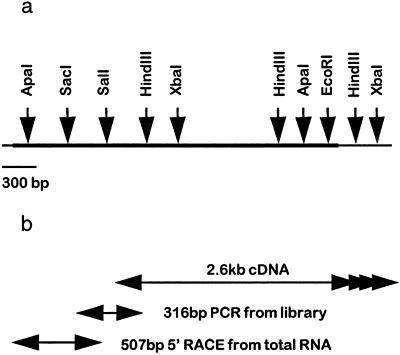
Exhaustive screening of the libraries yielded a cDNA of approximately 2.6 kb, but this was not the full-length cDNA. Subsequent 5′ primer extension from cDNA preparations and 5′ anchored PCR from aleurone layer RNA finally produced DNA fragments containing the 5′ region of the cDNA. The nucleotide sequences of all overlapping fragments were identical. A partial restriction map of the cDNA sequence and the relationships of the cDNA clones that were used to determine the full nucleotide sequence are shown in Figure 1.
Figure 1.
a, Partial restriction map of the 3429-bp cDNA encoding barley limit dextrinase. b, Overlapping cDNAs and PCR products used to obtain the complete nucleotide sequence of the cDNA for barley limit dextrinase. The 2.6-kb 3′ cDNA was screened from a library of GA3-treated aleurone layers. The 316-bp “central” sequence was obtained as a PCR product amplified from cDNA inserts excised from the same aleurone cDNA library. The 507-bp 5′ fragment was prepared by 5′ RACE PCR from aleurone total RNA. The arrows at the 3′ end indicate multiple polyadenylation sites.
The complete nucleotide sequence of the nearly full-length, 3429-bp cDNA and the deduced amino acid sequence are presented in Figure 2. Confirmation that the cDNA encoded a barley limit dextrinase was provided by the exact match of the sequence of the 20 NH2-terminal amino acids determined directly from purified enzyme (Fig. 2). In addition, deduced amino acid sequences corresponding to four endoproteinase Asp-N fragments generated from the purified enzyme were found (data not shown).
Figure 2.
Nucleotide sequence of the overlapping cDNAs for barley limit dextrinase and the deduced amino acid sequence for the enzyme. The NH2-terminal Ala of the mature polypeptide is indicated by the vertical arrow, and the amino acid sequence obtained directly from the purified enzyme is underlined and in bold. Potential N-glycosylation sites are underlined and amino acid residues that are likely to be involved in catalysis are in bold. The TGA stop codon is also shown in bold and the four large arrows near the 3′ end show the various positions of polyadenylic acid tails in different cDNAs isolated. The putative polyadenylation signal AATAAA is underlined. The cDNA accession number is AF122049.
The cDNA has an open reading frame extending from nucleotides 8 to 2893, and the sequence corresponding to the NH2-terminal amino acid sequence of the enzyme itself begins at nucleotide 242 (Fig. 2). The nucleotide sequence that encodes a putative transit peptide has five in-frame ATG-Met codons beginning at nucleotides 8, 14, 20, 29, and 179. The consensus sequence for translation start codons in plant genes is AACAATGGC, in which the most crucial requirements are believed to be a purine at position −3 and a G at position 4; the A of the ATG codon is designated as position +1 (Joshi, 1987; Lutcke et al., 1987; Cavener and Ray, 1991). None of the ATG codons in the limit dextrinase cDNA has both a purine at −3 and a G at position +4, but sequence context around the first ATG codon from the transcription start point beginning at nucleotide 8 reasonably matches the consensus sequence and is therefore considered to be the most likely translation start point (Fig. 2).
The coding region for the mature enzyme has a relatively balanced pattern of codon usage. Of the 884 codons in this region, 48% have C or G in the wobble-base position, and all codons are used. The coding region for the mature enzyme is followed by a TGA stop codon, which forms part of a 3′ untranslated region that ranges from approximately 260 to 530 bp in length. Polyadenylic acid tails were detected at four different sites in the cDNA clones studied (Fig. 2). A potential polyadenylation signal, AATAAA, begins 88 nucleotides downstream from the stop codon.
Isolation and Analysis of the Limit Dextrinase Gene
From approximately 3 × 106 plaques screened, nine positive genomic DNA clones were isolated when a 654-bp limit dextrinase cDNA was used as a probe. Restriction digestion and Southern-blot analyses showed that the gene was very large and was dispersed over two cloned genomic DNA fragments. Overlap of the clones themselves was performed by PCR from genomic DNA preparations, using primers corresponding to a sequence in the exons immediately 5′ and 3′ to the ends of the original genomic clones (nucleotides 1912–1936 and 1996–1971 in Fig. 2). A fragment of approximately 900 bp was amplified from DNA from four individual varieties and sequenced (data not shown). Six fragments from the two genomic clones were subcloned into pBluescript SK(+) for sequencing. Subsequent nucleotide sequence analyses confirmed that overlapping fragments were identical and that exon sequences were always identical to those determined from the cDNA. It was concluded therefore that the fragments represent part of a single limit dextrinase gene. The sequence of an 11.5-kb DNA fragment carrying the gene isolated in the present study can be found in the database under accession no. AF122050, and exhibits 98% positional identity with the nucleotide sequence of a barley limit dextrinase gene recently added to the database (accession no. AF022725).
The barley limit dextrinase gene consists of 27 exons separated by 26 introns. Exon/intron boundaries were identified by reference to the cDNA sequence shown in Figure 2. In all cases the 5′ and 3′ boundary sequences of the introns were closely related to the consensus sequences for monocotyledonous plants of AG↓GTAAGT for the 5′ boundary and TGCAG↓GT for the 3′ boundary (Simpson and Filipowicz, 1996).
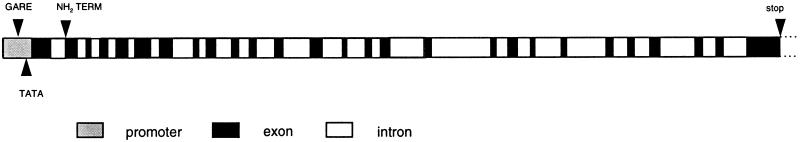
The structure of the barley limit dextrinase gene is represented diagrammatically in Figure 3. The exact positions of intron insertions, together with their lengths, are shown in Table I. The introns range in length from 93 to 822 bp: 3 are less than 100 bp, 11 are between 100 and 200 bp, 6 are between 200 to 299 bp, 4 are between 300 and 500 bp, and 2 are more than 500 bp (Table I).
