Abstract
Clathrin-mediated endocytosis was previously implicated as one of the cellular pathways involved in filoviral glycoprotein mediated viral entry into target cells. Here we have further dissected the requirements for different components of this pathway in Ebola versus Marburg virus glycoprotein (GP) mediated viral infection. Although a number of these components were involved in both cases; Ebola GP-dependent viral entry specifically required the cargo recognition proteins Eps15 and DAB2 as well as the clathrin adaptor protein AP-2. In contrast, Marburg GP-mediated infection was independent of these three proteins and instead required beta-arrestin 1 (ARRB1). These findings have revealed an unexpected difference between the clathrin pathway requirements for Ebola GP- versus Marburg GP pseudovirion infection. Anthrax toxin entry also uses a clathrin-, and ARRB1-dependent pathway for cellular entry, indicating that the mechanism used by Marburg GP pseudovirions may be more generally important for pathogen entry.
Keywords: Filoviral GP, pseudotyped virus, entry, clathrin-mediated endocytosis, Eps15, AP-2, DAB2, ARRB1, AP-1
Introduction
The Filoviridae family comprises of Marburg virus (MARV) and Ebola virus (EBOV), the causative agents of viral hemorrhagic fever (Schnittler and Feldmann, 2003). There have been several sporadic outbreaks of these virus infections since the late 1960’s, the most recent occurring in 2008 in Uganda (MARV), and in 2009 in the Democratic Republic of the Congo (EBOV). The high fatality rates associated with these viruses represents a potential global health challenge and also makes them ideal candidates for use as biological weapons. Consequently, these viruses have been classified as Category A Bioterrorism Agents by the US Centers for Disease Control and Prevention (CDC). There are currently no effective drugs or licensed vaccines to protect humans against filovirus infection (Sullivan et al., 2009). Therefore, there is an urgent need to better understand the mechanisms that control filovirus replication for designing effective therapeutic measures.
The target cells for filoviral infection are monocytes, macrophages, dendritic cells and endothelial cells (Geisbert et al., 2003) and (Connolly et al., 1999). Several cell surface proteins have been implicated in filovirus entry including folate receptor alpha (Chan et al., 2001), lectins (Marzi et al., 2006), (Ji et al., 2005), (Takada et al., 2004), (Simmons et al., 2003a) and (Alvarez et al., 2002), beta 1 integrins (Takada et al., 2000) and TAM receptors (Shimojima et al., 2006). T-cell immunoglobulin and mucin domain 1 (TIM-1) was recently reported to be a receptor for Ebola as well as Marburg virus (Kondratowicz et al., 2011), suggesting that these viruses bind to a common receptor.
Filovirus entry is mediated by the virus-encoded glycoprotein (GP), located on the viral surface lipid bilayer. The filoviral GP is a homotrimeric, class I viral fusion protein, expressed as a precursor that is post-translationally cleaved in the Trans Golgi Network (TGN) by a cellular proprotein convertase furin into the disulfide-linked GP1 (140kD) and GP2 (26kD) subunits (Jeffers, Sanders, and Sanchez, 2002) and (Volchkov et al., 1998). GP1 is primarily involved in receptor binding whereas GP2 facilitates virus-cell membrane fusion (White et al., 2008).
Following cell surface receptor binding, filoviruses are taken up by endocytosis. Using multiple approaches and cell lines we have previously shown that Ebola GP pseudovirus uses clathrin-mediated endocytosis as an entry pathway (Bhattacharyya et al., 2010). We also found that treatment with chlorpromazine, which was previously reported to block clathrin-dependent entry (Wang, Rothberg, and Anderson, 1993), inhibited wild type (WT) Ebola. However, it is now known that chlorpromazine also inhibits macropinocytosis and hence is not a specific inhibitor of the clathrin pathway (Ivanov, 2008). Clathrin and macropinocytic pathways have also been implicated in filovirus infection by other groups (Sanchez, 2007) and (Quinn et al., 2009); while the role of caveolae pathway has been both implicated and refuted (Empig and Goldsmith, 2002) and (Simmons et al., 2003b). It was recently suggested that while filoviral glycoprotein pseudotyped viruses enter via the clathrin pathway (Hunt et al., 2010); wild type filoviruses predominantly use macropinocytosis for entry (Saeed et al., 2010) and (Nanbo et al., 2010). These differences in entry pathway requirements could be due to the differences in size and shape of pseudovirions versus WT viruses (Cureton et al., 2010).
Upon entry, filoviruses are trafficked by cellular endocytic machinery to an acidic endosomal compartment, which is the site of virus-cell membrane fusion. The Ebola virus GP is activated to trigger fusion through proteolytic cleavage mediated by cellular lysosomal cysteine proteases, cathepsins B and L (Chandran et al., 2005), (Kaletsky, Simmons, and Bates, 2007), (Schornberg et al., 2006) and (Sanchez, 2007).
In this report we have investigated the specific requirements for different components of the clathrin endocytic machinery in Ebola GP versus Marburg GP pseudovirion entry. These studies have demonstrated that these two highly related glycoproteins exhibit differential requirements for several players of this pathway, uncovering critical differences in their entry mechanisms. Moreover, the factors required for Marburg GP mediated entry are very similar to those previously described for anthrax toxin entry (Abrami et al., 2010), suggesting that these components of the clathrin pathway may be broadly required by various pathogens to enter target cells.
Materials and methods
Cell lines, plasmids and chemicals
HEK293T cells, Human Osteo Sarcoma (HOS-CD4) cells and Human Microvascular Endothelial Cells (HMEC) were maintained as previously described (Bhattacharyya et al., 2010).
The plasmids pCB6-EbGP, VSVg, R7ΔEnvGFP, R73X4EnvGFP, mRFP-dominant-negative (DN) Eps15 (DIII), control Eps15 (D3Δ2), S15-mCherry and GFP-Vpr are previously described (Bhattacharyya et al., 2010) and (Campbell et al., 2007). The plasmid encoding Marburg GP, Musoke strain (pWRG7077) was received from Dr. M. Javad Aman (Bavari et al., 2002). The TagBFP and tdTomato plasmids were obtained from Dr. Michael W. Davidson and the latter plasmid was made in Dr. Roger Tsien’s lab (Shaner et al., 2004).
Polyethylenimine (PEI), Linear, MW 25,000 (Polysciences, Inc.) and chlorpromazine hydrochloride (Sigma) were dissolved in distilled water to make 1 mg/ml stock solutions. Sucrose (Fisher) was dissolved in distilled water to make a 5 M stock solution. Bafilomycin A1 (Baf A1) (Sigma) and FYdmk (Calbiochem) were dissolved in DMSO to make 20 μM and 10 mM stock solutions respectively.
Pseudotyped virus production
For preparing Marburg GP pseudotyped virus, HEK293T cells in a 15-cm plate were transiently co-transfected with 20 μg of pWRG7077 encoding Marburg GP and 30 μg of R7ΔEnvGFP HIV plasmids using 112 μl of PEI. 48 h post-transfection, the supernatant was centrifuged to pellet the cell debris and clarified by filtration through a 0.45 μm pore-size filter. EbGP and VSVg pseudotyped viruses and R73X4EnvGFP (wild type HIV) were prepared as previously described (Bhattacharyya et al., 2010). Virus titer was measured using a p24 ELISA kit (Perkin Elmer Lifesciences). The S15-mCherry and GFP-Vpr double-labeled virus was prepared as previously described (Campbell et al., 2007).
Flow cytometric analysis to measure viral transduction following drug treatments
HOS or HMEC cells in 12-well plates were pretreated with either 50 nM Baf A1 for 30 min or 0.45 M sucrose for 10 min or 10 μg/ml chlorpromazine for 45 min or a combination of 10μg/ml chlorpromazine and 0.45 M sucrose for 45 min for the various experiments. For the cathepsin L inhibitor experiment, HOS cells were pre-incubated with 10 μM cathepsin L inhibitor (FYdmk) for 4.5 h. The working concentration of FYdmk was prepared in DMEM containing 1 % DMSO. In each case, the cells were incubated with MARVGP, EbGP pseudotyped virus and wild type HIV overnight in the presence of drug. For the cathepsin L experiment, VSVg pseudotyped virus was used as a negative control and the viruses were added overnight in DMEM containing 1 % DMSO in the control cells. GFP fluorescence as a marker of infectivity was measured at 48 h post-infection by flow cytometry using FACS Calibur (Becton Dickinson). The cells were gated on forward and side scatter and identical gates were maintained for treated and control samples. Three independent infection samples were analyzed for each virus and the % GFP positive cells in the treated samples normalized to untreated samples was calculated.
Immunofluorescence analysis of viral transduction following transfection with DN and control mRFP-Eps15 plasmids
HOS cells were plated on coverslips and transiently transfected with DN or control mRFP-Eps15 plasmids using the Effectene transfection reagent (Qiagen). 24 h post-transfection, the cells were incubated with the various viruses for 4 h. 48 h post-infection, the cells were fixed and the DNA was stained with Hoechst. The coverslips were mounted onto glass slides with Gel Mount (Biomeda) and the dried slides were imaged as previously described (Bhattacharyya et al., 2010). The experiment was repeated three times and the % decline in infectivity in DN Eps15 transfected cells normalized to control Eps15 transfected cells was calculated using the following formula: (number of transfected and infected cells; yellow)/(number of transfected cells; red) X 100 %. The experimental scheme is shown in Supplemental Fig. 2.
