Abstract
Objective
Femoral neck geometric parameters (FNGPs), such as periosteal diameter (W), cross-sectional area (CSA), cortical thickness (CT), buckling ratio (BR), and section modulus (Z), are highly genetically correlated with body lean mass. However, the specific SNPs/genes shared by these phenotypes are largely unknown.
Methods
To identify the specific SNPs/genes shared between FNGPs and appendicular lean mass (ALM), we performed an initial bivariate genome-wide association study (GWAS) by scanning ∼690,000 SNPs in 1,627 unrelated Han Chinese adults (802 males and 825 females) and a follow-up replicate study in 2,286 unrelated US Caucasians.
Results
We identified 13 interesting SNPs that may be important for both FNGPs and ALM. Two SNPs, rs681900 located in the HK2 (hexokinase 2) gene and rs11859916 in the UMOD (uromodulin) gene, were bivariately associated with FNGPs and ALM (p = 7.58×10−6 for ALM-BR and p = 2.93×10−6 for ALM-W, respectively). The associations were then replicated in Caucasians, with corresponding p values of 0.024 for rs681900 and 0.047 for rs11859916. Meta-analyses yielded combined p values of 3.05×10−6 and 2.31×10−6 for rs681900 and rs11859916, respectively. Our findings are consistent with previous biological studies that implicated HK2 and UMOD in both FNGPs and ALM. Our study also identified a group of 11 contiguous SNPs, which spanned a region of ∼130 kb, were bivariately associated with FNGPs and ALM, with p values ranging from 3.06×10−7 to 4.60×10−6 for ALM-BR. The region contained two neighboring miRNA coding genes, MIR873 (MicroRNA873) and MIR876 (MicroRNA876).
Conclusion
Our study implicated HK2, UMOD, MIR873 and MIR876, as pleiotropic genes underlying variation of both FNGPs and ALM, thus suggesting their important functional roles in co-regulating both FNGPs and ALM.
Introduction
Osteoporosis is a common disease, particularly among the elderly, characterized by decreased bone strength and increased fracture risk [1], [2]. Hip fracture is the most common and serious type of osteoporotic fracture, often producing prolonged or permanent disability, or even death, for some patients [1], [3]. The musculoskeletal system, however, contains both bone and muscle, and these two tissue types are highly interdependent. Bones sustain mechanical loads and provide load points for muscles, and muscles keep bones in place and are responsible for major mechanical loading of bones [4].
Due to the morbidity, mortality, and health care costs associated with osteoporotic fractures, a variety of phenotypic characteristics have been analyzed for their associations with bone strength, and fracture risk. Bone mineral density (BMD) is considered to be an important, but not exclusive, determining factor for bone strength, and is also associated with fracture risk [5], [6]. Bone geometry, independent of BMD, is another important factor, that determines bone strength and is directly associated with osteoporotic fractures [7]. Several recent studies have reported that femoral neck geometric parameters (FNGPs) such as periosteal diameter (W), cross-sectional area (CSA), cortical thickness (CT), buckling ratio (BR), and section modulus (Z), can be used to improve the accuracy of identifying people at high risk of hip fracture [3], [8], [9], [10].
The muscular component of the musculosketal system, as defined by body lean mass, is also closely associated with human health. Low body lean mass is associated with a series of health problems, such as sarcopenia, impaired protein balance, obesity, and osteoporosis [3], [11]. Not surprisingly, body lean mass and FNGPs are closely related phenotypes. It has been demonstrated that bone geometry can serve as a useful index that represents adaptive responses of bone to altered mechanical loading [12]. Body lean mass, in turn, has been shown to contribute to variations of bone geometry at the femoral neck (FN) [13], [14], potentially due to genetic, mechanical, hormonal and nutritional factors. For example, dynamic strains provided by muscle may be an important stimulus of bone adaptation [14]. From the genetic perspective, previous bivariate quantitative genetic analyses have shown that body lean mass was significantly correlated with FNGPs, and that these phenotypic traits might share some common genetic factors [15]. Subsequently, bivariate whole genome linkage analysis reported that several genomic regions, such as 3q12 and 20q13, were linked with both FNGPs and body lean mass [16]. However, the specific SNPs/genes that are shared between these two phenotypes are largely unknown.
Bivariate GWAS is a newly developed effective approach to detect pleiotropic genes for complex traits [17]. To identify the specific pleiotropic SNPs/genes that contribute to both FNGPs and body lean mass, we performed an initial bivariate GWAS in a large Chinese sample, and a follow-up replicate study in Caucasians. We utilized appendicular lean mass (ALM) as our determinant of lean mass, as several studies have suggested that ALM is a better proxy measure of body skeletal muscle mass than total body lean mass for assessing exercise capacity and predicting related diseases [18], [19], [20]. ALM is calculated as the sum of lean mass in the arms and legs.
Results
The basic characteristics of the study subjects are shown in Table 1. Generally, FNGPs and ALM in men were higher than those in women. All study parameters, except CT in men, were higher in Caucasians than in Chinese subjects.
