Abstract
Genes that are expressed only in the young zygote are considered to be of great importance in the development of an isogamous green alga, Chlamydomonas reinhardtii. Clones representing the Zys3 gene were isolated from a cDNA library prepared using zygotes at 10 min after fertilization. Sequencing of Zys3 cDNA clones resulted in the isolation of two related molecular species. One of them encoded a protein that contained two kinds of protein-to-protein interaction motifs known as ankyrin repeats and WW domains. The other clone lacked the ankyrin repeats but was otherwise identical. These mRNA species began to accumulate simultaneously in cells beginning 10 min after fertilization, and reached maximum levels at about 4 h, after which time levels decreased markedly. Genomic DNA gel-blot analysis indicated that Zys3 was a single-copy gene. The Zys3 proteins exhibited parallel expression to the Zys3 mRNAs at first, appearing 2 h after mating, and reached maximum levels at more than 6 h, but persisted to at least 1 d. Immunocytochemical analysis revealed their localization in the endoplasmic reticulum, which suggests a role in the morphological changes of the endoplasmic reticulum or in the synthesis and transport of proteins to the Golgi apparatus or related vesicles.
Chlamydomonas reinhardtii is a unicellular, isogamous green alga, the sexual life cycle of which is controlled by genetically determined mating types consisting of two kinds of haploid cells that are morphologically very similar, but contain a distinct locus on their nuclear genome (Ferris and Goodenough, 1994). In sexual reproduction, the gametes are induced independently from corresponding vegetative cells in a nitrogen-starved environment. When they encounter cells of the opposite mating type, they recognize their partner, begin to agglutinate, and then fuse to become zygotes. After zygote formation, a number of events ensue, including preferential digestion of male-derived chloroplast nuclei (Kuroiwa et al., 1982), nuclear fusion (Cavalier-Smith, 1970; Kuroiwa et al., 1982), flagellar degeneration, and zygospore formation (Cavalier-Smith, 1976). All functional proteins and their mRNAs directly involved in these phenomena are thought to be synthesized only after cell fusion (Kuroiwa et al., 1983; Kuroiwa, 1991). Therefore, genes expressed specifically and relatively early in zygotes should play important roles in the regulation of this complex series of events.
Fertilization has been intensively studied in organisms such as the sea urchin, the newt, and mammals. The gametes of these animals highly differentiate to form sperm and eggs, and the egg already has the full complement of mRNA necessary for the very early stage of embryonic development, because the blocking of RNA synthesis has no effect on the embryo until it reaches the blastula stage (Gilbert, 1988). In contrast, a C. reinhardtii zygote undergoes a burst of gene expression immediately after cell fusion. Zygote-specific genes of C. reinhardtii have been isolated using differential screening by several groups (Ferris and Goodenough, 1987; Wegener and Beck, 1991; Uchida et al., 1993). Uchida et al. (1993) employed a cDNA library prepared from mRNAs of zygotes 10 min after cell fusion, so their clones may include fragments of essential genes that function from a very early stage and regulate the developmental system of a zygote. We report here molecular-biological and immunocytochemical characterization of one of these genes previously denoted as Zys3 (Uchida et al., 1993).
The deduced amino acid sequence of a full-length Zys3 cDNA clone contained two ankyrin repeats and two WW domains, both of which are known to be functional protein-to-protein interaction sites. The ankyrin repeat was originally noted in the CDC10 gene of Shizosaccharomyces pombe by Aves et al. (1985). CDC10 and its homologs SWI6 and SWI4 in Saccharomyces cerevisiae function in cell proliferation and mating-type switching as transcription complexes (Breeden and Nasmyth, 1987; Andrews and Herskowitz, 1989). A number of related genes have since been isolated, including Fem-1, a sex determinant in the nematode (Spence et al., 1990); Lin-12, Glp-1, and Notch, intrinsic membrane proteins (Wharton et al., 1985; Yochem et al., 1988; Yochem and Greenwald, 1989); GABPβ, NF-κB/p105, IκBα (MAD-3), bcl-3, and Arabidopsis AKRP, transcription-factor subunits or regulators of transcriptional systems (Bours et al., 1990, 1993; Ghosh et al., 1990; Kieran et al., 1990; Ohno et al., 1990; Haskill et al., 1991; Lamarco et al., 1991; Thompson et al., 1991; Zhang et al., 1992); and ankyrin, a cytoskeletal protein found in mammals, Drosophila, and nematodes (Lux et al., 1990; Bennet, 1992).