Immunofluorescence study to examine viral transduction following siRNA-mediated knockdown of clathrin endocytic pathway components
The siRNA duplexes (Dharmacon) were synthesized and used as previously described (Huang et al., 2004). Two siRNAs against each gene were included to rule out any potential off-target effects caused by a single siRNA. The siRNA duplexes were diluted to 20 μM before transfection. The target sequences and catalog numbers for the siRNAs are described in Supplemental Table 2.
HOS cells were plated on coverslips in 12-well plates and incubated in DMEM containing 10 % FBS without antibiotics for 24 h. The cells were transfected twice at 24 h intervals with 4 μl of each siRNA, 0.25 μg tdTomato plasmid (as a transfection marker) and 3 μl of Lipofectamine 2000 reagent (Invitrogen) in OPTI-MEM I medium. Control cells were similarly transfected with the non-targeting siRNA. The cells were incubated with the viruses for 4 h at 24 h after the second transfection for PICALM; at 48 h after the second transfection for CHC, epsin 1, ITSN1, DYN2, DAB2, LDLRAP1, INPPL1, ARRB1, HIP1, NUMB, REPS1 and REPS2 and at 72 h after the second transfection for AP2B and AP1M1. The cells were fixed at 48 h post-infection and the DNA was stained with Hoechst. The coverslips were mounted on glass slides and imaged with a Zeiss Axioskop 2 microscope. Several panels of images were collected from each coverslip and the number of transfected only as well as transfected and infected cells in each sample was counted. A minimum of 100 transfected cells was counted for each sample. The experiment was repeated three times and % decline in viral infectivity in specific siRNA treated cells normalized to non-targeting siRNA treated cells was calculated.
Quantitative PCR (qPCR) to measure mRNA expression levels following siRNA-mediated knockdown of clathrin endocytic pathway components
The siRNA-mediated knockdowns were performed as described in the previous section. RNA was isolated from the cells with the RNeasy mini kit (Qiagen) and cDNA was prepared with the QuantiTect reverse transcription kit (Qiagen). The qPCR reactions were normalized to cellular beta-actin mRNA expression levels and performed on an ABI Prism 7900HT Sequence Detection System using Power SYBRGreen PCR master mix (Applied Biosystems). The experiment was repeated three times and a dissociation curve analysis was performed for each reaction with the SDS software (Applied Biosystems). The results represent % mRNA expression levels in specific siRNA treated cells normalized to non-targeting siRNA treated cells. The primer sequences are described in Supplemental Table 3.
Cell viability following siRNA-mediated knockdown of clathrin endocytic pathway components
The siRNA-mediated knockdowns were performed as described in the previous section and cell viability was measured using Cell Titer Glo reagent (Promega) as per the manufacturer’s guidelines.
Immunofluorescence analysis to examine MARVGP pseudotyped virus fusion following siRNA-mediated knockdown of ARRB1
HOS cells on coverslips were co-transfected with the two ARRB1 siRNAs as described in the previous section using pTagBFP as a transfection marker and at 48 h after the second transfection; the cells were spinoculated with the S15-mCherry and GFP-Vpr double-labeled MARVGP pseudotyped virus for 1.5 h at 17 °C. The cells were incubated at 37 °C for 1 h, 2 h, 3 h or 4 h and then fixed. The coverslips were mounted on glass slides and imaged using a Zeiss LSM 710 laser scanning confocal microscope. Several panels of images were collected from each coverslip and the total numbers of double-positive virus particles as well as GFP-Vpr single-positive virus particles were counted in the ARRB1 siRNA-transfected versus control siRNA-transfected, BFP positive cells.
Statistical analysis of experimental data
p Values were determined by comparing the treated versus control samples using a paired student t test with the GraphPad InStat3 software. For the Eps15 experiment, the p Value was determined by comparing the % decline in infectivity of EbGP with respect to MARVGP and HIV in cells transfected with DN Eps15 normalized to control Eps15 using the one-way analysis of variance (ANOVA) test.
Results
To examine virus entry mediated by MARVGP and EbGP, we exploited a previously used envelope protein-deficient lentiviral (HIV-1) vector system (Bhattacharyya et al., 2010) pseudotyped with the Ebola or Marburg GP. As expected, infection of HOS cells by both types of filovirus GP pseudovirions was dependent upon low endosomal pH as well as cathepsin (Cat) L cleavage since it was blocked by treatment with the vacuolar ATPase inhibitor Bafilomycin A1 and the Cat L inhibitor FYdmk, respectively (Supplemental Fig. 1). For control purposes, we showed that these treatments did not impair infection with an isogenic HIV vector carrying the wild-type envelope protein, which directs pH-independent cellular entry (Miyauchi et al., 2009) and (Stein et al., 1987) or HIV pseudotyped with VSVg, which mediates low-pH dependent but Cat L independent entry (Chandran et al., 2005) (Supplemental Fig. 1).
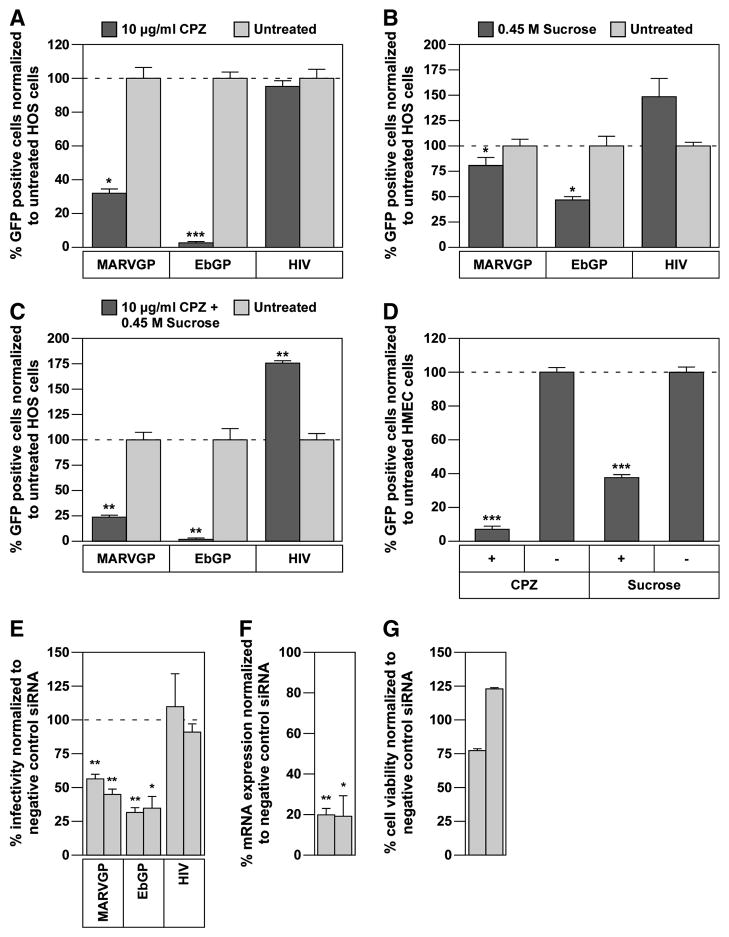
Previously, we showed that EbGP-dependent virus infection was blocked by treatment with two chemical inhibitors of the clathrin endocytic pathway; chlorpromazine and sucrose, and by RNAi-mediated knockdown of the clathrin heavy chain (CHC) (Bhattacharyya et al., 2010). To assess the role of clathrin-mediated endocytosis in MARVGP pseudovirion entry, we evaluated the effects of these treatments on infection. Similar to EbGP-mediated virus infection, MARVGP-mediated infection of HOS cells was blocked by treatment with either 10 μg/ml chlorpromazine (Fig. 1A) or 0.45 M sucrose (Fig. 1B), or by treatment with both inhibitors (Fig. 1C). These inhibitors also blocked MARVGP-dependent infection in physiologically relevant human microvascular endothelial cells (HMEC) (Fig. 1D). MARVGP-mediated infection was also blocked specifically by two independent siRNAs, which significantly knocked down the levels of CHC mRNA without causing any overt cytotoxicity (Fig. 1E–G). As expected, none of these treatments inhibited infection by the control HIV-1 virus. Taken together, these results demonstrate that MARVGP pseudovirions utilize a clathrin-dependent pathway for cellular entry.
Fig. 1. Chlorpromazine (CPZ), sucrose and RNAi-knockdown of clathrin heavy chain (CHC) inhibit Marburg GP mediated viral entry.