Table 1. Basic characteristics of study subjects.
| Traits | Chinese sample | US sample | |||
| Male (N = 802) | Female (N = 825) | Male (N = 558) | Female (N = 1728) | ||
| Age (years) | 31.44±11.98 | 37.44±13.78 | 50.19±16.03 | 51.13±13.00 | |
| W (cm) | 3.54±0.27 | 3.12±0.33 | 3.82±0.33 | 3.30±0.34 | |
| CSA (cm2) | 2.92±0.50 | 2.25±0.37 | 3.08±0.59 | 2.45±0.47 | |
| CT (cm) | 0.17±0.03 | 0.14±0.02 | 0.16±0.03 | 0.15±0.03 | |
| Z (cm3) | 1.84±0.38 | 1.26±0.29 | 2.12±0.49 | 1.45±0.36 | |
| BR | 10.97±2.08 | 11.08±2.76 | 12.29±2.57 | 11.46±2.43 | |
| ALM (kg) | 23.95±3.20 | 15.75±2.08 | 29.89±4.85 | 20.22±3.53 | |
Note: all the values are means ± SD.
Correlation analysis using our phenotypic data have shown that body lean mass was significantly correlated with W, CSA, CT and Z (Table 2), which is generally consistent with previous studies [15], [16].
Table 2. Results of bivariate GWAS for ALM and five FNGPs (p<1×10−5).
| Phenotypes pair | SNP | Chr. | Position | Gene | Allele1 | MAF2 | MAF3 | Bivariate p | Replication p |
| ALM-W | rs6789283 | 3 | 21996951 | - | C/T | 0.041 | 0.034 | 1.34×10−7 | 0.774 |
| (0.573**)4 | rs854140 | 5 | 56985279 | - | G/A | 0.462 | 0.467 | 3.50×10−6 | 0.825 |
| (0.501**)5 | rs17037864 | 4 | 160113841 | C4orf45 | A/G | 0.364 | 0.378 | 3.93×10−6 | 0.865 |
| rs2062713 | 12 | 113660935 | - | T/C | 0.289 | 0.333 | 7.33×10−6 | 0.592 | |
| rs681900 | 2 | 74928475 | HK2 | G/A | 0.162 | 0.133 | 7.58×10−6 | 0.024 | |
| ALM-CSA | rs4804662 | 19 | 7481408 | - | G/A | 0.231 | 0.211 | 4.72×10−7 | 0.216 |
| (0.729**) | rs10928979 | 2 | 127136698 | - | G/A | 0.371 | 0.433 | 6.46×10−7 | 0.441 |
| (0.642**) | rs6789283 | 3 | 21996951 | - | C/T | 0.041 | 0.011 | 2.50×10−6 | 0.531 |
| rs12098712 | 10 | 5601911 | - | T/C | 0.110 | 0.122 | 9.56×10−6 | 0.117 | |
| ALM-CT | rs4507747 | 8 | 141288307 | TRAPPC9 | T/C | 0.013 | 0.000 | 2.32×10−6 | 0.429 |
| (0.524**) | rs10928979 | 2 | 127136698 | - | G/A | 0.371 | 0.433 | 3.74×10−6 | 0.766 |
| (0.450**) | rs4804662 | 19 | 7481408 | - | G/A | 0.231 | 0.211 | 3.76×10−6 | 0.116 |
| ALM-Z | rs6789283 | 3 | 21996951 | - | C/T | 0.041 | 0.011 | 7.46×10−8 | 0.656 |
| (0.768**) | rs10928979 | 2 | 127136698 | - | G/A | 0.371 | 0.433 | 9.58×10−7 | 0.491 |
| (0.679**) | rs2081106 | 5 | 58514356 | PDE4D | T/A | 0.076 | 0.056 | 4.91×10−6 | 0.650 |
| rs4804662 | 19 | 7481408 | - | G/A | 0.231 | 0.211 | 6.19×10−6 | 0.222 | |
| rs12098712 | 10 | 5601911 | - | T/A | 0.110 | 0.122 | 8.72×10−6 | 0.075 | |
| ALM-BR | rs1368998 | 9 | 28869953 | MIR873 | T/G | 0.115 | 0.111 | 3.06×10−7 | 0.961 |
| (-0.095**) | rs12005658 | 9 | 28858243 | MIR876 | A/G | 0.116 | 0.111 | 3.96×10−7 | 0.976 |
| (-0.113**) | rs16913782 | 9 | 28866158 | MIR876 | T/C | 0.116 | 0.111 | 4.66×10−7 | 0.970 |
| rs969715 | 9 | 28810807 | MIR876 | C/T | 0.117 | 0.122 | 5.49×10−7 | 0.943 | |
| rs11998784 | 9 | 28865332 | MIR876 | G/A | 0.116 | 0.111 | 8.12×10−7 | 0.997 | |
| rs1389728 | 9 | 28807957 | MIR876 | C/T | 0.116 | 0.122 | 9.74×10−7 | 0.569 | |
| rs13299777 | 9 | 28929554 | MIR873 | C/T | 0.116 | 0.122 | 9.83×10−7 | 0.407 | |
| rs3849874 | 9 | 28859369 | MIR876 | C/G | 0.116 | 0.111 | 1.13×10−6 | 0.968 | |
| rs16913751 | 9 | 28843125 | MIR876 | T/C | 0.119 | 0.111 | 1.64×10−6 | 0.939 | |
| rs854140 | 5 | 56985279 | - | G/A | 0.462 | 0.467 | 2.10×10− 6 | 0.773 | |
| rs10491629 | 9 | 28845155 | MIR876 | T/G | 0.117 | 0.116 | 2.91×10−6 | 0.911 | |
| rs11859916 | 16 | 20258732 | UMOD | A/G | 0.227 | 0.200 | 2.93×10−6 | 0.047 | |
| rs10491633 | 9 | 28933695 | MIR873 | T/C | 0.115 | 0.122 | 4.60×10−6 | 0.293 | |
| rs4804662 | 19 | 7481408 | - | G/A | 0.231 | 0.211 | 8.10×10− 6 | 0.160 |
Note:
Bold font: the identified 13 SNPs.