The WW domain is found in a wide range of cytoskeletal, regulatory, and signaling molecules, including dystrophin, a cytoskeletal protein (Ahn and Kunkel, 1993) that stabilizes the membrane and generates contractile force; the human Yap protein, a mediator of cell-growth signals (Sudol et al., 1995); IQGAP1, a human GTPase-mediated, cytoskeleton-regulating protein (Hart et al., 1996); rat FE65, a transcription-factor activator (Bork and Sudol, 1994); and tobacco DB10, a RNA helicase (Itadani et al., 1994). Although these genes may have a diverse range of functions, they all probably work via protein-to-protein interactions, and in the case of the WW domain, the sequences of the specific ligands have also been determined (Einbond and Sudol, 1996). To our knowledge, Zys3 is the first gene that encodes sequences of both of these motifs. Another Zys3 mRNA that contains no complete ankyrin repeat was also present, but its temporal expression pattern was almost the same as that described above. These different mRNA species were suggested to be transcribed from the same single-copy gene.
Immunoblotting with two polyclonal antibodies indicated that the Zys3 gene products were expressed within 2 h of fertilization and persisted for at least 1 d. The Zys3 gene products began to accumulate at the extending ER of zygotes at 3 h after cell fusion. This localization pattern suggested their role in the control of the ER systems in synthesis, sorting, or transport of proteins required for further zygote development.
MATERIALS AND METHODS
Strains and Culture Conditions
Chlamydomonas reinhardtii wild-type strain 137c, mt+, and mt− were cultured as described by Uchida et al. (1993). Vegetative cells were maintained on Snell's medium II (Snell, 1982) supplemented with 1.0% (w/v) agar. Cells were harvested and suspended in distilled water at a concentration of 1.0 × 107 cells/mL. One-hundred-milliliter volumes of cell suspensions were incubated in Petri dishes under light for about 3 h, with occasional shaking to induce differentiation into gametes. After confirmation of gametogenesis, suspensions of mt+ and mt− gametes were mixed and sampled at the appropriate times.
Construction and Screening of a cDNA Library
For preparation of a cDNA library, total RNA was extracted by ultracentrifugation through a CsCl cushion (Sambrook et al., 1989). Poly(A+) was obtained using latex beads with immobilized oligo(dT)-cellulose (Oligotex, Nippon Roche, Kamakura, Japan).
Double-stranded cDNA was synthesized from the poly(A+) RNA of zygotes at 10 min after conjugation using a cDNA-synthesis kit (TimeSaver, Pharmacia Biotech) according to the manufacturer's instructions. An EcoRI adaptor-linker containing cleavage sites for NotI was ligated to the cDNA and then to λgt 10 DNA that had been digested with EcoRI and dephosphorylated using a cDNA rapid-cloning module (no. RPN1257, Amersham). The DNA was packaged into phage particles using an in vitro-packaging module (no. RPN1258, Amersham), and propagated using Escherichia coli NM514 as a host bacterium, which does not support growth of insert-free λgt 10. Approximately 1.0 × 105 independent recombinant phages were screened, and inserts in positive plaques were subcloned according to the method of Sambrook et al. (1989).
Sequence Analysis
Unidirectional deletions in the cloned fragments were produced using an exo/mung bean nuclease deletion kit (Stratagene). Single-stranded DNAs from selected deletion clones were purified from PEG-precipitated helper phage R408. Nucleotide sequences were determined with the dideoxyribonucleotide chain-termination method (Sanger, 1981) using a DNA-sequencing system (373S, Applied Biosystems, Foster City, CA) and a Taq terminator cycle-sequencing kit (DyeDeoxy, Applied Biosystems) according to the manufacturer's instructions. Sequencing data were analyzed with DNASIS software (Hitachi Software Engineering, Yokohama, Japan) and the BLAST program (Altshul et al., 1990).