(A) HOS cells were pre-treated with 10 μg/ml chlorpromazine for 45 min followed by incubation with a Marburg GP (MARVGP) or Ebola GP (EbGP) pseudotyped HIV-1 vector encoding GFP or wild type (WT) HIV overnight in the presence of the drug. 48 h post-infection, viral infectivity was measured by flow cytometry. Error bars represent SEM for three independent samples. * p Value for MARVGP < 0.05 and EbGP < 0.001. (B) HOS cells were pre-treated with 0.45 M sucrose for 10 min followed by incubation with the viruses described in panel (A) overnight in the presence of the drug. 48 h post-infection, viral infectivity was measured by flow cytometry. Error bars represent SEM for three independent samples. * p Values for MARVGP and EbGP < 0.05. (C) HOS cells were pre-treated with a combination of 10 μg/ml chlorpromazine and 0.45 M sucrose for 45 min followed by incubation with the viruses described in panel A overnight in the presence of the drugs. 48 h post-infection, viral infectivity was measured by flow cytometry. Error bars represent SEM for three independent samples. * p Values for MARVGP, EbGP and HIV < 0.01. (D) HMEC cells were pre-treated with 10 μg/ml chlorpromazine for 45 min or 0.45 M sucrose for 10 min followed by incubation with the MARVGP pseudotyped virus overnight in the presence of the drug. Viral infectivity was measured at 48 h post-infection by flow cytometry. Error bars represent SEM for three independent samples. * p Values for CPZ and sucrose < 0.001. (E) HOS cells were co-transfected with two siRNAs against CHC and a tdTomato plasmid (as a transfection marker) followed by infection with the viruses as described in panel (A). The data is represented as % infectivity in specific siRNA-transfected cells normalized to non-targeting siRNA transfected cells. Individual bars represent single siRNAs. Error bars represent SEM for three independent experiments. * p Values for MARVGP siRNAs < 0.01 and for EbGP: siRNA 1 < 0.01, siRNA 2 < 0.05. (F) HOS cells were transfected with two siRNAs against CHC as described in the materials and methods section. mRNA expression levels were measured by qPCR and normalized to cellular beta-actin mRNA expression levels. Individual bars represent single siRNAs. Error bars represent SEM for three independent experiments. * p Values for siRNA 1 < 0.01 and siRNA 2 < 0.05. (G) HOS cells were transfected with two siRNAs against CHC as described in the materials and methods section and cell viability was measured using the Cell Titer Glo reagent (Promega). Data from one representative experiment is shown in the figure. Individual bars represent single siRNAs. Error bars represent standard deviation for eight replicates.
Eps15 and AP-2 are specifically required for EbGP mediated entry
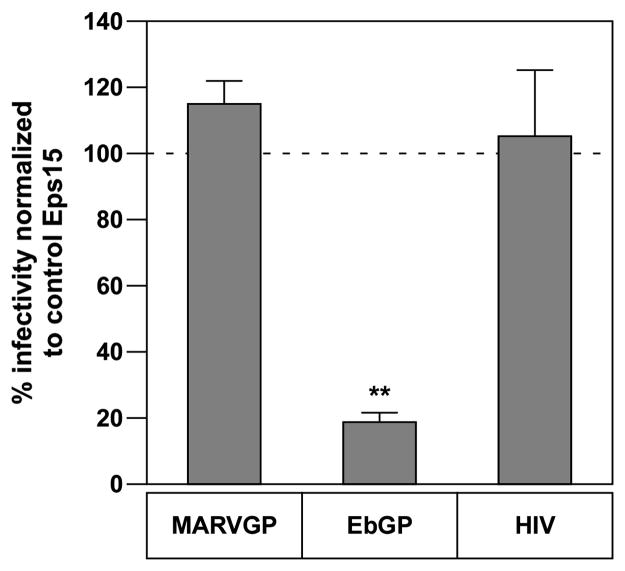
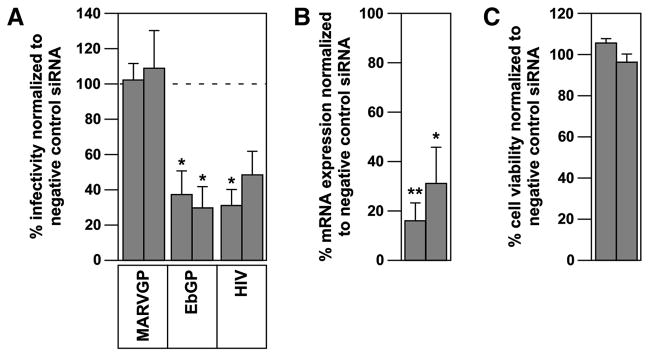
The cellular protein Eps15 links ubiquitinated cargo to clathrin through its binding to the alpha-subunit of AP-2 (Traub, 2003). Previously, we showed that expression of a dominant-negative version of Eps15 (DIII) blocks infection by EbGP-pseudotyped virus (Bhattacharyya et al., 2010). We tested the effect of this molecular inhibitor on MARVGP pseudotyped virus and surprisingly, it had no impact on infection (Fig. 2). Since Eps15 is known to constitutively associate with AP-2 (Benmerah et al., 1995), we then examined the effect of RNAi-mediated knockdown of AP-2 on both EbGP and MARVGP-dependent entry. As anticipated, EbGP-dependent infection was blocked by two siRNAs that knocked down the mRNA expression levels of AP-2 without altering cell viability, whereas MARVGP-mediated infection was not blocked by these siRNAs (Fig. 3A–C). We conclude that the mechanism of EbGP-mediated infection is dependent on both Eps15 and AP-2 whereas MARVGP-mediated infection is independent of these two cellular factors.
Fig. 2. Dominant-negative Eps15 does not inhibit Marburg GP mediated viral entry.
HOS cells were plated on coverslips and transfected with mRFP-DIII (dominant-negative, DN) or D3Δ2 (control) Eps15 plasmids. 24 h post-transfection, EbGP, MARVGP pseudotyped viruses or WT-HIV was added to the cells for 4 h. 48 h post-infection, the cells were fixed and the DNA was stained with Hoechst. Several panels of images were collected from each coverslip. The number of transfected only and transfected and infected cells was counted in each panel and the percentage of transfected and infected cells was determined. Graph represents % decline in viral infectivity in DN Eps15 transfected cells normalized to control Eps15 transfected cells. Error bars represent SEM for three independent experiments. * p Value for % decline in EbGP infectivity compared to MARVGP and HIV is < 0.01.
Fig. 3. Ebola GP requires AP-2 for entry but Marburg GP does not.
(A) HOS cells were co-transfected with two siRNAs against AP-2 and a tdTomato plasmid (as a transfection marker) followed by infection with the viruses as described in the materials and methods section. The data is represented as % infectivity in specific siRNA-transfected cells compared to non-targeting siRNA transfected cells. Individual bars represent single siRNAs. Error bars represent SEM for three independent experiments. * p Values for EbGP siRNAs and for HIV siRNA 1 < 0.05. (B) HOS cells were transfected with two siRNAs against AP-2 as described in the materials and methods section. The mRNA expression levels were measured by qPCR and normalized to cellular beta-actin mRNA expression levels. Individual bars represent single siRNAs. Error bars represent SEM for three independent experiments. * p Value for siRNA 1 < 0.01 and siRNA 2 < 0.05. (C) HOS cells were transfected with two siRNAs against AP-2 as described in the materials and methods section and cell viability was measured using the Cell Titer Glo reagent (Promega). Data from one representative experiment is shown in the figure. Individual bars represent single siRNAs. Error bars represent standard deviation for eight replicates.
Differential requirements for ARRB1 and DAB2 in Marburg GP versus Ebola GP mediated entry
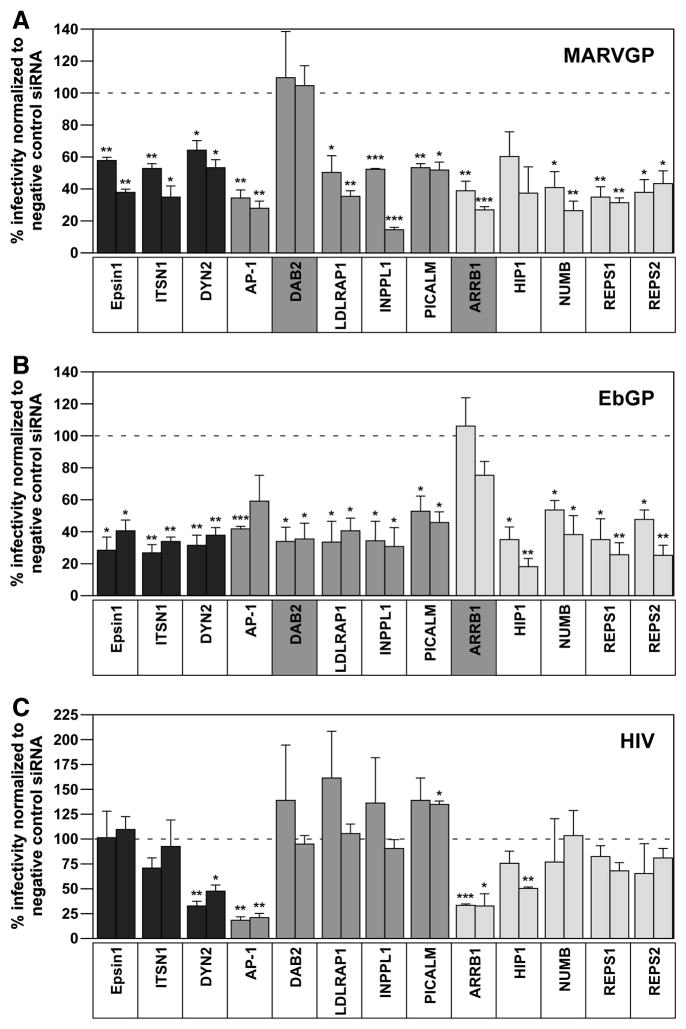
To further investigate the clathrin pathway requirements for EbGP and MARVGP-dependent entry, we tested the roles played by several members of this pathway including epsin 1, intersectin 1 (ITSN1), dynamin 2 (DYN2), adaptor-related protein complex 1, mu 1 subunit (AP1M1), disabled homolog 2 (DAB2), low density lipoprotein receptor adaptor protein 1 (LDLRAP1), inositol polyphosphate phosphatase-like 1 (INPPL1), phosphatidylinositol binding clathrin assembly protein (PICALM), beta-arrestin 1 (ARRB1), huntingtin interacting protein 1 (HIP1), Drosophila numb homolog (NUMB), RALBP1 associated Eps domain containing 1 (REPS1) and RALBP1 associated Eps domain containing 2 (REPS2). The roles of these factors in the clathrin pathway are described in the Discussion Section.