The first allele represents the minor allele of each locus.
Minor allele frequency calculated in our own Chinese subjects.
Minor allele frequency reported for Chinese in the public HapMap HCB database.
Phenotype correlation for Chinese sample. ** p≤0.01.
Phenotype correlation for US data. ** p≤0.01.
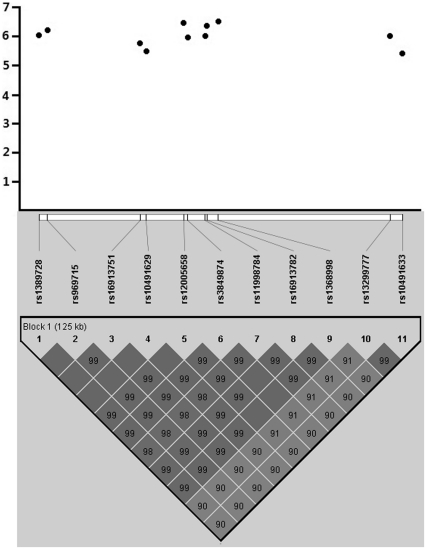
We identified 13 interesting SNPs that were bivariately associated with ALM and FNGPs. Among them, SNP rs681900 located in intron 1 of the hexokinase 2 gene (HK2), ranked among the top 5 SNPs for bivariate association with ALM-W (p = 7.58×10−6). Another SNP, rs11859916 of the uromodulin gene (UMOD) ranked among the top 3 loci for associations with ALM-BR (p = 2.93×10−6). We also found replicate association for these two SNPs in Caucasians, with corresponding p values of 0.024 for rs681900 and 0.047 for rs11859916 (Table 2). Meta-analyses yielded stronger associations, with pooled p values of 3.05×10−6 and 2.31×10−6 for rs681900 and rs11859916, respectively. A group of 11 contiguous SNPs spanning ∼130 kb region harboring the MIR876 (MicroRNA876) and MIR873 (MicroRNA873) genes were strongly associated with ALM-BR, with p values ranging from 3.06×10−7 to 4.60×10−6. Among them, two SNPs, rs12005658 and rs3849874, were located in the promoter region of the MIR876 gene. As shown in Figure 1, LD signals within this SNP group were very strong and all 11 SNPs were within one haplotype block.
Figure 1. Bivariate associations of 11 contiguous SNPs with ALM-BR in the region of the MIR876 and MIR873 genes.
Notes: The Y axis is the negative Log10p values. The LD between two SNPs is standardized D' (D/Dmax).
Since the five bone geometry parameters are closely related, we compared the results from the five phenotype pairs for the 13 interesting SNPs to detect common and/or specific SNPs for the five phenotype pairs. As shown in Table 3, three phenotype pairs (ALM-W, ALM-CT, and ALM-BR) generally have stronger association signals than other two (ALM-CSA and ALM-Z) for the 13 interesting SNPs. For SNP rs681900, there were consistently strong association signals across the five phenotype pairs, with p values ranging from 1.57×10−4 to 7.58×10−6, probably suggesting that this SNP has a common effect on all five phenotype pairs.
Table 3. Bivariately/univariate associations for 13 interesting SNPs.
| SNP | Role | Bivariate p values | univariate p values | ||||||||||
| ALM-W | ALM-CSA | ALM-CT | ALM-Z | ALM-BR | ALM | W | CSA | CT | Z | BR | |||
| rs681900 | Intron1 | 7.58×10−6 | 1.57×10−4 | 5.44×10−5 | 9.39×10−5 | 3.01×10−5 | 0.59 | 0.06 | 0.90 | 0.61 | 0.95 | 0.46 | |
| rs11859916 | Intron7 | 1.58×10−2 | 6.01×10−2 | 3.67×10−4 | 4.12×10−1 | 2.93×10−6 | 0.72 | 0.03 | 0.09 | 0.55 | 0.03 | 0.68 | |
| rs12005658 | Promoter | 4.60×10−5 | 6.42×10−3 | 4.14×10−5 | 4.61×10−2 | 3.96×10−7 | 0.84 | 0.87 | 0.36 | 0.45 | 0.76 | 0.98 | |
| rs16913782 | Upstream | 3.37×10−5 | 7.96×10−3 | 6.55×10−5 | 4.18×10−2 | 4.66×10−7 | 0.93 | 0.87 | 0.40 | 0.52 | 0.80 | 0.93 | |
| rs969715 | Downstream | 3.06×10−5 | 1.97×10−2 | 9.41×10−5 | 8.58×10−2 | 5.49×10−7 | 0.82 | 0.97 | 0.64 | 0.44 | 0.89 | 0.78 | |
| rs11998784 | Upstream | 9.41×10−5 | 9.25×10−3 | 7.37×10−5 | 6.61×10−2 | 8.12×10−7 | 0.96 | 1.00 | 0.37 | 0.44 | 0.82 | 0.87 | |
| rs1389728 | Downstream | 4.16×10−5 | 2.31×10−2 | 1.61×10−4 | 8.96×10−2 | 9.74×10−7 | 0.80 | 0.99 | 0.63 | 0.44 | 0.87 | 0.79 | |
| rs3849874 | Promoter | 8.96×10−5 | 7.99×10−3 | 9.57×10−5 | 4.95×10−2 | 1.13×10−6 | 0.89 | 0.88 | 0.39 | 0.50 | 0.79 | 0.96 | |
| rs16913751 | Downstream | 1.20×10−4 | 1.07×10−2 | 1.42×10−4 | 6.53×10−2 | 1.64×10−6 | 0.93 | 0.98 | 0.55 | 0.38 | 0.81 | 0.76 | |
| rs10491629 | Downstream | 1.37×10−4 | 1.04×10−2 | 2.43×10−4 | 4.82×10−2 | 2.91×10−6 | 0.91 | 0.96 | 0.57 | 0.36 | 0.85 | 0.73 | |
| rs1368998 | Downstream | 3.18×10−5 | 6.29×10−3 | 3.33×10−5 | 4.82×10−2 | 3.06×10−7 | 0.94 | 0.81 | 0.28 | 0.37 | 0.64 | 0.96 | |
| rs13299777 | Upstream | 8.76×10−5 | 1.51×10−3 | 1.91×10−5 | 3.27×10−2 | 9.83×10−7 | 0.65 | 0.81 | 0.93 | 0.68 | 0.70 | 0.71 | |
| rs10491633 | Upstream | 1.95×10−4 | 3.21×10−3 | 8.28×10−5 | 4.42×10−2 | 4.60×10−6 | 0.58 | 0.77 | 0.79 | 0.80 | 0.60 | 0.76 | |
As shown in Table 3, the bivariate association signals were generally stronger than the univariate association signals, suggesting that bivariate analysis has a higher power to detect shared genetic factors for related phenotype pairs [17]. For example, with SNP rs681900, the univariate associations were not significant for either ALM or W, but bivariate associations were consistently significant for all phenotypic pairs, with p values ranging from 1.57×10−4 to 7.58×10−6. To provide readers more details about the bivariate GWAS, all the SNPs with p value less than 10−4 were demonstrated in Appendix S1.
Discussion