Northern-Blot Hybridization and RNase Protection Assay
Total RNA was extracted according to the method of Kirk and Kirk (1985). RNA (10 μg/lane) was glyoxylated and electrophoresed in 1.1% (w/v) agarose gel (Agarose NA, Pharmacia Biotech) in 10 mm sodium phosphate buffer, pH 7.0, at 3 V/cm for 4 h with rapid circulation. After electrophoresis, RNA was transferred to a nylon membrane (Biodyne B, Pall Corporation, Port Washington, NY) with a vacuum-blotting apparatus (Vacugene, Pharmacia Biotech), and the membrane was treated as described by Sambrook et al. (1989).
Fragments from positions 225 to 507 of clone T91 and 232 to 590 of clone T106 were cut with SacII (Toyo Boseki, Osaka, Japan) and BamHI (Takara Biomedicals, Kyoto, Japan) and then subcloned into the pBluescript SKII+ to generate riboprobes specific for those clones. Extra parts of the multicloning site were deleted, and the resultant plasmid vectors were designated as pRPZ01 and pRPZ02, respectively. In vitro transcription was performed on 2 μg of template vector linearized with HincII (Takara Biomedicals) using T7-RNA polymerase (Stratagene) in a 20-μL reaction system consisting of T7 reaction buffer (Stratagene), RNase inhibitor (Toyo Boseki), 1 mm ATP, CTP, and GTP (Boehringer Manheim), 7.5 μm UTP (Boehringer Manheim), and 12.5 μm [α-32P]UTP (Amersham). Subsequent hybridization and digestion were performed according to the manufacturer's instructions for the RPAII kit (Ambion, Austin, TX) using 2 μg of C. reinhardtii total RNA. Samples were electrophoresed through a denaturing 5% (w/v) acrylamide gel using a Mini-PROTEAN II Cell (Bio-Rad) at 250 V for 30 min. The gels were sealed in a polypropylene bag and exposed to radiographic film (X-Omat AR, Kodak) for 2 to 6 h at −80°C.
Genomic Southern-Blot Analysis
Total DNA extraction was performed by the modified method of Ohta et al. (1992). The DNA (10 μg/lane) was digested with the restriction enzymes AvaI, PstI, and PvuII. Each restriction fragment was separated in a 1.1% (w/v) agarose gel, transferred onto a nylon membrane, and incubated with a labeled probe complementary to the full-length cDNA clone, as described previously (Sambrook et al., 1989). The membrane was then washed twice in 300 mm NaCl, 30 mm trisodium citrate, 0.1% (w/v) SDS at 65°C for 15 min, twice in 15 mm NaCl, 1.5 mm trisodium citrate, 0.1% (w/v) SDS at 65°C for 15 min, and then autoradiographed.
Preparation of Anti-Zys3 Protein Antibodies and Western-Blot Analysis
Residues 262 to 275 of the deduced amino acid sequence of Zys3 proteins, MHPNRRWYNTATRE, were selected as an antigen for the preparation of rabbit anti-Zys3 protein antibody.
A fragment from 116 to 1115 of clone T106 was isolated with Nae I (Takara Biomedicals), ligated in the SmaI site of the pQE32 expression vector (Qiagen, Chatsworth, CA), and introduced into E. coli XL-1-Blue competent cells according to the method of Inoue et al. (1991). Subsequent extraction and purification of the fusion protein were carried out as described in the manufacturer's instructions. Two milligrams of purified fusion protein was used for the immunization of rats.
The sera were used without further purification in the following experiments and their specific antibodies were designated as α-Zyspept3 and α-Zysfuse3.
Sampling of total proteins for western-blot analysis was performed essentially according to the method of Arnburst et al. (1993). About 1.0 × 107 cells of each sample were dissolved in 50 μL of 1× sample buffer (62.5 mm Tris-Cl, pH 6.8, 10% [v/v] glycerol, 2% [w/v] SDS, 5% [v/v] β-mercaptethanol, and 0.01% [w/v] bromphenol blue) with boiling for 5 min, and 5 μL was used for western-blot analysis.
Samples were fractionated by SDS-PAGE through a 12% Laemmli gel (Laemmli, 1970) at 200 V for 42 min using a Mini-PROTEAN II Cell (Bio-Rad). We used a miniature transblot apparatus (Bio-Rad) for electroblotting onto nitrocellulose membranes, in which proteins within a gel could be transferred at 100 V for 1 h in the cooled transfer buffer (20% [v/v] methanol, 25 mm Tris-HCl, and 192 mm Gly). Labeled proteins were detected with an assay kit (Immune-Blot, Bio-Rad).