Two independent siRNAs were employed that significantly knocked down mRNA expression levels of each of these factors without adversely impacting cell viability (Supplemental Fig. 3). Some of these siRNAs were extensively validated in a previous study (Huang et al., 2004), while the remaining siRNAs were chosen from a well-characterized Dharmacon library. For practical purposes, the genes were tested in 3 groups: group 1 genes are shown in black bars, group 2 in dark grey bars and group 3 in light grey bars (Fig. 4).
Fig. 4. Ebola and Marburg GP mediated entry have differential requirements for DAB2 and ARRB1.
(A) HOS cells were co-transfected with two siRNAs each against epsin 1, ITSN1, DYN2, AP1M1, DAB2, LDLRAP1, INPPL1, PICALM, ARRB1, HIP1, NUMB, REPS1 and REPS2 and a tdTomato plasmid (as a transfection marker) followed by infection with the MARVGP pseudotyped virus as described in the materials and methods section. The data is represented as % infectivity in specific siRNA-transfected cells normalized to non-targeting siRNA transfected cells. Individual bars represent single siRNAs for each gene. Error bars represent SEM for three independent experiments. * p Values for epsin1, AP1M1 and REPS1 siRNAs < 0.01; ITSN1: siRNA 1 < 0.01, siRNA 2 < 0.05; DYN2 and REPS2 siRNAs < 0.05; LDLRAP1: siRNA 1 < 0.05, siRNA 2 < 0.01; INPPL1 siRNAs < 0.001; PICALM: siRNA 1 < 0.01, siRNA 2 < 0.05; ARRB1: siRNA 1 < 0.01, siRNA 2 < 0.001 and NUMB: siRNA 1 < 0.05, siRNA 2 < 0.01. (B) HOS cells were co-transfected with the siRNAs described in panel (A) and a tdTomato plasmid followed by infection with the EbGP pseudotyped virus as described in the materials and methods section. The data is represented as % infectivity in specific siRNA-transfected cells normalized to non-targeting siRNA transfected cells. Individual bars represent single siRNAs for each gene. Error bars represent SEM for three independent experiments. * p Values for epsin1, DAB2, LDLRAP1, INPPL1, PICALM and NUMB siRNAs < 0.05; ITSN1 and DYN2 siRNAs < 0.01; AP1M1 siRNA 1 = 0.001; HIP1: siRNA 1 < 0.05, siRNA 2 < 0.01; REPS1: siRNA 1 < 0.05, siRNA 2 < 0.01 and REPS2: siRNA 1 < 0.05, siRNA 2 < 0.01. (C) HOS cells were co-transfected with the siRNAs described in panel (A) and a tdTomato plasmid followed by infection with WT-HIV as described in the materials and methods section. The data is represented as % infectivity in specific siRNA-transfected cells normalized to non-targeting siRNA transfected cells. Individual bars represent single siRNAs for each gene. Error bars represent SEM for three independent experiments. * p Values for DYN2: siRNA1 < 0.01, siRNA 2 < 0.05; AP1M1 siRNAs < 0.01; PICALM: siRNA 2 < 0.05; ARRB1: siRNA 1 < 0.001, siRNA 2 < 0.05 and HIP1: siRNA 2 < 0.01.
These studies revealed that epsin 1, ITSN1, LDLRAP1, INPPL1, PICALM, NUMB, REPS1 and REPS2 were required by both MARVGP and EbGP suggesting a conserved requirement for these factors in filoviral GP mediated entry. The siRNAs targeting these factors had no impact on infection by the isogenic vector containing the HIV-1 envelope protein, indicating that they do not influence a post-entry step of infection (Fig. 4). By contrast, DYN2 and AP-1 also inhibited infection by HIV-1, suggesting that these proteins may be required for the endocytosis-dependent infection of the viruses. DYN2 could possibly mediate HIV-1 entry via macropinocytosis (Liu et al., 2008) and (Marechal et al., 2001) and endosomal fusion of HIV-1 particles (Miyauchi et al., 2009). AP-2 and beta-arrestins are known to mediate endocytosis of G protein-coupled receptors (GPCRs) (Shimizu et al., 2009) and HIV-1 co-receptors CXCR4 and CCR5 are GPCRs (Unutmaz, KewalRamani, and Littman, 1998), which could explain the inhibitory effects of the AP-2 and ARRB1 siRNAs on HIV-1 infection. siRNA-mediated knockdown of HIP-1 also reduced infection by all three of these viruses, although statistically significant levels of inhibition with both siRNAs were obtained only with the EbGP pseudotyped virus (Fig. 4). Most importantly, these studies demonstrated that EbGP and MARVGP specifically employ DAB2 and ARRB1, respectively for entry (Fig. 4).
ARRB1 is involved in pre-fusion step(s) of MARVGP mediated virus trafficking
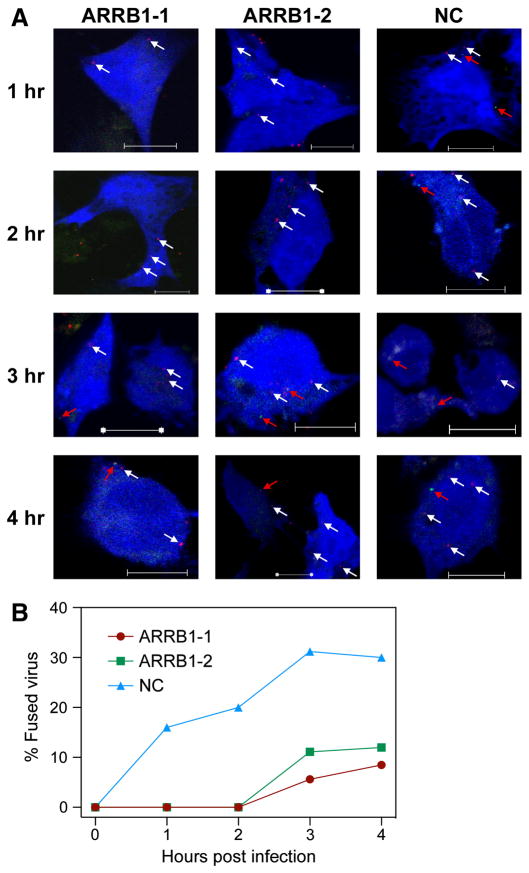
We previously used S15-mCherry and GFP-Vpr double-labeled, VSVg pseudotyped HIV to monitor virus fusion by deconvolution microscopy; S15-mCherry is very efficiently incorporated into the viral lipid membrane and the loss of mCherry signal during infection corresponds to viral fusion events (Campbell et al., 2007). Therefore, we generated a similar double-labeled MARVGP pseudotyped HIV to examine the step during virus entry affected by ARRB1 RNA knockdown. Previous studies demonstrated that the halftime of MARVGP pseudotyped HIV entry and fusion varies from ~ 50 mins in HeLa cells (Yonezawa, Cavrois, and Greene, 2005) to ~ 3 to 4 h in HEK293T cells (Empig and Goldsmith, 2002). Therefore, we monitored MARVGP pseudotyped HIV entry at 1, 2, 3 and 4 h post-infection in HOS cells co-transfected with ARRB1 siRNAs and pTagBFP (as a transfection marker), which encodes a blue fluorescent protein. Several panels of images were collected for each sample at every time point and the total numbers of S15-mCherry and GFP-Vpr double-positive virus particles as well as the GFP-Vpr single-positive virus particles in the BFP positive cells were counted. The raw data is shown in Supplemental Table 1. These studies demonstrated that virus fusion is significantly delayed in cells where ARRB1 is knocked down wherein compared to control siRNA transfected cells, no virus fusion was observed at 1–2 hours post-infection and there were fewer fused virions at 3–4 hours post-infection (Fig. 5A and B). These results show that there is no obvious defect in virus binding to cells deficient in ARRB1 but instead the block lies at a post-binding step leading to virus fusion.
Fig. 5. ARRB1 is required for efficient MARVGP pseudovirion fusion.