Using the novel multivariate approach, we performed a bivariate GWAS for ALM and FNGPs, and found that 13 interesting SNPs located within or near four genes, HK2, UMOD, MIR876 and MIR873, showed strong associations with both femoral neck bone geometry and ALM. The present study represents the first effort to detect shared genetic factors for these closely related phenotypes (ALM and FNGPs).
GWAS provides impressive statistical power for detecting novel genetic variants that underlie common human diseases. To date, however, most published GWAS's utilize a univariate framework to analyze different phenotypes separately. Although these univariate GWA analyses have led to the discovery of novel genes for several complex diseases, this approach often lacks sufficient power to detect pleiotropic genes that may influence multiple phenotypes. Newly developed methodologies for bivariate GWAS's have higher power for detecting pleiotropic genes than univariate approaches [17], [21]. The results shown in Table 3 clearly demonstrate the advantage of bivariate association studies for identifying pleiotropic genes.
The importance of the 13 interesting SNPs is supported by both the statistical evidence provided in this manuscript, and by the known functions of the four genes at the genomic regions containing these 13 interesting SNPs. HK2 encodes the protein hexokinase 2, the predominant form of hexokinase presented in skeletal muscle. Hexokinase is responsible for phosphorylating glucose to produce glucose-6-phosphate, the first step in most glucose metabolism pathways. Hexokinase 2 is one of the key enzymes involved in regulating glucose metabolism for muscle tissue [22], [23]. Glucose metabolism, one of the most basic cellular biochemical reactions, provides energy and material for fundamental cellular activities such as protein metabolism, cell growth, and proliferation [24], [25], [26], [27]. These activities are essential for normal muscle growth, and may influence lean mass in human [28]. Glucose metabolism is also associated with bone development, as elevated glucose levels have been shown to inhibit calcium uptake and bone mineralization [29]. Moreover, bone resorption is dependent on glucose concentrations. Reduced expression of HK2 is also associated with non-insulin-dependent diabetes, which may lead to osteoporosis and losses in lean mass [30], [31]. This collective information strongly supports the conclusion that HK2 is involved in both muscle and bone metabolism.
The UMOD gene encodes uromodulin, the most abundant protein in mammalian urine. Uromodulin may influence both bone and muscle by regulating renal excretion of metabolites. Uromodulin is involved in calcium metabolism, and acts as a regulator of calcium oxalate crystallization [32]. Missense mutation of UMOD in mice causes moderate increases in plasma calcium concentrations, and significant decreases in bone mineral density and bone mineral content [33]. Uromodulin may also influence lean mass through creatinine, energy, and protein metabolism, as mice with UMOD mutations have reduced body weight, including body lean mass, and significant increases in plasma creatinine and urea levels, compared with normal mice [33].
Of particular interest, a group of 11 contiguous SNPs locates within genomic regions containing two miRNA encoding genes (MIR876 and MIR873) were strongly associated with ALM-BR in Chinese. Two of the 11 SNPs, rs12005658 and rs3849874 were located in the promoter region of MIR876. MicroRNAs (miRNAs) are involved in post-transcriptional regulation of gene expression in multicellular organisms by affecting both the stability and translation of mRNA. We used the TargetScan platform (http://www.targetscan.org release 5.1), web-based software with a low false positive rate, to identify predicted miRNA target genes. MIR876 encodes two MicroRNAs: miR-876–3p and miR-876–5p. Using TargetScan software, 93 and 148 genes were predicted to be targets for miR-876–3p and miR-876–5p, respectively. Among them, ACTN4 (actinin, alpha 4) [34], MBNL1 (muscleblind-like) [35], and SEPN1 (selenoprotein N, 1) [36] are involved in muscle metabolism, and ANKH (ankylosis, progressive homolog) [37], TAPT1 (transmembrane anterior posterior transformation 1) [38], and TRPS1 (trichorhinophalangeal syndrome I) [39] are related to bone metabolism. For MIR873, 176 target genes were identified using the TargetScan platform. Several of these genes are also involved in muscle or bone metabolism. For example, DMD (dystrophin) and BMP7 (bone morphogenetic protein 7) are associated with muscle and bone metabolism, respectively [40]. However, the exact mechanisms by which MIR876 and MIR873 are involved in co-regulating bone and muscle metabolism are unclear. Functional studies are being planned to validate our findings.