Immunofluorescence Microscopy and Electron Microscopy
About 1 × 107 C. reinhardtii cells were fixed according to the method of Armbrust et al. (1993). After blocking in 5% (w/v) BSA, 0.05% (v/v) Tween 20 in 1× PBS for 1 h at 37°C, a 1:100 dilution of α-Zysfuse3 was added to specimens attached to coverslips, which were then incubated for 12 h or more at room temperature. The sample was washed twice in 1× PBS for 10 min, and then incubated again at 37°C for 1 h with 5% (w/v) BSA, 0.05% (v/v) Tween 20 in PBS containing a 1:100 dilution of FITC-conjugated goat anti-rat IgG antibody. After subsequent washing with 1× PBS twice for 10 min, the coverslip was placed on a glass slide onto which DAPI and 50% (v/v) glycerol containing n-propylgallate had been dropped. The signal was observed by an epifluorescence microscope (model BHSRFC, Olympus, Tokyo, Japan).
For electron microscopy, cells were fixed in 2% (v/v) glutaraldehyde for 2 h at 4°C and with 2% (w/v) OsO4 for 2 h at room temperature, dehydrated through an ethanol series, and substituted for propylene oxide. The samples were embedded in Spurr's resin (Spurr, 1969), as described by Nozaki et al. (1994). After polymerization, sections were cut with a diamond knife on an ultramicrotome (model MT-6000 XL, RMC-Eiko, Kawasaki, Japan), and then stained with uranyl acetate and lead citrate. Samples for immunodetection were fixed only with 2% (v/v) glutaraldehyde and embedded in London Resin White (London Resin Co., Hampshire, UK), as described previously (Johnson and Rosenbaum, 1990).
Following incubation for 2 d at 50°C, samples were sliced and collected on Formvar-coated nickel grids. Each mesh was inverted onto drops of blocking solution (PBS, pH 7.4, and 1% [w/v] BSA) for 30 min, incubated on drops of anti-Zys3 protein antibody (diluted in blocking solution) overnight at 4°C, and washed with PBS, pH 7.4, containing 0.01% (v/v) Tween 20 (Bio-Rad). They were then incubated on drops of gold goat anti-rat IgG antibody (10-nm EM grade, Zymed, San Francisco, CA) diluted to 1:100 in blocking solution, pH 8.2, for 2 h, washed with PBS, pH 8.2, plus 0.01% (v/v) Tween 20, and finally with distilled water before being air dried. Sections were contrasted with uranyl acetate and viewed on an electron microscope (JEM-1200 EX, Jeol).
RESULTS
Nucleotide Sequences of Zys3 cDNAs
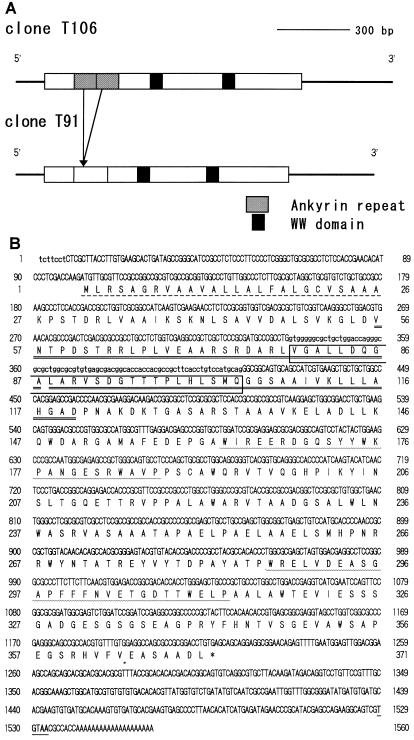
To obtain longer cDNA inserts with greater efficiency, a new cDNA library was constructed using mRNA prepared from zygotes 10 min after conjugation. This library was estimated to contain about 3.5 × 106 independent clones, and a total of 2 × 105 phages was screened. Thirty-nine positive clones were isolated at the first screening, eight of which were arbitrarily selected for a second round of screening. Based on insert sizes, three inserts were expected to contain nearly full-length Zys3 cDNA (data not shown) and were further processed for sequence analysis. The restriction maps of two different clones are shown in Figure 1A.
Figure 1.