(A) HOS cells were co-transfected with siRNAs against ARRB1 or negative control (NC) and TagBFP plasmid (as a transfection marker) and challenged with S15-mCherry and GFP-Vpr double-labeled MARVGP pseudotyped virus. At various time points, the cells were fixed and several panels of images were collected for each sample. The total numbers of S15-mCherry and GFP-Vpr double-positive virus particles (yellow) as well as GFP-Vpr single-positive virus particles (green) were counted in the BFP positive (blue) cells. Representative images from each time point are shown. The white arrows indicate S15-mCherry and GFP-Vpr double-positive virus particles (unfused virus) and the red arrows point towards GFP-Vpr single-positive virus particles (fused virus). Scale bars represent 10 μm. (B) The total numbers of S15-mCherry and GFP-Vpr double-positive virions as well as GFP-Vpr single-positive virions in BFP positive cells were counted in the various panels of images collected from each sample above. The loss of S15-mCherry signal corresponding to fused virions was calculated for every sample for all the time points. Graph represents percentage of fused virions for each sample at various time points. The initial viral input level was normalized to 100 %.
Discussion
In this report we have shown that like Ebola GP pseudovirions, Marburg GP pseudovirion entry also involves clathrin-mediated endocytosis. Furthermore, a number of components of the clathrin endocytic pathway are generally required by both Ebola and Marburg GP pseudovirions. However, whereas Ebola GP pseudovirions specifically require Eps15, AP-2, and DAB2, Marburg GP pseudovirions specifically require ARRB1 (Fig. 6A).
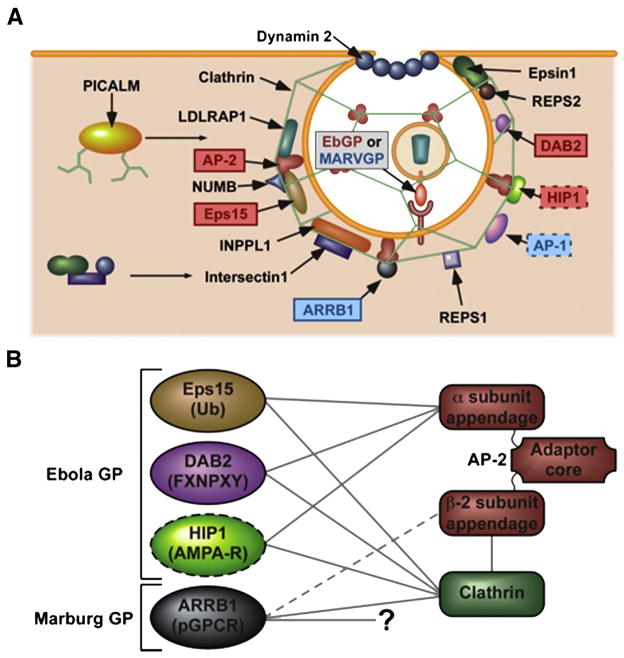
Fig. 6. Schematic representation of the differential requirements for various components of the clathrin pathway in Ebola GP versus Marburg GP pseudotyped viral entry.
(A) Ebola GP requires Eps15, AP-2, DAB2 and possibly HIP1 for its entry. In contrast, Marburg GP mediated entry is independent of these factors but instead requires ARRB1 and AP-1. Both viruses require CHC, epsin 1, ITSN1, DYN2, LDLRAP1, INPPL1, PICALM, NUMB, REPS1 and REPS2. The interactions of the factors with each other and also their roles in the formation of clathrin vesicles are depicted. (B) Eps15, DAB2 and HIP1, which are required by EbGP mediated virus entry are known to bind to the alpha-appendage of AP-2. In contrast ARRB1, which is required by MARVGP mediated virus entry is known to bind to the beta-appendage of AP-2. The target sequences recognized by these proteins are shown in parentheses. Figure adapted from (Traub, 2003) and (Traub, 2009).
These differential requirements point to unexpected differences between the entry mechanisms used by EbGP and MARVGP pseudovirions. Eps15 and DAB2 are involved in cargo recognition, interacting with different labels on cell surface proteins that are destined for internalization by the clathrin pathway. The EH domains of Eps15 interact with NPF motifs and recognize ubiquitin residues on its cargo (Salcini et al., 1997) and (Traub, 2003), whereas DAB2 recognizes an NPXY internalization sequence through its PTB domain (Mishra et al., 2002). Both of these factors bind to the α-adaptin subunit of AP-2 (Fig. 6B), an interaction that appears to be critically important for EbGP-mediated virus entry. By contrast, ARRB1, which is selectively important for MARVGP-mediated infection, recognizes phosphorylated serine and threonine residues on GPCRs (Oakley et al., 2001) and binds both to the beta-adaptin subunit of AP-2 as well as directly to clathrin through its C-terminus (Traub, 2003) and (Traub, 2009). Also, blocking interaction of ARRB1 with AP-2 was shown to enhance its interaction with clathrin (Schmid et al., 2006). Therefore, since AP-2 is not required for MARVGP-mediated virus entry, our results suggest that perhaps the direct interaction between ARRB1 and clathrin is important in this case. Furthermore, our results also demonstrate that ARRB1 is involved in post-binding and pre-fusion step(s) of MARVGP pseudotyped virus trafficking.
In addition to Eps15, other proteins involved in clathrin-mediated endocytosis such as Epsin 1, ITSN1, REPS1 and REPS2 are known to possess Eps homology (EH) domains (Polo et al., 2003), (Naslavsky and Caplan, 2005) and (Hussain et al., 1999). Epsin 1 is involved in clathrin-mediated endocytosis of influenza virus (Chen and Zhuang, 2008), whose entry is known to be AP-2 independent (Lakadamyali, Rust, and Zhuang, 2006). Also, epsin 1 binds to both ITSN1 (Yamabhai et al., 1998) and Eps15 (Chen et al., 1998) through the same EH-domain binding sequence and therefore, simultaneous binding of Eps15 and ITSN1 is unlikely. The Src homology 3 (SH3) domains of ITSN bind to dynamin and synaptojanin (Sengar et al., 1999). Dynamin assembles at the neck of the coated pits and functions as a constrictase to pinch off the pits (McNiven et al., 2000). DAB2 is known to function independently of AP-2 and LDLRAP1 in sorting LDL receptor (Maurer and Cooper, 2006). LDLRAP1 binds to clathrin and AP-2 (He et al., 2002). INPPL1 recruits ITSN1 to clathrin-coated pits on the plasma membrane (Xie, Vandenbroere, and Pirson, 2008). PICALM promotes assembly of clathrin triskelia into cages (Prasad and Lippoldt, 1988).β-arrestin can bind to clathrin directly (Goodman et al., 1996) and interaction of ARRB1 and 2 with AP-2 is required for endocytosis of GPCRs (Hamdan et al., 2007). HIP1 localizes with clathrin at the plasma membrane, is involved in the formation of the coated vesicle (Gottfried, Ehrlich, and Ashery, 2009) and (Legendre-Guillemin et al., 2005) and can bind to AP-2 (Metzler et al., 2001). NUMB can bind to the α-adaptin subunit of AP-2 (Santolini et al., 2000), Eps15 (Iannolo et al., 1997) and EHD4 (Smith et al., 2004). REPS1 can form complexes with two adaptor proteins Crk and Grb2 of the clathrin pathway (Yamaguchi et al., 1997). REPS2 binds directly to epsin through its EH domain (Morinaka et al., 1999). Thus, all these factors play important roles in the clathrin endocytic pathway.
TIM-1 was recently shown to be a common receptor for filoviruses (Kondratowicz et al., 2011); therefore future studies characterizing the interactions of TIM-1 with these factors of the clathrin endocytic machinery would provide further insights into the roles played by these factors in EbGP- and MARVGP-dependent viral entry. It would also be interesting to investigate if there are any differential requirements for host cell factors in filoviral entry through macropinocytosis.
Some other viruses that, like MARVGP pseudovirions have Eps15 and AP-2 independent entry mechanisms include influenza virus and mouse hepatitis virus type 2 (Lakadamyali, Rust, and Zhuang, 2006) and (Pu and Zhang, 2008). A key finding from our study is that the clathrin pathway requirements for MARVGP-mediated entry are strikingly similar to those recently reported for anthrax toxin entry (Abrami et al., 2010). In each case, cellular entry requires ARRB1, AP-1 and DYN2 but is independent of Eps15 and AP-2. AP-1 has also been shown to play a role in AP-2 independent, clathrin-mediated endocytosis of Listeria monocytogenes (Cossart and Veiga, 2008). These findings raise the intriguing possibility that a clathrin endocytic pathway, which is defined by ARRB1, AP-1 and DYN2 dependence is more generally important for pathogen and/or microbial virulence factor entry into cells. Studies are ongoing to further characterize this ARRB1 and AP-1 dependent pathway of virus entry.
Supplementary Material
Supplemental Fig. 1. Validation of the pseudotyping system and transduction assay for Ebola and Marburg GP pseudotyped HIV.
(A) HOS cells were pre-treated with 50 nM Bafilomycin A1 (Baf A1) for 30 min followed by incubation with the viruses described in the materials and methods section, overnight in the presence of the drug. 48 h post-infection, viral infectivity was measured by flow cytometry. Error bars represent SEM for three independent samples. * p Values for MARVGP < 0.001 and EbGP < 0.01. (B) HOS cells were pre-treated with 10 μM cathepsin L inhibitor (FYdmk) for 4.5 h followed by incubation with the viruses described in the materials and methods section, overnight in the presence of the drug. 48 h post-infection, viral infectivity was measured by flow cytometry. Error bars represent SEM for three independent samples. * p Values for MARVGP and EbGP < 0.01.