The association results from our initial Chinese sample were replicated in a Caucasian population. Much of the genetic background across the two ethnic groups is similar, which indicates that mutual replication between different ethnic groups is feasible. We found consistent associations across the two ethnic groups for two SNPs (rs681900 and rs11859916), suggesting the consistent effects of these two SNPs on both Chinese and Caucasians. However, ethnic genetic heterogeneity has also been observed among different races when studying specific phenotypes or diseases [41], [42]. Our study found a group of 11 contiguous SNPs ranked at the top for bivariate association with ALM and BR in the Chinese sample, but these findings were not replicated in Caucasians. It is possible that the failure to replicate these findings in Caucasians was attributable to genetic diversity in this region between Chinese and Caucasians.
In conclusion, we used a novel bivariate GWAS approach in a large Chinese sample, and a follow-up replication study in Caucasians, combined with functional evidence, to identify two genes, HK2 and UMOD, that appear to co-regulate FNGPs and ALM. Two additional MicroRNA genes (MIR873 and MIR876) were also associated with bone geometry and ALM in Chinese, but these findings were not replicated in Caucasians. These findings enhance our knowledge of genetic associations between bone geometry and ALM, and provide a rationale for subsequent functional studies of these implicated genes in the pathophysiology of diseases related to these phenotypes, such as hip fracture and sarcopenia.
Materials and Methods
Subjects and phenotypes
The study was approved by the institutional review boards of Hunan Normal University, Xian Jiao Tong University and University of Missouri-Kansas City. All study participants signed informed consent documents before they entered the project. Two independent samples were included in this study: a sample of 1,627 unrelated adult Han Chinese (802 males and 825 females) recruited from Changsha and Xi'an and their surroundings areas, and another sample of 2,286 unrelated homogeneous US Caucasians (including 558 males and 1,728 females) recruited from the Midwestern US in Kansas City, Missouri and Omaha, Nebraska. Anthropometric measures and a structured questionnaire including diet, lifestyle, medical history, family information and others were obtained for all subjects.
The five FNGPs, such as W and CSA are calculated based on the BMD (g/cm2) and bone size (cm2) at the FN. Detailed calculation formulas for the five parameters have been described elsewhere [43], [44], [45]. ALM (g) was calculated as the sum of lean soft tissue (nonfat, non-bone) mass in the arms and legs. BMD and bone size at the FN and ALM were measured by dual-energy X-ray absorptiometry (DXA) with Hologic densitometers (Hologic Inc., Waltham, MA, USA) that were calibrated daily. For Chinese subjects, the coefficient of variation (CV) values of DXA measurements for BMD, bone size at the FN and ALM were 1.87%, 1.94%, and 1.0%, respectively. Similar CV values were obtained with US Caucasians.
Genotyping
Genomic DNA was extracted from peripheral blood leukocytes using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN, USA). All subjects were genotyped using the Human mapping SNP 6.0 assay kit (Affymetrix, Inc, Santa Clara, CA), following the standard protocol recommended by the manufacturer. For quality control (QC) of SNPs, we set the default value of greater than 0.4 as the contrast QC threshold. The final average contrast QC across the entire sample reached the high level of 2.62. In the initial stage, 909,622 SNPs were genotyped for the Chinese subjects. After excluding 17,888 SNP with allele frequencies deviating extremely from Hardy-Weinberg equilibrium (p<0.01) and 202,984 SNPs with minor allele frequencies (MAF) <1% (618 SNP were included by both exclusion criteria), a final total of 689,368 SNPs were retained for subsequent analyses, yielding an average marker spacing of ∼4 kb throughout the human genome.
Statistical Analyses
Although previous studies have reported that FNGPs and body lean mass are two related phenotypes [15], [16], we re-estimated their phenotype correlation used our Chinese data. The bivariate correlation analysis was performed using the statistical package SPSS version 17.0.
We adopted similar statistical analyses in the initial GWAS and replicate study. Before association analyses, raw phenotypes of FNGPs and ALM were adjusted for age and sex. Principal component analysis (PCA) was performed [46] to calculate the principal components, and the ten default main eigenvectors were used as covariates to adjust raw phenotypic data for correction of population stratification.