A, Restriction maps of cDNA clones T106 and T91. Solid lines and rectangles indicate the noncoding and coding regions of these clones, respectively. The two ankyrin repeats in T106 are indicated as shaded boxes and the WW domains as black boxes. The corresponding region of T91 was partially deleted, and no ankyrin repeat was present. B, Nucleotide (above) and deduced amino acid (below) sequences of cDNA clones T106 and T91. Lowercase letters indicate the sequences present only in clone T106. The amino acids composing the N-terminal putative signal peptide, ankyrin repeats, and WW domains are represented with broken lines, double underlines, and wavy lines, respectively. Clone T91 lacks the amino acid sequences in the open box, and thus contains no ankyrin repeat. A stop codon, TGA, is represented with an asterisk. The putative polyadenylation signal of C. reinhardtii (Youngblom et al., 1984) is underlined. Accession numbers for clones T106 and T91 are AB004042 and AB004043, respectively.
Figure 1B shows the sequences of cDNA clones that contain the putative full-length Zys3-coding region. In clone T106, 101 bp of the 5′ untranslated region was followed by 1113 bp of an open reading frame, 326 bp of a 3′ noncoding sequence, and 20 bp of poly(A+) tail. A consensus polyadenylation signal of C. reinhardtii (Youngblom et al., 1984) could be found in the 3′ untranslated region at position 1529. The open reading frame corresponding to a polypeptide of 371 amino acid residues with a predicted molecular mass of 39.3 kD was recognized. A putative signal peptide of 25 residues was found using the pSORT program (http://psort.nibb.ac.jp/) at the N terminus of the proteins. A repeated amino acid motif, ----D--G-TPLH-AA-------V--LL--GA-, which has been described in a number of genes, was located from residues 56 to 89 and 90 to 120 of clone T106, although one residue of the second repeat was deleted. This domain sequence is called an ankyrin repeat (Bennet, 1992) and is supposed to be involved in protein-to-protein interactions. The alignment of ankyrin repeats in various genes is shown in Figure 2A. In ankyrin repeat 1 of clone T106, 10 amino acids were well conserved relative to the ankyrin consensus sequence and, in ankyrin repeat 2, 11 amino acids were identical. A second DNA clone, T91, had extensive homology to T106 but lacked complete ankyrin repeats.
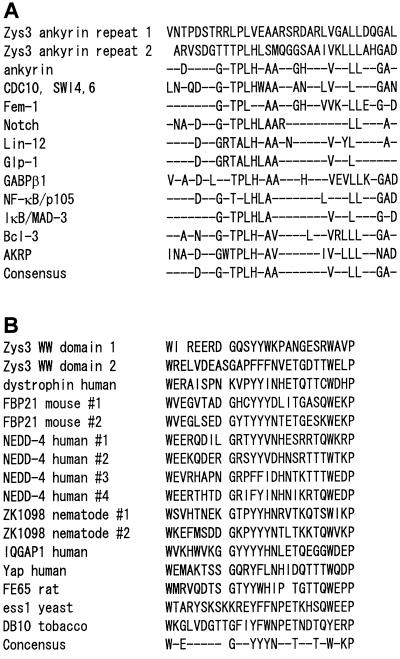
Figure 2.
A, Comparison of ankyrin repeats of a Zys3 protein and other gene products. Consensus amino acids within repeats of a particular gene product are shown on the right. Consensus amino acids of all repeats are indicated in the bottom row. The sequence data (except those of Zys3) are cited from Spence et al. (1990), Haskill et al. (1991), and Zhang et al. (1992). B, Comparison of the WW domains of Zys3 with other proteins described by Sudol et al. (1995) or Chan et al. (1996).
Two WW domains (Sudol et al., 1995) were apparent at residues 163 to 187 and 287 to 313 of clone T106 (Figs. 1 and 2B), matching the consensus sequences W--------(-) (-)V(F,Y)F(Y,W)------(-)G(S,T,N,E)G(S,T,Q,C,R)F(Y,W)--P (BEAUTY search; Worley et al., 1995).