Figure 3.
Structure of an 11.5-kb genomic DNA fragment carrying the barley limit dextrinase gene, showing the 27 exons (black), the 26 introns (white), and the promoter region (shaded). Positions of the GA3 response elements (GARE), the putative TATA box of the promoter, the NH2 terminus of the mature protein, and the stop codon are indicated.
Table I.
Exons and introns of a barley limit dextrinase gene
| Exons
|
Introns
|
||||
|---|---|---|---|---|---|
| No. | Position | Length | No. | Position | Length |
| bp | bp | ||||
| 1 | 1,718 –1,948 | 230 | 1 | 1,949 –2,133 | 184 |
| 2 | 2,134 –2,278 | 144 | 2 | 2,279 –2,388 | 109 |
| 3 | 2,393 –2,445 | 52 | 3 | 2,446 –2,561 | 115 |
| 4 | 2,562 –2,670 | 108 | 4 | 2,671 –2,780 | 109 |
| 5 | 2,781 –2,917 | 136 | 5 | 2,918 –3,017 | 99 |
| 6 | 3,018 –3,214 | 196 | 6 | 3,215 –3,321 | 106 |
| 7 | 3,322 –3,471 | 149 | 7 | 3,472 –3,731 | 259 |
| 8 | 3,732 –3,792 | 60 | 8 | 3,793 –3,886 | 93 |
| 9 | 3,887 –4,007 | 120 | 9 | 4,008 –4,201 | 193 |
| 10 | 4,202 –4,292 | 90 | 10 | 4,293 –4,423 | 130 |
| 11 | 4,424 –4,517 | 93 | 11 | 4,518 –4,752 | 234 |
| 12 | 4,753 –4,848 | 95 | 12 | 4,849 –5,234 | 385 |
| 13 | 5,235 –5,375 | 140 | 13 | 5,376 –5,626 | 250 |
| 14 | 5,627 –5,677 | 50 | 14 | 5,678 –5,907 | 229 |
| 15 | 5,908 –5,984 | 76 | 15 | 5,985 –6,084 | 99 |
| 16 | 6,085 –6,171 | 86 | 16 | 6,172 –6,631 | 459 |
| 17 | 6,632 –6,722 | 90 | 17 | 6,723 –7,545 | 822 |
| 18 | 7,546 –7,617 | 71 | 18 | 7,618 –7,772 | 154 |
| 19 | 7,773 –7,898 | 125 | 19 | 7,899 –8,079 | 180 |
| 20 | 8,080 –8,136 | 56 | 20 | 8,137 –8,512 | 375 |
| 21 | 8,513 –8,587 | 74 | 21 | 8,588 –9,123 | 535 |
| 22 | 9,124 –9,212 | 88 | 22 | 9,213 –9,419 | 206 |
| 23 | 9,420 –9,497 | 77 | 23 | 9,498 –9,663 | 165 |
| 24 | 9,664 –9,802 | 138 | 24 | 9,803 –10,281 | 478 |
| 25 | 10,282 –10,391 | 109 | 25 | 10,392 –10,587 | 195 |
| 26 | 10,588 –10,687 | 99 | 26 | 10,688 –10,930 | 242 |
| 27 | 10,931 –11,459+ | 528+ | |||
Promoter Region of the Gene
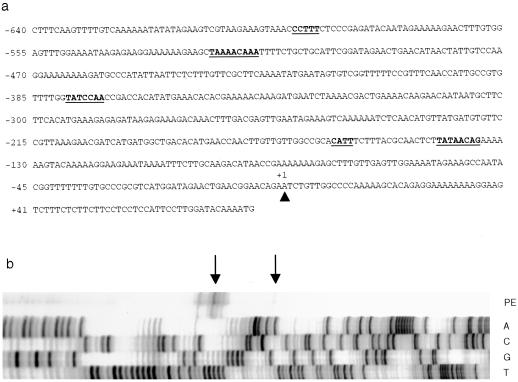
The nucleotide sequence of the promoter region of the barley limit dextrinase gene is shown in Figure 4a. To define the transcription start point of the gene, oligonucleotide-primed synthesis of cDNA from aleurone RNA preparations was performed. Termination products were detected in three main positions (Fig. 4b). The position that most closely matched the transcription start point consensus sequence CTCATCA for plant genes (Fig. 4a; compare Joshi, 1987) was the AGAATCT sequence farthest from the translation start codon (Fig. 2). We have therefore numbered the gene sequence from that point.
Figure 4.
Promoter region of the barley limit dextrinase gene. a, Nucleotide sequence of the gene upstream from the translation start ATG codon. The putative transcription start point is indicated by the arrowhead and is designated nucleotide +1. Underlined sequences starting from the 5′ end of the sequence include the CCTTT pyrimidine box, the TAAAACAAA box and the TATCCAA box of the putative GA3 response element, and the CATT box and the proposed TATA box of the gene. b, Primer extension from total RNA of GA3-treated aleurone layers using a primer specific for the 5′ region of the cDNA. The top two lanes show the major primer extension (PE) termination products (arrows), whereas the lower four lanes represent the nucleotide sequence of the gene, primed with the same oligonucleotide used for the primer extension.
A putative TATA box begins 142 bp upstream from the transcription start point, but it only approximately matches the consensus sequence ACTATATATAG for plant genes and is located farther upstream than most TATA boxes (Fig. 4a; compare Joshi, 1987). A CATT sequence begins approximately 20 bp 5′ to the possible TATA box. In view of the relatively poor correspondence of these promoter sequences with plant consensus sequences, and because of the possibility that the 5′ region of the gene might be interrupted by introns, approximately 4 kb of sequence upstream from that shown in Figure 4a was determined. However, no additional transcription start point or potential TATA box sequences were detected (data not shown). Furthermore, the promoter of the single limit dextrinase gene from rice has similarly ill-defined promoter sequences (Y. Nakamura, personal communication).