We performed bivariate GWAS to detect associations between each SNP and two phenotypes. An additive genetic model was applied to both univariate and bivariate association analyses. Based on a linear model, bivariate regression analyses were conducted using the R software package (available at http://www.r-project.org). This method is expressed as follows: for an individual i, yi is a vector of a length of 2 and coding the individual's bivariate phenotype, which can be modeled as
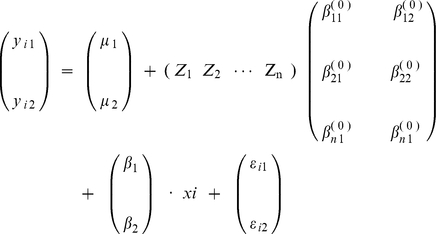 |
In this model: 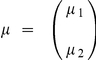 is the grand mean vector; Z = (Z1 Z2···Zn) is a vector coding for covariates, that may include other risk factors and confounding factors; β's are the corresponding effects of covariates or the SNP under test; xi is the genotype score at the locus of interest for individual i, and εi is the vector of random error. We compared the likelihood of the model under the null hypothesis (SNP effects are restricted to 0), with that under the alternative hypothesis (the SNP effects are not 0), to test the alternative hypothesis. Then the likelihood ratio can convert to an F-statistic, which follows an F-distribution under the null hypothesis. The bivariate p value was calculated based on the F-statistic.
is the grand mean vector; Z = (Z1 Z2···Zn) is a vector coding for covariates, that may include other risk factors and confounding factors; β's are the corresponding effects of covariates or the SNP under test; xi is the genotype score at the locus of interest for individual i, and εi is the vector of random error. We compared the likelihood of the model under the null hypothesis (SNP effects are restricted to 0), with that under the alternative hypothesis (the SNP effects are not 0), to test the alternative hypothesis. Then the likelihood ratio can convert to an F-statistic, which follows an F-distribution under the null hypothesis. The bivariate p value was calculated based on the F-statistic.
To compare the results from univariate and bivariate association analyses, we also conducted univariate association with each of the tested phenotypes using Plink (version 1.07, http://pngu.mgh.harvard.edu/~purcell/plink/) in our GWAS and replicate cohorts, where genotypic association analysis was performed under a linear regression framework. Genotype was treated as the independent variable, study phenotype (such as ALM, W) as the dependent variable, and phenotype was modeled as a linear function of alternative genotypes at a certain SNP.
To quantify overall evidence of association achieved in our GWAS and in the US replication cohort, we combined individual p values of the two cohorts using a Fisher's method[47] for meta-analysis. The calculation was performed using the MetaP web tool (http://people.genome.duke.edu/~dg48/metap.php). The linkage disequilibrium (LD) [standardized D′(D/Dmax)] patterns of interesting SNPs and the haplotype block map was analyzed using Haploview software (available at http://www.broad.mit.edu/mpg/haploview/).
Supporting Information
(XLS)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was partially supported by the Natural Science Foundation of China (NSFC) (30900810, 30771222, 31071097, and 30600364), the NSFC-Canadian Institutes of Health Research (CIHR) Joint Health Research Initiative Proposal (30811120436), the NSFC/RGC Joint Research Scheme (30731160618), the Shanghai Leading Academic Discipline Project (S30501) and startup fund from Shanghai University of Science and Technology. HWD was partially supported by grants from NIH (P50AR055081, R01AG026564, R01AR050496, RC2DE020756, R01AR057049, and R03TW008221) and the Franklin D. Dickson/Missouri Endowment. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Melton3rd LJ. Adverse outcomes of osteoporotic fractures in the general population. J Bone Miner Res. 2003;18:1139–1141. doi: 10.1359/jbmr.2003.18.6.1139. [DOI] [PubMed] [Google Scholar]
- 2.Peng Z, Tuukkanen J, Zhang H, Jamsa T, Vaananen HK. The mechanical strength of bone in different rat models of experimental osteoporosis. Bone. 1994;15:523–532. doi: 10.1016/8756-3282(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 3.Pulkkinen P, Partanen J, Jalovaara P, Jamsa T. Combination of bone mineral density and upper femur geometry improves the prediction of hip fracture. Osteoporos Int. 2004;15:274–280. doi: 10.1007/s00198-003-1556-3. [DOI] [PubMed] [Google Scholar]
- 4.Handoll HH, Gillespie WJ, Gillespie LD, Madhok R. Moving towards evidence-based healthcare for musculoskeletal injuries: featuring the work of the Cochrane Bone, joint and Muscle Trauma Group. J R Soc Promot Health. 2007;127:168–173. doi: 10.1177/1466424007079491. [DOI] [PubMed] [Google Scholar]
- 5.Stone KL, Seeley DG, Lui LY, Cauley JA, Ensrud K, et al. BMD at multiple sites and risk of fracture of multiple types: long-term results from the Study of Osteoporotic Fractures. J Bone Miner Res. 2003;18:1947–1954. doi: 10.1359/jbmr.2003.18.11.1947. [DOI] [PubMed] [Google Scholar]
- 6.Ammann P, Rizzoli R. Bone strength and its determinants. Osteoporos Int 14 Suppl. 2003;3:S13–18. doi: 10.1007/s00198-002-1345-4. [DOI] [PubMed] [Google Scholar]