Characterization of the Zys3 Gene
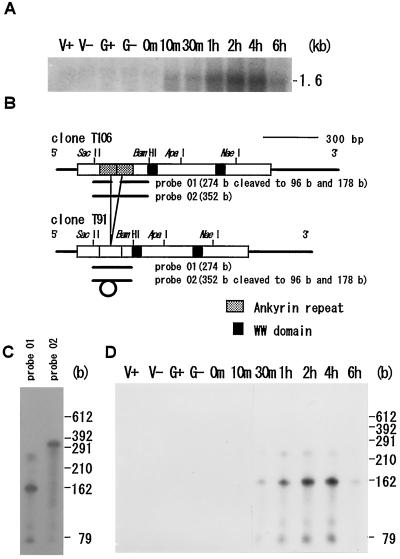
Northern-blot hybridization carried out with a full-length Zys3 cDNA probe showed that Zys3 was not expressed in the vegetative cells and gametes of either mating type, and that it began to accumulate at 10 min and reached a maximum level at 4 h after zygote formation. The signal then decreased dramatically after 6 h (Fig. 3A; Uchida et al., 1993). No additional signals could be found at other positions even with this full-length probe (Uchida et al., 1993). As mentioned above, screening of the cDNA library led to the isolation of two homologous mRNA species (Fig. 1A), both of which would be detected by this probe. The RNase protection assay was used to distinguish between the expression patterns of these two mRNAs. The T91-specific probe (305 bases) from pRPZ01 and the T106-specific probe (383 bases) from pRPZ02, which protected 274- and 352-base fragments, respectively (Fig. 3B), were applied to the total RNA samples from zygotes at 4 h (Fig. 3C). In both lanes the fragments of the expected sizes appeared with cleaved, smaller bands, indicating that both types of transcripts are present in vivo. The T91-specific probe was then used to examine the temporal expression of T91 (Fig. 3D). T91 transcripts were first detected 10 min after cell fusion, with the highest level of expression at 4 h and a rapid decrease at 6 h (Fig. 3D). This pattern was similar to that obtained by northern blotting, but in this case, more intense signals were visible at the sites of fragments 178 and 96 bases in length. These smaller fragments must represent the cleaved probe from pRPZ01, which was not protected from RNase A because of its incomplete hybridization with C. reinhardtii Zys3 mRNA of the T106 type, and showed the same temporal pattern of expression as T91. Another experiment with the T106-specific probe resulted in confirmation of the same pattern. Therefore, we conclude that both types of transcripts were synthesized simultaneously, but that T106 mRNA was far more abundant than T91 mRNA in the zygotes.
Figure 3.
A, Northern-blot hybridization using the full-length T106 probe of 10 μg of total RNA of vegetative mt+ (V+) and mt− (V−) cells, mt+ (G+) and mt− (G−) gametes, and zygotes at 0, 10, and 30 min and 1, 2, 4, and 6 h. B, Location of the ankyrin repeats on clone T106 and of the deleted sequence on clone T91. The region selected as specific probes for T91 or T106 and resultant fragments degraded by RNase A are indicated as solid lines. C, Difference in signal patterns in the RNase protection assay applied to total RNA of the zygote at 4 h with either specific probe. D, Survey of the temporal expression patterns of Zys3 mRNA by the RNase protection assay using the partial sequence specific for cDNA clone T91. HincII-digested φX174 DNA was used as a molecular marker.
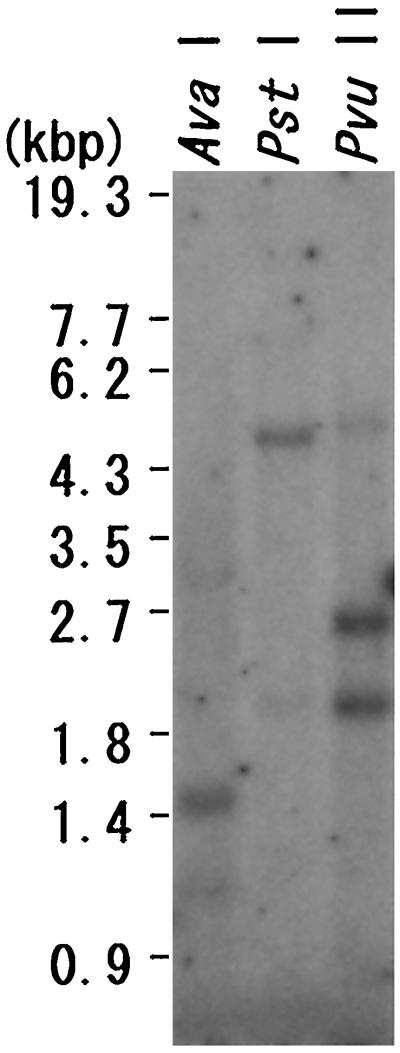
Genomic DNA of C. reinhardtii was digested with AvaI, PstI, or PvuII, and electrophoresed. Figure 4 shows an autoradiograph of the blotted membrane, which was incubated with the probe complementary to the full-length cDNA clone T106 and washed in high-stringency conditions. The probe selectively and strongly hybridized to fragments of about 1.5 kb of AvaI-digested total DNA, about 4.5 kb of PstI-digested total DNA, and about 2.5 and 1.9 kb of PvuII-digested total DNA. As indicated in the sequence data (Fig. 1B), there was one PvuII site (5′ CAG/CTG 3′) on the Zys3 cDNA at the middle position of 665 to 670 nucleotides. These results indicate that Zys3 exists as a single-copy gene. Other related sequences appeared in the digested C. reinhardtii nuclear genome when Southern-blot analysis was performed with moderate washing conditions (60°C; data not shown).