Northern-Blot Analysis
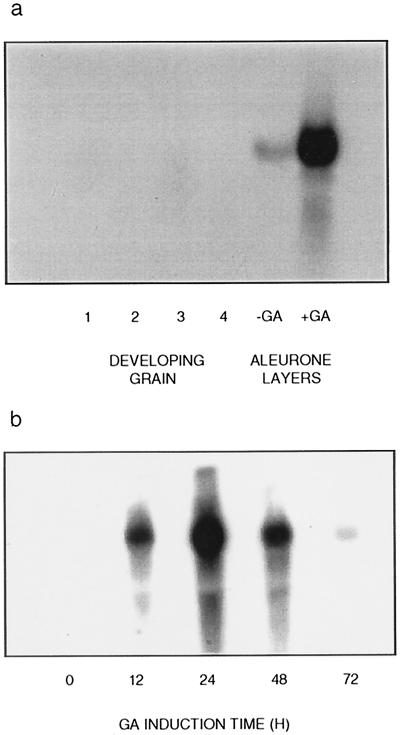
When RNA preparations from developing barley grain and from GA3-treated aleurone layers were subjected to northern-blot analysis, a 2-kb cDNA probe hybridized strongly with a single RNA species of approximately 3.4 kb in GA3-treated aleurone layers (Fig. 5a). No limit dextrinase gene transcripts were detected in the developing grain (Fig. 5a), leaves, roots, coleoptiles, or scutella (data not shown), and transcript levels in untreated aleurone layers were low (Fig. 5a).
Figure 5.
Northern blots of RNA preparations from developing barley grain and GA3-treated aleurone layers using limit dextrinase cDNA as a probe. a, Lanes 1 to 4, RNA from grain during the period 19 to 40 DPA (grain weight 27–70 mg); lanes 5 and 6, RNA from aleurone layers treated for 48 h with (+GA) or without (−GA) 2 μm GA3. b, Time course of GA3 induction of limit dextrinase mRNA transcripts in isolated barley aleurone layers.
In GA3-treated aleurone layers, limit dextrinase gene transcripts could be detected after 12 h, were most abundant at 24 h, and decreased thereafter (Fig. 5b). In view of the apparent induction of limit dextrinase gene transcripts by GA3 in the isolated barley aleurone layers, the promoter sequence of the gene (Fig. 4a) was scanned for the presence of cis-acting elements of the GA-response complex that are commonly detected in gene promoters induced by the hormone in cereal aleurone layers (Gubler et al., 1995). These elements include a pyrimidine box, a TAACAAA box, and a TATCCAC box (Gubler and Jacobsen, 1992). The barley limit dextrinase gene promoter has similar sequences, which include a CCCTTTCTCCC pyrimidine box beginning at nucleotide −594, a TAAAACAAA sequence at nucleotide −523, and a TATCCAA sequence at nucleotide −379 (Fig. 4a).
RT-PCR Analysis of mRNAs in Developing Grain
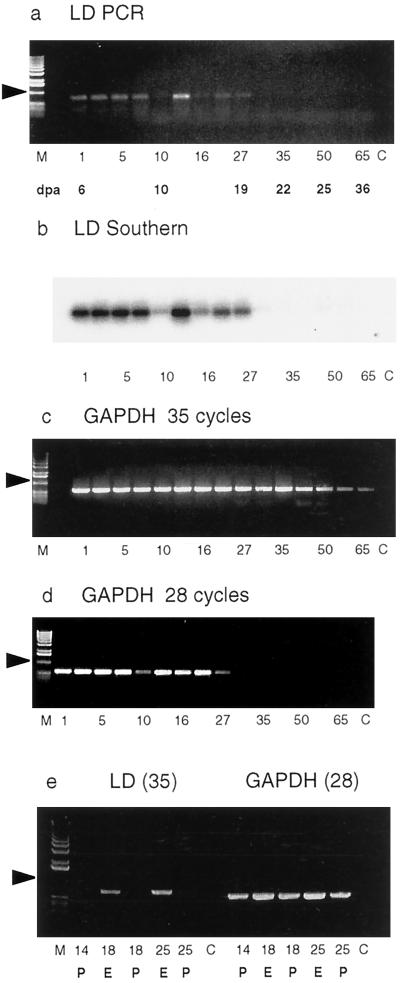
Although no limit dextrinase mRNA transcripts were detected by northern-blot analysis at a stage of grain development when starch synthesis was likely to be in progress (Fig. 5a; compare MacLeod and Duffus, 1988), Nakamura et al. (1996) isolated a rice limit dextrinase cDNA from a library generated from RNA of developing grain, and it has been widely suggested that the enzyme could be involved in starch synthesis during grain development (Nakamura et al., 1997). RT-PCR was therefore used in further attempts to detect limit dextrinase mRNA in developing grain, and, to ensure that transcripts were not overlooked, RNA preparations isolated over a broader time scale in starchy endosperm development (6–36 DPA) were also examined. The resultant amplification products are shown in Figure 6a, together with a Southern blot of the PCR products (Fig. 6b). Nucleotide sequence analysis of the PCR products confirmed that they corresponded to the limit dextrinase sequence (data not shown). It is therefore clear that mRNA encoding limit dextrinase is present at relatively low levels in developing endosperm until the grain achieves a mass of 25 to 27 mg (Fig. 6a; this corresponds to approximately 19 DPA). Thereafter, the levels of limit dextrinase mRNA transcripts decrease. The abundance of the control mRNA for GAPDH also decreases after this time, and this was especially apparent when PCR amplification of the GAPDH cDNA was reduced from 35 to 28 cycles (Fig. 6, c and d).
Figure 6.
RT-PCR analysis of limit dextrinase mRNA transcripts in the developing barley grain 6 to 36 DPA. Lane M, DNA molecular mass markers; lane C, control PCR reactions; lanes 1 to 65, weight of the grain (in milligrams) at the time of RNA extraction. Below are shown the corresponding times of RNA extraction (6–36 DPA). a, RT-PCR products amplified with limit dextrinase (LD) primers; b, Southern blot of the RT-PCR products shown after probing with a limit dextrinase cDNA; c, RT-PCR products amplified with primers for the constitutively expressed GAPDH after 35 cycles; d, RT-PCR products of GAPDH primers after 28 cycles; e, limit dextrinase (LD) primers and 35 PCR cycles of RNA preparations extracted from the pericarp (P) and endosperm (E) of developing grain, together with products amplified in 28 cycles using the GAPDH primers.