- 7.Peacock M, Turner CH, Liu G, Manatunga AK, Timmerman L, et al. Better discrimination of hip fracture using bone density, geometry and architecture. Osteoporos Int. 1995;5:167–173. doi: 10.1007/BF02106096. [DOI] [PubMed] [Google Scholar]
- 8.Crabtree N, Lunt M, Holt G, Kroger H, Burger H, et al. Hip geometry, bone mineral distribution, and bone strength in European men and women: the EPOS study. Bone. 2000;27:151–159. doi: 10.1016/s8756-3282(00)00300-8. [DOI] [PubMed] [Google Scholar]
- 9.Crabtree NJ, Kroger H, Martin A, Pols HA, Lorenc R, et al. Improving risk assessment: hip geometry, bone mineral distribution and bone strength in hip fracture cases and controls. The EPOS study. European Prospective Osteoporosis Study. Osteoporos Int. 2002;13:48–54. doi: 10.1007/s198-002-8337-y. [DOI] [PubMed] [Google Scholar]
- 10.Melton3rd LJ, Beck TJ, Amin S, Khosla S, Achenbach SJ, et al. Contributions of bone density and structure to fracture risk assessment in men and women. Osteoporos Int. 2005;16:460–467. doi: 10.1007/s00198-004-1820-1. [DOI] [PubMed] [Google Scholar]
- 11.Bethesda . Proceedings. J Gerontol A Biol Sci Med Sci 50 Spec No:; 1995. Workshop on sarcopenia: muscle atrophy in old age. pp. 1–161. [PubMed] [Google Scholar]
- 12.Beck TJ, Oreskovic TL, Stone KL, Ruff CB, Ensrud K, et al. Structural adaptation to changing skeletal load in the progression toward hip fragility: the study of osteoporotic fractures. J Bone Miner Res. 2001;16:1108–1119. doi: 10.1359/jbmr.2001.16.6.1108. [DOI] [PubMed] [Google Scholar]
- 13.Wu S, Lei SF, Chen XD, Tan LJ, Jian WX, et al. The contributions of lean tissue mass and fat mass to bone geometric adaptation at the femoral neck in Chinese overweight adults. Ann Hum Biol. 2007;34:344–353. doi: 10.1080/03014460701275749. [DOI] [PubMed] [Google Scholar]
- 14.Ferretti JL, Cointry GR, Capozza RF, Frost HM. Bone mass, bone strength, muscle-bone interactions, osteopenias and osteoporoses. Mech Ageing Dev. 2003;124:269–279. doi: 10.1016/s0047-6374(02)00194-x. [DOI] [PubMed] [Google Scholar]
- 15.Sun X, Lei SF, Deng FY, Wu S, Papacian C, et al. Genetic and environmental correlations between bone geometric parameters and body compositions. Calcif Tissue Int. 2006;79:43–49. doi: 10.1007/s00223-006-0041-3. [DOI] [PubMed] [Google Scholar]
- 16.Deng FY, Xiao P, Lei SF, Zhang L, Yang F, et al. Bivariate whole genome linkage analysis for femoral neck geometric parameters and total body lean mass. J Bone Miner Res. 2007;22:808–816. doi: 10.1359/jbmr.070303. [DOI] [PubMed] [Google Scholar]
- 17.Liu YZ, Pei YF, Liu JF, Yang F, Guo Y, et al. Powerful bivariate genome-wide association analyses suggest the SOX6 gene influencing both obesity and osteoporosis phenotypes in males. PLoS One. 2009;4:e6827. doi: 10.1371/journal.pone.0006827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heymsfield SB, Smith R, Aulet M, Bensen B, Lichtman S, et al. Appendicular skeletal muscle mass: measurement by dual-photon absorptiometry. Am J Clin Nutr. 1990;52:214–218. doi: 10.1093/ajcn/52.2.214. [DOI] [PubMed] [Google Scholar]
- 19.Kim J, Wang Z, Heymsfield SB, Baumgartner RN, Gallagher D. Total-body skeletal muscle mass: estimation by a new dual-energy X-ray absorptiometry method. Am J Clin Nutr. 2002;76:378–383. doi: 10.1093/ajcn/76.2.378. [DOI] [PubMed] [Google Scholar]
- 20.Marcora S, Casanova F, Williams E, Jones J, Elamanchi R, et al. Preliminary evidence for cachexia in patients with well-established ankylosing spondylitis. Rheumatology (Oxford) 2006;45:1385–1388. doi: 10.1093/rheumatology/kel127. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Pei Y, Papasian CJ, Deng HW. Bivariate association analyses for the mixture of continuous and binary traits with the use of extended generalized estimating equations. Genet Epidemiol. 2009;33:217–227. doi: 10.1002/gepi.20372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang PY, Jensen J, Printz RL, Granner DK, Ivy JL, et al. Overexpression of hexokinase II in transgenic mice. Evidence that increased phosphorylation augments muscle glucose uptake. J Biol Chem. 1996;271:14834–14839. [PubMed] [Google Scholar]
- 23.Fueger PT, Heikkinen S, Bracy DP, Malabanan CM, Pencek RR, et al. Hexokinase II partial knockout impairs exercise-stimulated glucose uptake in oxidative muscles of mice. Am J Physiol Endocrinol Metab. 2003;285:E958–963. doi: 10.1152/ajpendo.00190.2003. [DOI] [PubMed] [Google Scholar]
- 24.Streffer C. Glucose-, energy-metabolism and cell proliferation in tumors. Adv Exp Med Biol. 1994;345:327–333. doi: 10.1007/978-1-4615-2468-7_43. [DOI] [PubMed] [Google Scholar]
- 25.Liechty EA, Denne SC, Lemons JA, Kien CL. Effects of glucose infusion on leucine transamination and oxidation in the ovine fetus. Pediatr Res. 1991;30:423–429. doi: 10.1203/00006450-199111000-00006. [DOI] [PubMed] [Google Scholar]
- 26.Movcessian SG, Boghosian AA, Kamalian RG, Urganjian MK. Effect of Krebs cycle components and glucose on the deamination of mono- and dinucleotides in brain mitochondrial fractions. Vopr Biokhim Mozga. 1972;7:11–17. [PubMed] [Google Scholar]