Supplemental Fig. 2. Schematic representation of the experimental setup for the Eps15 and siRNA experiments.
HOS cells on coverslips were transiently transfected with either the mRFP-Eps15 constructs or co-transfected with the different siRNAs and a tdTomato plasmid (as a transfection marker) for various time points described in the materials and methods section. The cells were then incubated with the GFP reporter viruses described in materials and methods section, for 4 h. 48 h post-infection, the cells were fixed and the DNA was stained with Hoechst. Several panels of images were collected from each coverslip and the number of transfected only (red) and transfected and infected (yellow) cells were counted. A minimum of 100 transfected cells was counted for each sample. The % of transfected and infected cells in the total population of transfected cells was calculated in each case.
Supplemental Fig. 3. Control experiments to determine the efficiency and specificity of the siRNAs.
(A) HOS cells were transfected with two siRNAs each against epsin 1, ITSN1, DYN2, AP1M1, DAB2, LDLRAP1, INPPL1, PICALM, ARRB1, HIP1, NUMB, REPS1 and REPS2 as described in the materials and methods section and the mRNA expression levels were measured by qPCR and normalized to cellular beta-actin mRNA expression levels. Individual bars represent single siRNAs for each gene. Error bars represent SEM for three independent experiments. * p Values for epsin 1: siRNA 1 < 0.05, siRNA 2 < 0.01; ITSN1: siRNA 1 < 0.001, siRNA 2 < 0.01; DYN2 DAB2, LDLRAP1 and ARRB1 siRNAs < 0.01; AP1M1: siRNA 1 < 0.001, siRNA 2 < 0.01; INPPL1: siRNA 1 < 0.05, siRNA 2 < 0.001; PICALM: siRNA 1 < 0.05, siRNA 2 < 0.001; HIP1: siRNA 1 < 0.01, siRNA 2 < 0.001; NUMB: siRNA 1 < 0.001, siRNA 2 < 0.05; REPS1: siRNA 1 < 0.05, siRNA 2 < 0.01 and REPS2: siRNA 1 < 0.001, siRNA 2 < 0.01. (B) HOS cells were transfected with the siRNAs described in panel (A) and cell viability was measured using the Cell Titer Glo reagent (Promega). Data from one representative experiment is shown in the figure. Individual bars represent single siRNAs for each gene. Error bars represent standard deviation for eight replicates.
Supplemental Table 1. Raw data from Immunofluorescence experiment to examine kinetics of S15-mCherry and GFP-Vpr double-labeled MARVGP pseudotyped virus fusion following ARRB1 knockdown.
HOS cells on coverslips were co-transfected with siRNAs against ARRB1 or negative control (NC) and a TagBFP plasmid (as a transfection marker) followed by incubation with S15-mCherry and GFP-Vpr double-labeled MARVGP pseudotyped virus for various time points. The cells were fixed and several panels of images were collected from each coverslip. The total numbers of S15-mCherry and GFP-Vpr double-positive virus particles as well as GFP-Vpr single-positive virus particles were counted in the BFP positive cells. The raw numbers from each image are shown.
Supplemental Table 2. Target sequences and catalog numbers (Dharmacon) of the siRNAs used in this study.
Supplemental Table 3. Sequences of the primers used to measure mRNA expression levels in this study.
Acknowledgments
We are grateful to Drs. M. Javad Aman (USAMRIID), Paul Bates (University of Pennsylvania), Mark Muesing (Rockefeller University), Roger Tsien (UCSD), Michael W. Davidson (Florida State University) and the National Institutes of Health (NIH) AIDS reagent program for providing the plasmids used in this study. We also thank Dr. Samuel L. Pfaff (Salk Institute) for allowing usage of the Zeiss Axioskop 2 microscope in his laboratory, The Waitt Advanced Biophotonics Center Core Facility for usage of the Zeiss LSM 710 laser scanning confocal microscope and John Naughton for help with figure preparation.
This work was supported by a National Institutes of Health (NIH) grant AI052051 to T.J.H and funding from the Nomis Foundation, the James B. Pendleton Charitable Trust and the Rodney Dean and Cornelia Hayes Mackey Endowment Fund to J.A.T.Y. T.J.H is also an Elizabeth Glaser Scientist.
References
- Abrami L, Bischofberger M, Kunz B, Groux R, van der Goot FG. Endocytosis of the anthrax toxin is mediated by clathrin, actin and unconventional adaptors. PLoS Pathog. 2010;6(3):e1000792. doi: 10.1371/journal.ppat.1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez CP, Lasala F, Carrillo J, Muniz O, Corbi AL, Delgado R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J Virol. 2002;76(13):6841–4. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavari S, Bosio CM, Wiegand E, Ruthel G, Will AB, Geisbert TW, Hevey M, Schmaljohn C, Schmaljohn A, Aman MJ. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J Exp Med. 2002;195(5):593–602. doi: 10.1084/jem.20011500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmerah A, Gagnon J, Begue B, Megarbane B, Dautry-Varsat A, Cerf-Bensussan N. The tyrosine kinase substrate eps15 is constitutively associated with the plasma membrane adaptor AP-2. J Cell Biol. 1995;131(6 Pt 2):1831–8. doi: 10.1083/jcb.131.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Warfield KL, Ruthel G, Bavari S, Aman MJ, Hope TJ. Ebola virus uses clathrin-mediated endocytosis as an entry pathway. Virology. 2010;401(1):18–28. doi: 10.1016/j.virol.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EM, Perez O, Melar M, Hope TJ. Labeling HIV-1 virions with two fluorescent proteins allows identification of virions that have productively entered the target cell. Virology. 2007;360(2):286–93. doi: 10.1016/j.virol.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SY, Empig CJ, Welte FJ, Speck RF, Schmaljohn A, Kreisberg JF, Goldsmith MA. Folate receptor-alpha is a cofactor for cellular entry by Marburg and Ebola viruses. Cell. 2001;106(1):117–26. doi: 10.1016/s0092-8674(01)00418-4. [DOI] [PubMed] [Google Scholar]
- Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308(5728):1643–5. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zhuang X. Epsin 1 is a cargo-specific adaptor for the clathrin-mediated endocytosis of the influenza virus. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0803711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Fre S, Slepnev VI, Capua MR, Takei K, Butler MH, Di Fiore PP, De Camilli P. Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature. 1998;394(6695):793–7. doi: 10.1038/29555. [DOI] [PubMed] [Google Scholar]
- Connolly BM, Steele KE, Davis KJ, Geisbert TW, Kell WM, Jaax NK, Jahrling PB. Pathogenesis of experimental Ebola virus infection in guinea pigs. J Infect Dis. 1999;179(Suppl 1):S203–17. doi: 10.1086/514305. [DOI] [PubMed] [Google Scholar]
- Cossart P, Veiga E. Non-classical use of clathrin during bacterial infections. J Microsc. 2008;231(3):524–8. doi: 10.1111/j.1365-2818.2008.02065.x. [DOI] [PubMed] [Google Scholar]
- Cureton DK, Massol RH, Whelan SP, Kirchhausen T. The length of vesicular stomatitis virus particles dictates a need for actin assembly during clathrin-dependent endocytosis. PLoS Pathog. 2010;6(9) doi: 10.1371/journal.ppat.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Empig CJ, Goldsmith MA. Association of the caveola vesicular system with cellular entry by filoviruses. J Virol. 2002;76(10):5266–70. doi: 10.1128/JVI.76.10.5266-5270.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Hensley LE, Larsen T, Young HA, Reed DS, Geisbert JB, Scott DP, Kagan E, Jahrling PB, Davis KJ. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am J Pathol. 2003;163(6):2347–70. doi: 10.1016/S0002-9440(10)63591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman OB, Jr, Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, Keen JH, Benovic JL. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 1996;383(6599):447–50. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- Gottfried I, Ehrlich M, Ashery U. HIP1 exhibits an early recruitment and a late stage function in the maturation of coated pits. Cell Mol Life Sci. 2009;66(17):2897–911. doi: 10.1007/s00018-009-0077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Rochdi MD, Breton B, Fessart D, Michaud DE, Charest PG, Laporte SA, Bouvier M. Unraveling G protein-coupled receptor endocytosis pathways using real-time monitoring of agonist-promoted interaction between beta-arrestins and AP-2. J Biol Chem. 2007;282(40):29089–100. doi: 10.1074/jbc.M700577200. [DOI] [PubMed] [Google Scholar]
- He G, Gupta S, Yi M, Michaely P, Hobbs HH, Cohen JC. ARH is a modular adaptor protein that interacts with the LDL receptor, clathrin, and AP-2. J Biol Chem. 2002;277(46):44044–9. doi: 10.1074/jbc.M208539200. [DOI] [PubMed] [Google Scholar]