To ensure that the limit dextrinase mRNA did not originate from the pericarp or other maternal tissues of the grain, the endosperm was squeezed out from the developing grain before to RNA isolation, and RT-PCR analysis was repeated on RNA extracted from both the endosperm and the remaining pericarp and other tissues. The majority of mRNA encoding limit dextrinase was shown to be in the endosperm, as noted in previous immunological studies (Sissons et al., 1993), although some could be detected in the pericarp (Fig. 6e). It should be noted that a thin layer of aleurone tissue could be detected surrounding the starchy endosperm that was squeezed out from the grain. However, the aleurone layer could not be easily dissected away from the starchy endosperm, and the results reported here therefore relate to the endosperm as a whole. The RT-PCR procedure also allowed the detection of limit dextrinase mRNA in young leaves, young roots, and the scutellum of 1-d-germinated grain; apparent levels in these tissues were relatively low (data not shown).
Enzyme Activity
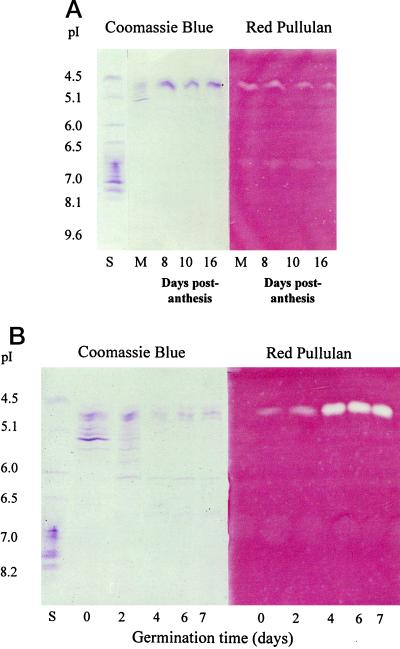
In situ detection of activity in IEF gels confirmed that active enzyme was present during grain development, albeit at low levels (Fig. 7A). Similarly, active enzyme could be detected in extracts of germinated grain (Fig. 7B). The faint bands that are seen in the pI-6.5 to -7.0 region of the Red Pullulan gel in Figure 7A and more faintly in Figure 7B are believed to be artifacts of the gel system rather than another limit dextrinase isoform, because the band was also present in blank lanes where no extract was loaded (e.g. in the far left lane of the Red Pullulan panel in Fig. 7B). Quantitative measurements of limit dextrinase activity in developing endosperm and germinated grain are compared in Table II.
Figure 7.
IEF of extracts of a barley developing grain (A) and a germinated grain (B). The left panels show proteins stained with Coomassie brilliant blue and the right panels show limit dextrinase activity revealed by clearing of a Red Pullulan overlay gel. Lanes S, pI protein standards; lanes M, mature grain and other extracts from developing endosperm 8, 10, and 16 DPA. The number of days after the initiation of germination are indicated in B.
Table II.
Limit dextrinase activity in tissue extracts from developing endosperm and germinated grain
| Tissue | Activity |
|---|---|
| milliunits mg−1 protein h−1 | |
| Developing endosperm | |
| 8 DPA | 65 ± 7 |
| 10 DPA | 44 ± 5 |
| 16 DPA | 34 ± 5 |
| Mature grain | 23 ± 4 |
| Germinated grain | |
| 0 d | 44 ± 7 |
| 2 d | 75 ± 7 |
| 4 d | 571 ± 52 |
| 6 d | 627 ± 69 |
Assays were performed according to the method of McCleary (1992) and MacGregor et al. (1994a).
Properties of the Encoded Enzyme
The barley limit dextrinase appears to have a relatively long presequence before the NH2-terminal Ala residue of the mature enzyme (Fig. 2). This sequence is 78 amino acid residues in length and contains 13 Ala residues (17% of the presequence on a molar basis), 12 Pro residues (15%), 12 Arg residues (15%), and 9 Ser residues (12%). Thus, these four residues account for nearly 60% of the amino acids in the putative transit peptide (Fig. 2), and the Ala, Pro, and Arg residues are two to three times more abundant in the transit peptide than in the mature polypeptide (data not shown). Furthermore, the major amino acids are asymmetrically distributed along the transit peptide. The Arg and Pro residues are concentrated toward the NH2-terminal end, and clusters of two or three contiguous residues are apparent. Positively charged amino acid residues, sometimes in clusters, are found at relatively regular intervals along the NH2-terminal part of the transit peptide (Fig. 2). Similarly, clusters of Ser residues are observed in the NH2-terminal region. An internal Met residue is present and, toward the COOH-terminus of the peptide, negatively charged Glu residues are also present. Although the net charge of the transit peptide at neutral pH is +10, there is a clear delineation between positive charges at the NH2 terminus and negative charges at the COOH terminus. No extended hydrophobic regions are seen.
The region of the cDNA from the codon specifying the NH2-terminal Ala residue at nucleotide 242 to the TGA stop codon at nucleotide 2894 encodes a polypeptide of 884 amino acid residues. Seven potential N-glycosylation sites begin at amino acid residues 229, 440, 524, 529, 642, 816, and 851 in the deduced sequence (Fig. 2).
When the deduced amino acid sequence for the barley limit dextrinase was compared with sequences in the DNA and protein databases, the best matches were with the rice R-enzyme (81% positional identity; Nakamura et al., 1996), a pullulanase from spinach (63% positional identity; accession no. X83969; A. Renz, R. Schmid, J. Kossmann, and E. Beck, unpublished data), a pullulanase from the bacterium Klebsiella aerogenes (39% identity; Katsuragi et al., 1987), the sugary1 gene of maize (24% identity; James et al., 1995), and an isoamylase from Pseudomonas amyloderamosa (25% identity; Amemura et al., 1988). The alignment of these sequences, shown in Figure 8, is restricted to the catalytic site region, but blocks of approximately 2 to 10 identical amino acids are also distributed along the rest of the polypeptide chain. The blocks of highly conserved amino acid sequences noted in starch-debranching enzymes of diverse origin by Nakamura et al. (1997) are also conserved in the barley enzyme (data not shown).
Figure 8.
Amino acid sequence alignments of the catalytic domains of debranching enzymes from barley (Fig. 2), rice (Nakamura et al., 1996), spinach (A. Renz, R. Schmid, J. Kossmann, and E. Beck, unpublished data), and K. aerogenes (Katsuragi et al., 1987), using the PileUp and PrettyBox programs (Devereux et al., 1984). Shaded and hatched boxes indicate identical and homologous residues, respectively.