- 27.Vander Heiden MG, Plas DR, Rathmell JC, Fox CJ, Harris MH, et al. Growth factors can influence cell growth and survival through effects on glucose metabolism. Mol Cell Biol. 2001;21:5899–5912. doi: 10.1128/MCB.21.17.5899-5912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leidy HJ, Carnell NS, Mattes RD, Campbell WW. Higher protein intake preserves lean mass and satiety with weight loss in pre-obese and obese women. Obesity (Silver Spring) 2007;15:421–429. doi: 10.1038/oby.2007.531. [DOI] [PubMed] [Google Scholar]
- 29.Balint E, Szabo P, Marshall CF, Sprague SM. Glucose-induced inhibition of in vitro bone mineralization. Bone. 2001;28:21–28. doi: 10.1016/s8756-3282(00)00426-9. [DOI] [PubMed] [Google Scholar]
- 30.Braithwaite SS, Palazuk B, Colca JR, Edwards3rd CW, Hofmann C. Reduced expression of hexokinase II in insulin-resistant diabetes. Diabetes. 1995;44:43–48. doi: 10.2337/diab.44.1.43. [DOI] [PubMed] [Google Scholar]
- 31.Vestergaard H, Bjorbaek C, Hansen T, Larsen FS, Granner DK, et al. Impaired activity and gene expression of hexokinase II in muscle from non-insulin-dependent diabetes mellitus patients. J Clin Invest. 1995;96:2639–2645. doi: 10.1172/JCI118329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carvalho M, Mulinari RA, Nakagawa Y. Role of Tamm-Horsfall protein and uromodulin in calcium oxalate crystallization. Braz J Med Biol Res. 2002;35:1165–1172. doi: 10.1590/s0100-879x2002001000009. [DOI] [PubMed] [Google Scholar]
- 33.Kemter E, Rathkolb B, Rozman J, Hans W, Schrewe A, et al. Novel missense mutation of uromodulin in mice causes renal dysfunction with alterations in urea handling, energy, and bone metabolism. Am J Physiol Renal Physiol. 2009;297:F1391–1398. doi: 10.1152/ajprenal.00261.2009. [DOI] [PubMed] [Google Scholar]
- 34.Goffart S, Franko A, Clemen CS, Wiesner RJ. Alpha-actinin 4 and BAT1 interaction with the cytochrome c promoter upon skeletal muscle differentiation. Curr Genet. 2006;49:125–135. doi: 10.1007/s00294-005-0043-0. [DOI] [PubMed] [Google Scholar]
- 35.Dansithong W, Paul S, Comai L, Reddy S. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J Biol Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- 36.Okamoto Y, Takashima H, Higuchi I, Matsuyama W, Suehara M, et al. Molecular mechanism of rigid spine with muscular dystrophy type 1 caused by novel mutations of selenoprotein N gene. Neurogenetics. 2006;7:175–183. doi: 10.1007/s10048-006-0046-0. [DOI] [PubMed] [Google Scholar]
- 37.Kim HJ, Minashima T, McCarthy EF, Winkles JA, Kirsch T. Progressive ankylosis protein (ANK) in osteoblasts and osteoclasts controls bone formation and bone remodeling. J Bone Miner Res. 2010;25:1771–1783. doi: 10.1002/jbmr.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howell GR, Shindo M, Murray S, Gridley T, Wilson LA, et al. Mutation of a ubiquitously expressed mouse transmembrane protein (Tapt1) causes specific skeletal homeotic transformations. Genetics. 2007;175:699–707. doi: 10.1534/genetics.106.065177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Izumi K, Takagi M, Parikh AS, Hahn A, Miskovsky SN, et al. Late manifestations of tricho-rhino-pharangeal syndrome in a patient: Expanded skeletal phenotype in adulthood. Am J Med Genet A. 2010;152A:2115–2119. doi: 10.1002/ajmg.a.33511. [DOI] [PubMed] [Google Scholar]
- 40.Roldan JC, Jepsen S, Miller J, Freitag S, Rueger DC, et al. Bone formation in the presence of platelet-rich plasma vs. bone morphogenetic protein-7. Bone. 2004;34:80–90. doi: 10.1016/j.bone.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 41.Kochi Y, Suzuki A, Yamada R, Yamamoto K. Genetics of rheumatoid arthritis: underlying evidence of ethnic differences. J Autoimmun. 2009;32:158–162. doi: 10.1016/j.jaut.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 42.Palmer CG, Martinez A, Fox M, Sininger Y, Grody WW, et al. Ethnic differences in parental perceptions of genetic testing for deaf infants. J Genet Couns. 2008;17:129–138. doi: 10.1007/s10897-007-9134-z. [DOI] [PubMed] [Google Scholar]
- 43.Zhao LJ, Liu XG, Liu YZ, Liu YJ, Papasian CJ, et al. Genome-wide association study for femoral neck bone geometry. J Bone Miner Res. 2010;25:320–329. doi: 10.1359/jbmr.090726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beck T. Measuring the structural strength of bones with dual-energy X-ray absorptiometry: principles, technical limitations, and future possibilities. Osteoporos Int 14 Suppl. 2003;5:S81–88. doi: 10.1007/s00198-003-1478-0. [DOI] [PubMed] [Google Scholar]
- 45.Chen Y, Xiong DH, Guo YF, Pan F, Zhou Q, et al. Pathway-based genome-wide association analysis identified the importance of EphrinA-EphR pathway for femoral neck bone geometry. Bone. 2010;46:129–136. doi: 10.1016/j.bone.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou F, Lee S, Knowles MR, Wright FA. Quantification of population structure using correlated SNPs by shrinkage principal components. Hum Hered. 2010;70:9–22. doi: 10.1159/000288706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fisher RA. Combining independent tests of significance. American Statistician. 1948;2:30. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)