- Huang F, Khvorova A, Marshall W, Sorkin A. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J Biol Chem. 2004;279(16):16657–61. doi: 10.1074/jbc.C400046200. [DOI] [PubMed] [Google Scholar]
- Hunt CL, Kolokoltsov AA, Davey RA, Maury W. The Tyro3 receptor kinase Axl enhances macropinocytosis of Zaire ebolavirus. J Virol. 2010 doi: 10.1128/JVI.01278-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain NK, Yamabhai M, Ramjaun AR, Guy AM, Baranes D, O’Bryan JP, Der CJ, Kay BK, McPherson PS. Splice variants of intersectin are components of the endocytic machinery in neurons and nonneuronal cells. J Biol Chem. 1999;274(22):15671–7. doi: 10.1074/jbc.274.22.15671. [DOI] [PubMed] [Google Scholar]
- Iannolo G, Salcini AE, Gaidarov I, Goodman OB, Jr, Baulida J, Carpenter G, Pelicci PG, Di Fiore PP, Keen JH. Mapping of the molecular determinants involved in the interaction between eps15 and AP-2. Cancer Res. 1997;57(2):240–5. [PubMed] [Google Scholar]
- Ivanov AI. Pharmacological inhibition of endocytic pathways: is it specific enough to be useful? Methods Mol Biol. 2008;440:15–33. doi: 10.1007/978-1-59745-178-9_2. [DOI] [PubMed] [Google Scholar]
- Jeffers SA, Sanders DA, Sanchez A. Covalent modifications of the ebola virus glycoprotein. J Virol. 2002;76(24):12463–72. doi: 10.1128/JVI.76.24.12463-12472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Olinger GG, Aris S, Chen Y, Gewurz H, Spear GT. Mannose-binding lectin binds to Ebola and Marburg envelope glycoproteins, resulting in blocking of virus interaction with DC-SIGN and complement-mediated virus neutralization. J Gen Virol. 2005;86(Pt 9):2535–42. doi: 10.1099/vir.0.81199-0. [DOI] [PubMed] [Google Scholar]
- Kaletsky RL, Simmons G, Bates P. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J Virol. 2007;81(24):13378–84. doi: 10.1128/JVI.01170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratowicz AS, Lennemann NJ, Sinn PL, Davey RA, Hunt CL, Moller-Tank S, Meyerholz DK, Rennert P, Mullins RF, Brindley M, Sandersfeld LM, Quinn K, Weller M, McCray PB, Jr, Chiorini J, Maury W. From the Cover: T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc Natl Acad Sci U S A. 2011;108(20):8426–31. doi: 10.1073/pnas.1019030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M, Rust MJ, Zhuang X. Ligands for clathrin-mediated endocytosis are differentially sorted into distinct populations of early endosomes. Cell. 2006;124(5):997–1009. doi: 10.1016/j.cell.2005.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre-Guillemin V, Metzler M, Lemaire JF, Philie J, Gan L, Hayden MR, McPherson PS. Huntingtin interacting protein 1 (HIP1) regulates clathrin assembly through direct binding to the regulatory region of the clathrin light chain. J Biol Chem. 2005;280(7):6101–8. doi: 10.1074/jbc.M408430200. [DOI] [PubMed] [Google Scholar]
- Liu YW, Surka MC, Schroeter T, Lukiyanchuk V, Schmid SL. Isoform and splice-variant specific functions of dynamin-2 revealed by analysis of conditional knockout cells. Mol Biol Cell. 2008;19(12):5347–59. doi: 10.1091/mbc.E08-08-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal V, Prevost MC, Petit C, Perret E, Heard JM, Schwartz O. Human immunodeficiency virus type 1 entry into macrophages mediated by macropinocytosis. J Virol. 2001;75(22):11166–77. doi: 10.1128/JVI.75.22.11166-11177.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi A, Akhavan A, Simmons G, Gramberg T, Hofmann H, Bates P, Lingappa VR, Pohlmann S. The signal peptide of the ebolavirus glycoprotein influences interaction with the cellular lectins DC-SIGN and DC-SIGNR. J Virol. 2006;80(13):6305–17. doi: 10.1128/JVI.02545-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer ME, Cooper JA. The adaptor protein Dab2 sorts LDL receptors into coated pits independently of AP-2 and ARH. J Cell Sci. 2006;119(Pt 20):4235–46. doi: 10.1242/jcs.03217. [DOI] [PubMed] [Google Scholar]
- McNiven MA, Cao H, Pitts KR, Yoon Y. The dynamin family of mechanoenzymes: pinching in new places. Trends Biochem Sci. 2000;25(3):115–20. doi: 10.1016/s0968-0004(99)01538-8. [DOI] [PubMed] [Google Scholar]
- Metzler M, Legendre-Guillemin V, Gan L, Chopra V, Kwok A, McPherson PS, Hayden MR. HIP1 functions in clathrin-mediated endocytosis through binding to clathrin and adaptor protein 2. J Biol Chem. 2001;276(42):39271–6. doi: 10.1074/jbc.C100401200. [DOI] [PubMed] [Google Scholar]
- Mishra SK, Keyel PA, Hawryluk MJ, Agostinelli NR, Watkins SC, Traub LM. Disabled-2 exhibits the properties of a cargo-selective endocytic clathrin adaptor. Embo J. 2002;21(18):4915–26. doi: 10.1093/emboj/cdf487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137(3):433–44. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaka K, Koyama S, Nakashima S, Hinoi T, Okawa K, Iwamatsu A, Kikuchi A. Epsin binds to the EH domain of POB1 and regulates receptor-mediated endocytosis. Oncogene. 1999;18(43):5915–22. doi: 10.1038/sj.onc.1202974. [DOI] [PubMed] [Google Scholar]
- Nanbo A, Imai M, Watanabe S, Noda T, Takahashi K, Neumann G, Halfmann P, Kawaoka Y. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010;6(9) doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naslavsky N, Caplan S. C-terminal EH-domain-containing proteins: consensus for a role in endocytic trafficking, EH? J Cell Sci. 2005;118(Pt 18):4093–101. doi: 10.1242/jcs.02595. [DOI] [PubMed] [Google Scholar]
- Oakley RH, Laporte SA, Holt JA, Barak LS, Caron MG. Molecular determinants underlying the formation of stable intracellular G protein-coupled receptor-beta-arrestin complexes after receptor endocytosis*. J Biol Chem. 2001;276(22):19452–60. doi: 10.1074/jbc.M101450200. [DOI] [PubMed] [Google Scholar]
- Polo S, Confalonieri S, Salcini AE, Di Fiore PP. EH and UIM: endocytosis and more. Sci STKE. 2003;2003(213):re17. doi: 10.1126/stke.2132003re17. [DOI] [PubMed] [Google Scholar]
- Prasad K, Lippoldt RE. Molecular characterization of the AP180 coated vesicle assembly protein. Biochemistry. 1988;27(16):6098–104. doi: 10.1021/bi00416a040. [DOI] [PubMed] [Google Scholar]
- Pu Y, Zhang X. Mouse hepatitis virus type 2 enters cells through a clathrin-mediated endocytic pathway independent of Eps15. J Virol. 2008;82(16):8112–23. doi: 10.1128/JVI.00837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn K, Brindley MA, Weller ML, Kaludov N, Kondratowicz A, Hunt CL, Sinn PL, McCray PB, Jr, Stein CS, Davidson BL, Flick R, Mandell R, Staplin W, Maury W, Chiorini JA. Rho GTPases modulate entry of Ebola virus and vesicular stomatitis virus pseudotyped vectors. J Virol. 2009;83(19):10176–86. doi: 10.1128/JVI.00422-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. Cellular Entry of Ebola Virus Involves Uptake by a Macropinocytosis-Like Mechanism and Subsequent Trafficking through Early and Late Endosomes. PLoS Pathog. 2010;6(9) doi: 10.1371/journal.ppat.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcini AE, Confalonieri S, Doria M, Santolini E, Tassi E, Minenkova O, Cesareni G, Pelicci PG, Di Fiore PP. Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes Dev. 1997;11(17):2239–49. doi: 10.1101/gad.11.17.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A. Analysis of filovirus entry into vero e6 cells, using inhibitors of endocytosis, endosomal acidification, structural integrity, and cathepsin (B and L) activity. J Infect Dis. 2007;196(Suppl 2):S251–8. doi: 10.1086/520597. [DOI] [PubMed] [Google Scholar]
- Santolini E, Puri C, Salcini AE, Gagliani MC, Pelicci PG, Tacchetti C, Di Fiore PP. Numb is an endocytic protein. J Cell Biol. 2000;151(6):1345–52. doi: 10.1083/jcb.151.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid EM, Ford MG, Burtey A, Praefcke GJ, Peak-Chew SY, Mills IG, Benmerah A, McMahon HT. Role of the AP2 beta-appendage hub in recruiting partners for clathrin-coated vesicle assembly. PLoS Biol. 2006;4(9):e262. doi: 10.1371/journal.pbio.0040262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittler HJ, Feldmann H. Viral hemorrhagic fever--a vascular disease? Thromb Haemost. 2003;89(6):967–72. [PubMed] [Google Scholar]
- Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol. 2006;80(8):4174–8. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengar AS, Wang W, Bishay J, Cohen S, Egan SE. The EH and SH3 domain Ese proteins regulate endocytosis by linking to dynamin and Eps15. Embo J. 1999;18(5):1159–71. doi: 10.1093/emboj/18.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22(12):1567–72. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shimizu N, Tanaka A, Oue A, Mori T, Ohtsuki T, Apichartpiyakul C, Uchiumi H, Nojima Y, Hoshino H. Broad usage spectrum of G protein-coupled receptors as coreceptors by primary isolates of HIV. AIDS. 2009;23(7):761–9. doi: 10.1097/QAD.0b013e328326cc0d. [DOI] [PubMed] [Google Scholar]