DISCUSSION
The nucleotide sequence of the mRNA encoding barley limit dextrinase has been obtained from overlapping cDNA clones and from PCR amplification products. The clones were screened from a barley aleurone library, which was generated from poly(A+) RNA isolated from aleurone layers treated for 48 h with 2 μm GA3 (Banik et al., 1996) and from a commercially available cDNA library from barley seedlings. The largest cDNA isolated was 2.6 kb in length, and, despite extensive screening of available cDNA libraries, full-length cDNAs could not be found. This is probably attributable to the large size of the mRNA and to the presence of secondary structures caused by GC-rich sequences in the 5′ region of the mRNA (Fig. 2). Complementary DNAs encoding the 5′ region were eventually obtained from RNA preparations by PCR procedures. Four independent polyadenylation sites were detected at the 3′ end of the cDNAs (Fig. 2).
The cDNA sequence has an open reading frame that encodes a putative transit peptide of 78 amino acid residues and a mature polypeptide of 884 amino acid residues. The calculated molcular mass of the mature polypeptide is 97,417 D and the calculated pI is 5.0. These values are comparable to a molecular mass of about 105 kD and a pI of 4.2 to 4.6 reported for purified barley limit dextrinase enzyme preparations (Sissons et al., 1992; Kristensen et al., 1998; MacGregor et al., 1994a). The apparent discrepancy between the deduced Mr value and the value obtained for the purified enzyme might result from glycosylation of the native enzyme; seven potential N-glycosylation sites are found in the deduced amino acid sequence (Fig. 2).
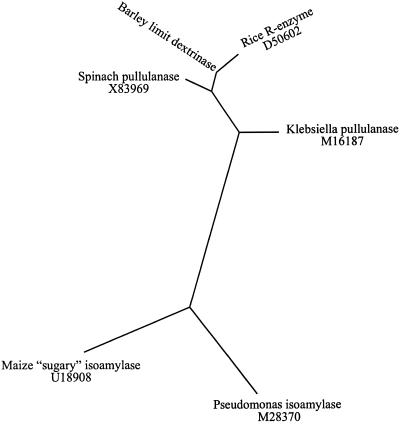
Alignment of the barley limit dextrinase amino acid sequence with other sequences in the protein and DNA databases reveals significant similarities with enzymes in the pullulanase and isoamylase groups of debranching enzymes from higher plants and from microbial sources (Fig. 8). When the amino acid sequences of similar debranching enzymes were examined for phylogenetic relatedness using the PileUp program (Devereux et al., 1984), two distinct groups could be distinguished (Fig. 9). Debranching enzymes of the pullulanase type, including representatives from higher plants and bacteria, were grouped together, whereas the isoamylase type fell into a second group (Fig. 9).
Figure 9.
Unrooted, radial phylogenetic tree of selected starch-debranching enzymes as determined using the PileUp, EprotDist, and EFitch programs of the University of Wisconsin package (version 8; Devereux et al., 1984). The two major branches distinguish the pullulanase type (upper branch) from the isoamylase type (lower branch). The accession numbers of the sequences are shown.
Glycosyl hydrolases have been classified into distinct families based on amino acid sequence similarities and hydrophobic cluster analyses (Henrissat and Bairoch, 1993), which suggest that the barley limit dextrinase is a member of the family 13 group of glycosyl hydrolases. On this basis, the catalytic nucleophile is likely to be Asp-472 and the catalytic acid Glu-509. In common with other family 13 enzymes, the barley limit dextrinase would be expected to retain anomeric configuration during hydrolysis of substrates (Jesperson et al., 1991).
A gene encoding the barley limit dextrinase was also isolated, and the sequence of an 11.5-kb genomic DNA fragment has been determined (Fig. 3). In an independent study, the cDNAs isolated here have been used to show that there is only one limit dextrinase gene in barley and that the gene is located on the long arm of chromosome 4H, where it has now been mapped (C.-D. Li, X.-Q. Zhang, R.C.M. Lance, L.C. MacLeod, G.B. Fincher, and P. Langridge, unpublished data). The most striking structural feature of the barley limit dextrinase gene is its highly fragmented nature: 26 introns are found in the coding region of the nascent polypeptide (Fig. 3), ranging in size from fewer than 100 nucleotides to more than 800 bp (Table I). Whether the highly fragmented nature of the barley limit dextrinase gene has evolutionary or functional significance is not known, but the transcriptional unit is certainly more complex than others that are expressed in aleurone layers of germinated barley (Fincher, 1989).
A cDNA encoding the barley limit dextrinase was used in northern-blot analyses to examine transcriptional patterns of the gene in various tissues. Limit dextrinase mRNA transcripts could not be detected by northern-blot analysis in developing grain (Fig. 5a), nor could they be detected in coleoptiles, young leaves, young roots, or scutellum of 1-d-germinated grain (data not shown). Limit dextrinase mRNA was observed only in northern blots of RNA preparations from isolated aleurone layers (Fig. 5a). The abundance of limit dextrinase mRNA was greatly enhanced by treatment of the aleurone layers with 2 μm GA3. The mRNA levels increased in the first 24 h after treatment and thereafter decreased (Fig. 5b). The addition of ABA abolished the GA3 induction of limit dextrinase mRNA (data not shown). The induction patterns of limit dextrinase gene transcription by GA3 in isolated barley aleurone layers are similar to those observed for (1→3,1→4)-β-glucanases (Mundy and Fincher, 1986), (1→4)-β-xylan endohydrolases (Banik et al., 1996), and α-amylases (Chandler et al., 1984). The GA3 induction of the limit dextrinase gene is consistent with the presence of nucleotide sequence elements in the promoter region of the gene that have previously been implicated in the GA3 induction of gene expression in barley aleurone layers (Fig. 5; Gubler and Jacobsen, 1992; Banik et al., 1997).
The expression patterns described above must be reconciled with the likely functions of limit dextrinase in barley, each of which involve the hydrolysis of (1→6)-α-glucosyl linkages in amylopectin or derived oligosaccharides. It is widely assumed that in germinated grain the enzyme is required for the complete depolymerization of stored starch to Glc, and the induction of limit dextrinase mRNA transcripts by GA3 in isolated aleurone layers (Fig. 5a) is consistent with this function. A second role that has been suggested for limit dextrinase is in starch synthesis in the developing grain. This could involve the processing of preamylopectin (Martin and Smith, 1995; Mouille et al., 1996; Nakamura et al., 1996; Rahman et al., 1998) or the provision of oligomeric primers for starch synthases (Duffus and Cochrane, 1993). Any role in starch synthesis would presumably be performed in amyloplasts of the developing grain, but could also occur in chloroplasts of leaves and other photosynthetic tissues. A third possible function of limit dextrinase is in the turnover of starch that accumulates transiently in chloroplasts and in amyloplasts in the parenchyma cells of the scutellum in germinated grain (Smart and O'Brien, 1973).
The apparent absence of significant levels of limit dextrinase mRNA in developing grains in the scutellum of germinated grain and in young vegetative tissues, as shown by northern-blot analyses, was therefore unexpected, particularly because the homologous limit dextrinase (R-enzyme) from rice was purified from the starchy endosperm of developing grain and the cDNA encoding the rice enzyme was generated from RNA prepared from the same tissue (Nakamura et al., 1996). When more sensitive RT-PCR methods were used, low levels of limit dextrinase mRNA were detected in the developing endosperm until about 20 DPA, when levels rapidly declined (Fig. 6a). These expression patterns correspond closely to the timing of starch synthesis in developing barley grains, although the latter is dependent to some extent on growth temperatures (MacLeod and Duffus, 1988). Given the relatively low levels of limit dextrinase mRNA in developing endosperm (Fig. 5), grain extracts were subsequently assayed for the enzyme itself. Activity in developing grain was detected at levels similar to those found initially in the germinated grain, and the results confirm earlier immunological studies suggesting that limit dextrinase protein is present in developing barley kernels (Sissons et al., 1993). The presence of low levels of limit dextrinase mRNA and enzyme activity in developing endosperm at times that coincide with the deposition of starch is consistent with the proposed role for the enzyme in amylopectin synthesis in cereals (Rahman et al., 1998), although it must be emphasized that the endosperm tissue used here did include some associated aleurone cells.
The RT-PCR method also confirmed that limit dextrinase mRNA was present in young leaves, young roots, and the scutellum of germinated grain (data not shown), but levels were low and we have not yet begun developmental studies of transcription in these tissues.
Clearly, the various functions that have been ascribed to limit dextrinases in starch synthesis and hydrolysis would require targeting of the enzyme to different subcellular compartments. Enzyme synthesized in the aleurone layer would be expected to be secreted into the starchy endosperm and to carry a signal peptide of the type that would target the nascent polypeptide to the ER for eventual secretion from the cell. On the other hand, targeting of the enzyme to amyloplasts in developing endosperm or to chloroplasts in leaves would require a transit-peptide-type targeting signal. Indeed, the 78-amino acid leader sequence on the limit dextrinase polypeptide (Fig. 2) is more typical of transit peptides that target polypeptides to plastids, and bears little resemblance to normal ER-type signal peptides (Fig. 2). Generally, transit peptides are longer than signal peptides, do not have extended hydrophobic regions, are rich in Ser, Thr, and Pro residues, and are highly charged (von Heijne et al., 1989; Baba et al., 1993; Edwards et al., 1995). These features are found in the barley limit dextrinase presequence (Fig. 2). The limit dextrinase transit peptide is also notable for the clear separation of positive charges and Pro residues toward the NH2 terminus, and negatively charged Glu residues toward the COOH terminus of the peptide (Fig. 2). These characteristics are also found in the rice R-enzyme presequence (Nakamura et al., 1996).
If a single limit dextrinase enzyme derived from a single gene were to participate in starch synthesis or turnover in chloroplasts or amyloplasts and were also secreted into the extracellular space from aleurone layer cells in germinated grain, one might anticipate that alternative splicing of pre-mRNA could be used to attach the appropriate targeting signal to the NH2 terminus of the nascent polypeptide. The highly fragmented nature of the barley limit dextrinase gene described here would allow such a mechanism. It is also noteworthy that a potential intron 3′ splicing site is located precisely adjacent to the NH2-terminal Ala residue of the mature enzyme (Fig. 2), where alternate splicing of introns encoding different targeting signals could occur. With this possibility in mind, we sequenced more than 4 kb upstream from the promoter sequence of the barley limit dextrinase gene shown in Figure 4 in an attempt to find signal-peptide sequences that would indicate targeting to the ER, but found no such sequences. Furthermore, when PCR and primer-extension techniques were applied to RNA from the GA3-treated aleurone cells, the presence of the transit-peptide sequence was always confirmed (data not shown).
Given that aleurone cells contain no starch (MacGregor and Fincher, 1993), are nonphotosynthetic, and that limit dextrinase is thought to be secreted into the starchy endosperm to catalyze debranching of limit dextrins, the presence of the transit peptide remains difficult to explain. Plastids have been reported in isolated barley aleurone layers after prolonged treatment with GA3 (Jones, 1969), and these could be the sites for limit dextrinase targeting. But how the enzyme would be released from the aleurone cells to be secreted into the starchy endosperm is not known. In a recent and comprehensive study of the development of limit dextrinase enzyme activity in de-embryonated barley half-grains and whole, germinated grains, Schroeder and MacGregor (1998) showed that GA3 caused a significant increase in total enzyme activity over the first 3 d of treatment, but little if any enzyme was secreted from the aleurone layer. Enzyme activity in the starchy endosperm remained low throughout and, because their extraction procedures were designed to release any limit dextrinase-inhibitor complexes, it seemed unlikely that much activity originated from the developing endosperm. The results of Schroeder and MacGregor (1998) are entirely consistent with those reported here.
It might be argued that limit dextrinase is not required in the starchy endosperm until the final stages of reserve mobilization, when the enzyme could mediate in the recovery of the last few glucosyl residues that are bound in the limit dextrins. Perhaps the enzyme might be released if, after their reserves were depleted, aleurone layer cells eventually burst. Morphological examinations of senescing aleurone layers from both wheat and barley show that the cells do indeed die after prolonged treatment with GA3 (Kuo et al., 1996; Wang et al., 1996), but this process is characteristic of programmed cell death rather than necrosis. In programmed cell death, DNA fragmentation occurs to produce oligonucleosome-sized fragments in a fashion similar to apoptosis, the cytoplasm and nuclei of dead and dying cells collapse and shrink, but membrane integrity appears to be maintained and there is little leakage of cell contents (Pennell and Lamb, 1997).
It has been proposed that the retention of membrane structure would offer some protection against pathogen attack, in contrast to necrotic cell death, in which cell leakage usually occurs and releases nutrients that could support microbial growth (Pennell and Lamb, 1997). The programmed cell death that is observed in cereal aleurone (Kuo et al., 1996; Wang et al., 1996) is therefore unlikely to result in the wholesale release of accumulated limit dextrinase. In support of this, Schroeder and MacGregor (1998) showed that only very small amounts of limit dextrinase were secreted from aleurone cells after 5 d of treatment with GA3, and at this stage programmed cell death would have been well advanced (Wang et al., 1996).