- Shimojima M, Takada A, Ebihara H, Neumann G, Fujioka K, Irimura T, Jones S, Feldmann H, Kawaoka Y. Tyro3 family-mediated cell entry of Ebola and Marburg viruses. J Virol. 2006;80(20):10109–16. doi: 10.1128/JVI.01157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G, Reeves JD, Grogan CC, Vandenberghe LH, Baribaud F, Whitbeck JC, Burke E, Buchmeier MJ, Soilleux EJ, Riley JL, Doms RW, Bates P, Pohlmann S. DC-SIGN and DC-SIGNR bind ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology. 2003a;305(1):115–23. doi: 10.1006/viro.2002.1730. [DOI] [PubMed] [Google Scholar]
- Simmons G, Rennekamp AJ, Chai N, Vandenberghe LH, Riley JL, Bates P. Folate receptor alpha and caveolae are not required for Ebola virus glycoprotein-mediated viral infection. J Virol. 2003b;77(24):13433–8. doi: 10.1128/JVI.77.24.13433-13438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Dho SE, Donaldson J, Tepass U, McGlade CJ. The cell fate determinant numb interacts with EHD/Rme-1 family proteins and has a role in endocytic recycling. Mol Biol Cell. 2004;15(8):3698–708. doi: 10.1091/mbc.E04-01-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein BS, Gowda SD, Lifson JD, Penhallow RC, Bensch KG, Engleman EG. pH-independent HIV entry into CD4-positive T cells via virus envelope fusion to the plasma membrane. Cell. 1987;49(5):659–68. doi: 10.1016/0092-8674(87)90542-3. [DOI] [PubMed] [Google Scholar]
- Sullivan NJ, Martin JE, Graham BS, Nabel GJ. Correlates of protective immunity for Ebola vaccines: implications for regulatory approval by the animal rule. Nat Rev Microbiol. 2009;7(5):393–400. doi: 10.1038/nrmicro2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada A, Fujioka K, Tsuiji M, Morikawa A, Higashi N, Ebihara H, Kobasa D, Feldmann H, Irimura T, Kawaoka Y. Human macrophage C-type lectin specific for galactose and N-acetylgalactosamine promotes filovirus entry. J Virol. 2004;78(6):2943–7. doi: 10.1128/JVI.78.6.2943-2947.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada A, Watanabe S, Ito H, Okazaki K, Kida H, Kawaoka Y. Downregulation of beta1 integrins by Ebola virus glycoprotein: implication for virus entry. Virology. 2000;278(1):20–6. doi: 10.1006/viro.2000.0601. [DOI] [PubMed] [Google Scholar]
- Traub LM. Sorting it out: AP-2 and alternate clathrin adaptors in endocytic cargo selection. J Cell Biol. 2003;163(2):203–8. doi: 10.1083/jcb.200309175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub LM. Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat Rev Mol Cell Biol. 2009;10(9):583–96. doi: 10.1038/nrm2751. [DOI] [PubMed] [Google Scholar]
- Unutmaz D, KewalRamani VN, Littman DR. G protein-coupled receptors in HIV and SIV entry: new perspectives on lentivirus-host interactions and on the utility of animal models. Semin Immunol. 1998;10(3):225–36. doi: 10.1006/smim.1998.0134. [DOI] [PubMed] [Google Scholar]
- Volchkov VE, Feldmann H, Volchkova VA, Klenk HD. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci U S A. 1998;95(10):5762–7. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, Rothberg KG, Anderson RG. Mis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J Cell Biol. 1993;123(5):1107–17. doi: 10.1083/jcb.123.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43(3):189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Vandenbroere I, Pirson I. SHIP2 associates with intersectin and recruits it to the plasma membrane in response to EGF. FEBS Lett. 2008;582(20):3011–7. doi: 10.1016/j.febslet.2008.07.048. [DOI] [PubMed] [Google Scholar]
- Yamabhai M, Hoffman NG, Hardison NL, McPherson PS, Castagnoli L, Cesareni G, Kay BK. Intersectin, a novel adaptor protein with two Eps15 homology and five Src homology 3 domains. J Biol Chem. 1998;273(47):31401–7. doi: 10.1074/jbc.273.47.31401. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Urano T, Goi T, Feig LA. An Eps homology (EH) domain protein that binds to the Ral-GTPase target, RalBP1. J Biol Chem. 1997;272(50):31230–4. doi: 10.1074/jbc.272.50.31230. [DOI] [PubMed] [Google Scholar]
- Yonezawa A, Cavrois M, Greene WC. Studies of ebola virus glycoprotein-mediated entry and fusion by using pseudotyped human immunodeficiency virus type 1 virions: involvement of cytoskeletal proteins and enhancement by tumor necrosis factor alpha. J Virol. 2005;79(2):918–26. doi: 10.1128/JVI.79.2.918-926.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. Validation of the pseudotyping system and transduction assay for Ebola and Marburg GP pseudotyped HIV.
(A) HOS cells were pre-treated with 50 nM Bafilomycin A1 (Baf A1) for 30 min followed by incubation with the viruses described in the materials and methods section, overnight in the presence of the drug. 48 h post-infection, viral infectivity was measured by flow cytometry. Error bars represent SEM for three independent samples. * p Values for MARVGP < 0.001 and EbGP < 0.01. (B) HOS cells were pre-treated with 10 μM cathepsin L inhibitor (FYdmk) for 4.5 h followed by incubation with the viruses described in the materials and methods section, overnight in the presence of the drug. 48 h post-infection, viral infectivity was measured by flow cytometry. Error bars represent SEM for three independent samples. * p Values for MARVGP and EbGP < 0.01.
Supplemental Fig. 2. Schematic representation of the experimental setup for the Eps15 and siRNA experiments.
HOS cells on coverslips were transiently transfected with either the mRFP-Eps15 constructs or co-transfected with the different siRNAs and a tdTomato plasmid (as a transfection marker) for various time points described in the materials and methods section. The cells were then incubated with the GFP reporter viruses described in materials and methods section, for 4 h. 48 h post-infection, the cells were fixed and the DNA was stained with Hoechst. Several panels of images were collected from each coverslip and the number of transfected only (red) and transfected and infected (yellow) cells were counted. A minimum of 100 transfected cells was counted for each sample. The % of transfected and infected cells in the total population of transfected cells was calculated in each case.
Supplemental Fig. 3. Control experiments to determine the efficiency and specificity of the siRNAs.
(A) HOS cells were transfected with two siRNAs each against epsin 1, ITSN1, DYN2, AP1M1, DAB2, LDLRAP1, INPPL1, PICALM, ARRB1, HIP1, NUMB, REPS1 and REPS2 as described in the materials and methods section and the mRNA expression levels were measured by qPCR and normalized to cellular beta-actin mRNA expression levels. Individual bars represent single siRNAs for each gene. Error bars represent SEM for three independent experiments. * p Values for epsin 1: siRNA 1 < 0.05, siRNA 2 < 0.01; ITSN1: siRNA 1 < 0.001, siRNA 2 < 0.01; DYN2 DAB2, LDLRAP1 and ARRB1 siRNAs < 0.01; AP1M1: siRNA 1 < 0.001, siRNA 2 < 0.01; INPPL1: siRNA 1 < 0.05, siRNA 2 < 0.001; PICALM: siRNA 1 < 0.05, siRNA 2 < 0.001; HIP1: siRNA 1 < 0.01, siRNA 2 < 0.001; NUMB: siRNA 1 < 0.001, siRNA 2 < 0.05; REPS1: siRNA 1 < 0.05, siRNA 2 < 0.01 and REPS2: siRNA 1 < 0.001, siRNA 2 < 0.01. (B) HOS cells were transfected with the siRNAs described in panel (A) and cell viability was measured using the Cell Titer Glo reagent (Promega). Data from one representative experiment is shown in the figure. Individual bars represent single siRNAs for each gene. Error bars represent standard deviation for eight replicates.
Supplemental Table 1. Raw data from Immunofluorescence experiment to examine kinetics of S15-mCherry and GFP-Vpr double-labeled MARVGP pseudotyped virus fusion following ARRB1 knockdown.
HOS cells on coverslips were co-transfected with siRNAs against ARRB1 or negative control (NC) and a TagBFP plasmid (as a transfection marker) followed by incubation with S15-mCherry and GFP-Vpr double-labeled MARVGP pseudotyped virus for various time points. The cells were fixed and several panels of images were collected from each coverslip. The total numbers of S15-mCherry and GFP-Vpr double-positive virus particles as well as GFP-Vpr single-positive virus particles were counted in the BFP positive cells. The raw numbers from each image are shown.
Supplemental Table 2. Target sequences and catalog numbers (Dharmacon) of the siRNAs used in this study.
Supplemental Table 3. Sequences of the primers used to measure mRNA expression levels in this study.