In conclusion, the results presented here are consistent with a role for limit dextrinase in starch synthesis in developing barley grain, but raise some questions as to the involvement of the enzyme in starch degradation during the mobilization of the starchy endosperm in germinated grain. Perhaps a previously undetected, isoamylase-like enzyme is more important for amylopectin degradation in germinated barley grain. Isoamylase has been described in maize (James et al., 1995; Rahman et al., 1998) and, although it could debranch amylopectin during starch degradation, it would go undetected in the pullulanase assays that are commonly used to quantitate limit dextrinase activity.
ACKNOWLEDGMENTS
We gratefully acknowledge the assistance and skill of Drs. Lin Chen, Ann MacGregor, and Peilin Xu with various aspects of the work.
Abbreviations:
- DPA
days postanthesis
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- RACE
rapid amplification of cDNA ends
- RT
reverse transcriptase
Footnotes
This work was supported by a grant from the South East Australian Malting Barley Quality Improvement Program, which includes contributions from Joe White Maltings, the Grains Research and Development Corporation, the Strategic Research Foundation, Barrett-Burston International, the Australian Associated Brewers, and the Adelaide Malting Company.
LITERATURE CITED
- Aman P, Hesselman K, Tilly A-C. The variation in chemical composition of Swedish barleys. J Cereal Sci. 1985;3:73–77. [Google Scholar]
- Amemura A, Chakraborty R, Fujita M, Noumi T, Futai M. Cloning and nucleotide sequence of the isoamylase gene from Pseudomonas amyloderamosa SB-15. J Biol Chem. 1988;263:9271–9275. [PubMed] [Google Scholar]
- Baba T, Nishihara M, Mizuno K, Kawasaki T, Shimada H, Kobayashi E, Ohnishi S, Tanaka K-I, Arai Y. Identification, cDNA cloning, and gene expression of soluble starch synthase in rice (Oryza sativa L.) immature seeds. Plant Physiol. 1993;103:565–573. doi: 10.1104/pp.103.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball S, Guan H-P, James M, Myers A, Keeling P, Mouille G, Buleon A, Colonna P, Preiss J. From glycogen to amylopectin: a model for the biogenesis of the plant starch granule. Cell. 1996;86:349–352. doi: 10.1016/s0092-8674(00)80107-5. [DOI] [PubMed] [Google Scholar]
- Banik M, Garrett TPJ, Fincher GB. Molecular cloning of cDNAs encoding (1→4)-β-xylan endohydrolases from the aleurone layer of germinated barley (Hordeum vulgare) Plant Mol Biol. 1996;31:1163–1172. doi: 10.1007/BF00040833. [DOI] [PubMed] [Google Scholar]
- Banik M, Li C-D, Langridge P, Fincher GB. Structure, hormonal regulation, and chromosomal location of genes encoding barley (1→4)-β-xylan endohydrolases. Mol Gen Genet. 1997;253:599–608. doi: 10.1007/s004380050362. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cavener DR, Ray SC. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991;19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler PM, Jacobsen JV. Primer extension studies on α-amylase mRNAs in barley aleurone. II. Hormonal regulation of expression. Plant Mol Biol. 1991;16:637–645. doi: 10.1007/BF00023428. [DOI] [PubMed] [Google Scholar]
- Chandler PM, Zwar JA, Jacobsen JV, Higgins TJV, Inglis AS. The effects of gibberellic acid and abscisic acid on α-amylase mRNA levels in barley aleurone layers studied using an α-amylase cDNA clone. Plant Mol Biol. 1984;3:407–418. doi: 10.1007/BF00033389. [DOI] [PubMed] [Google Scholar]
- Chen L, Fincher GB, Hoj PB. Evolution of polysaccharide hydrolase substrate specificity: catalytic amino acids are conserved in barley (1→3,1→4)-glucanase and (1→3)-β-glucanase. J Biol Chem. 1993;268:13318–13326. [PubMed] [Google Scholar]
- Chrispeels MJ, Varner JE. Gibberellic acid enhanced synthesis and release of α-amylase and ribonuclease by isolated barley aleurone layers. Plant Physiol. 1967;42:398–406. doi: 10.1104/pp.42.3.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffus CM, Cochrane MP (1993) Development, structure and composition of the barley kernel. In AW MacGregor, RS Bhatty, eds, Barley: Chemistry and Technology. American Association of Cereal Chemists, St. Paul, MN, pp 31–72
- Edwards A, Marshall J, Sidebottom C, Visser RGF, Smith AM, Martin C. Biochemical and molecular characterisation of a novel starch synthase from potato tubers. Plant J. 1995;8:283–294. doi: 10.1046/j.1365-313x.1995.08020283.x. [DOI] [PubMed] [Google Scholar]
- Fincher GB. Molecular and cellular biology associated with endosperm mobilization in germinating cereal grains. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:305–346. [Google Scholar]
- Frohman M, Dush M, Martin G. Rapid amplification of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Jacobsen JV. Gibberellin-responsive elements in the promoter of a barley high-pI α-amylase gene. Plant Cell. 1992;4:1435–1441. doi: 10.1105/tpc.4.11.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Kalla R, Roberts JK, Jacobsen JV. Gibberellin-regulated expression of a Myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell. 1995;7:1879–1891. doi: 10.1105/tpc.7.11.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann M-L, Geuss U, Fischer S, Kresse G-B. Peptidyl-Asp metalloendopeptidase. Methods Enzymol. 1995;20:782–787. doi: 10.1016/0076-6879(95)48053-6. [DOI] [PubMed] [Google Scholar]
- Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrmova M, Fincher GB. Purification and properties of three (1→3)-β-glucanase isoenzymes from young leaves of barley (Hordeum vulgare) Biochem J. 1993;289:453–461. doi: 10.1042/bj2890453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James MG, Robertson DS, Meyers AM. Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell. 1995;7:417–429. doi: 10.1105/tpc.7.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesperson HM, MacGregor EA, Sierks MR, Svensson B. Comparison of the domain-level organization of starch hydrolases and related enzymes. Biochem J. 1991;280:51–55. doi: 10.1042/bj2800051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RL. Gibberellic acid and the fine structure of barley aleurone cells. II. Changes during the synthesis and secretion of α-amylase. Planta. 1969;88:73–86. doi: 10.1007/BF00396117. [DOI] [PubMed] [Google Scholar]
- Joshi CP. An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res. 1987;15:6643–6653. doi: 10.1093/nar/15.16.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuragi N, Takizawa N, Murooka Y. Entire nucleotide sequence of the pullulanase gene of Klebsiella aerogenes W70. J Bacteriol. 1987;169:2301–2306. doi: 10.1128/jb.169.5.2301-2306.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen M, Planchot V, Abe J-i, Svensson B. Large scale purification and characterization of barley limit dextrinase, a member of the α-amylase structural family. Cereal Chem. 1998;75:473–479. [Google Scholar]
- Kuo A, Cappelluti S, Cervantes-Cervantes M, Rodriguez M, Bush DS. Okadaic acid, a protein phosphatase inhibitor, blocks calcium changes, gene expression, and cell death induced by gibberellin in wheat aleurone cells. Plant Cell. 1996;8:259–269. doi: 10.1105/tpc.8.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EY, Marshall JJ, Whelan WJ. The substrate specificity of amylopectin-debranching enzymes from sweet corn. Arch Biochem Biophys. 1971;143:365–374. doi: 10.1016/0003-9861(71)90223-2. [DOI] [PubMed] [Google Scholar]
- Longstaff MA, Bryce JH. Development of limit dextrinase in germinated barley (Hordeum vulgare L.). Evidence of proteolytic activation. Plant Physiol. 1993;101:881–889. doi: 10.1104/pp.101.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutcke HA, Chow KC, Mickel FS, Moss KA, Kern HF, Scheele GA. Selection of AUG initiation codons differs in plants and animals. EMBO J. 1987;6:43–48. doi: 10.1002/j.1460-2075.1987.tb04716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor AW, Fincher GB (1993) Carbohydrates of the barley grain. In AW MacGregor, RS Bhatty, eds, Barley: Chemistry and Technology. American Association of Cereal Chemists, St. Paul, MN, pp 73–130
- MacGregor AW, Macri LJ, Schroeder SW, Bazin SL. Limit dextrinase from malted barley: extraction, purification, and characterization. Cereal Chem. 1994a;71:610–617. [Google Scholar]
- MacGregor AW, Macri LJ, Schroeder SW, Bazin SL. Purification and characterisation of limit dextrinase inhibitors from barley. J Cereal Sci. 1994b;20:33–41. [Google Scholar]
- MacLeod LC, Duffus CM. Temperature effects on starch granules in developing barley grains. J Cereal Sci. 1988;8:29–37. [Google Scholar]
- Martin C, Smith AM. Starch synthesis. Plant Cell. 1995;7:971–985. doi: 10.1105/tpc.7.7.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleary BV. Measurement of the content of limit dextrinase in cereal flours. Carbohydr Res. 1992;227:257–268. [Google Scholar]
- Mouille G, Maddelein ML, Libessart N, Talaga P, Decq A, Delrue B, Ball S. Preamylopectin processing: a mandatory step for starch biosynthesis in plants. Plant Cell. 1996;8:1353–1366. doi: 10.1105/tpc.8.8.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy J, Fincher GB. Effects of gibberellic acid and abscisic acid on levels of translatable mRNA for (1→3,1→4)-β-glucanase in barley aleurone. FEBS Lett. 1986;198:349–352. [Google Scholar]
- Nakamura Y, Kubo A, Shimamune T, Matsuda T, Harada K, Satoh H. Correlation between activities of starch debranching enzyme and alpha-polyglucan structure in endosperms of sugary-1 mutants of rice. Plant J. 1997;12:143–153. [Google Scholar]
- Nakamura Y, Umemoto T, Ogata N, Kuboki Y, Yano M, Sasaki T. Starch debranching enzyme (R-enzyme or pullulanase) from developing rice endosperm: purification, cDNA and chromosomal localization of the gene. Planta. 1996;199:209–218. doi: 10.1007/BF00196561. [DOI] [PubMed] [Google Scholar]
- Pan D, Nelson OE. A debranching enzyme deficiency in endosperm of the sugary-1 mutants of maize (Zea mays) Plant Physiol. 1984;74:324–328. doi: 10.1104/pp.74.2.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell RI, Lamb C. Programmed cell death in plants. Plant Cell. 1997;9:1157–1168. doi: 10.1105/tpc.9.7.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Wong K, Jane J, Myers AM, James MG. Characterization of SU1 isoamylase, a determinant of storage starch structure in maize. Plant Physiol. 1998;117:425–435. doi: 10.1104/pp.117.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder SW, MacGregor AW. Synthesis of limit dextrinase in germinated barley kernels and aleurone tissues. J Am Soc Brew Chem. 1998;56:32–37. [Google Scholar]
- Simpson GG, Filipowicz W. Splicing of precursors to mRNA in higher plants: mechanism, regulation and sub-nuclear organization of the spliceosomal machinery [Review] Plant Mol Biol. 1996;32:1–41. doi: 10.1007/BF00039375. [DOI] [PubMed] [Google Scholar]
- Sissons MJ, Lance RCM, Sparrow DHB. Studies on limit dextrinase in barley. I. Purification of malt limit dextrinase and production of monospecific antibodies. J Cereal Sci. 1992;16:107–116. [Google Scholar]
- Sissons MJ, Lance RCM, Sparrow DHB. Studies on limit dextrinase in barley. 3. Limit dextrinase in developing kernels. J Cereal Sci. 1993;17:19–24. [Google Scholar]
- Slakeski N, Baulcombe DC, Devos KM, Doan DNP, Fincher GB. Structure and tissue-specific regulation of genes encoding barley (1→3,1→4)-β-glucan endohydrolases. Mol Gen Genet. 1990;224:437–449. doi: 10.1007/BF00262439. [DOI] [PubMed] [Google Scholar]
- Smart MG, O'Brien TP. Observations on the scutellum. II. Histochemistry and autofluorescence of the cell wall in mature grain and during germination of wheat, barley, oats and ryegrass. Aust J Bot. 1973;27:403–411. [Google Scholar]
- von Heijne G, Steppuhn J, Herrmann RG. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- Wang M, Oppedijk BJ, Lu X, Duijn BV, Schilperoort RA. Apoptosis in barley aleurone during germination and its inhibition by abscisic acid. Plant Mol Biol. 1996;32:1125–1134. doi: 10.1007/BF00041396. [DOI] [PubMed] [Google Scholar]