Figure 4.
Southern-blot hybridization analysis using the probe complementary to the full-length cDNA clone T106. Separated 10-μg total DNA samples that had been digested with AvaI, PstI, or PvuII were blotted onto nylon membranes and hybridized with the labeled T106 cDNA probe. StyI-digested λDNA was used as a molecular marker.
Expression Pattern of Zys3 Proteins
The sequence from position 116 to 1115 of clone T106 was ligated to pQE32, an expression vector that imposed six His residues on the N terminus of the recombinant protein for affinity purification. By inducing overexpression of the protein in bacteria, a distinct band emerged at the apparent molecular mass position of 38 kD in their lysate (Fig. 5). The amount of this protein increased as the interval between induction and sampling of E. coli was elongated (data not shown), and this protein also appeared in the fraction of affinity-purified sample, indicating that an expression system of Zys3 fusion protein was established.
Figure 5.
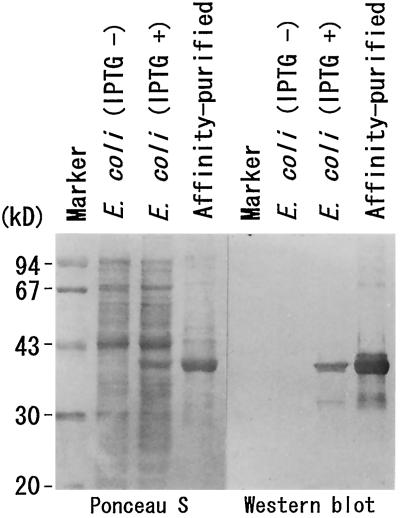
Specificity of α-Zyspept3 against the bacterially expressed fusion protein. Total proteins of noninduced E. coli (IPTG −), E. coli induced with IPTG (IPTG +), and affinity-purified fusion protein were electrophoresed, blotted, and visualized by staining with Ponceau S (left). The results of western blotting against α-Zyspept3 using the same membrane are shown on the right. α-Zyspept3 specifically reacted with the 38-kD fusion protein.
We constructed two polyclonal antibodies against Zys3 proteins. The first was designed against the partial polypeptide sequence of clone T106 (see Methods) of residues 262 to 275 and was named α-Zyspept3. The second was generated against the overexpressed fusion protein and was named α-Zysfuse3.
The α-Zyspept3 antibody specifically reacted with the 38-kD fusion protein in IPTG-induced E. coli cell lysate and the affinity-purified sample (Fig. 5, lanes 3 and 4), but no such signal could be seen in noninduced cells (lane 2).
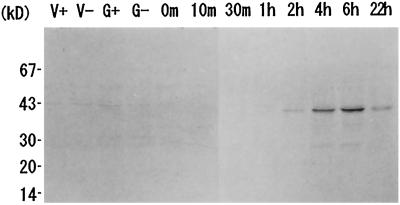
Against the fractionated total protein of C. reinhardtii vegetative cells, gametes, and zygotes sampled at the indicated times (Fig. 6), α-Zyspept3 antibody was allowed to react on the nitrocellulose membrane. This antibody recognized proteins of C. reinhardtii that had migrated to the apparent molecular mass site of 40 kD. Aside from the distinct main signal, a minor signal was also detected as a slightly heavier molecule. This may represent the Zys3 precursor still possessing the signal peptide (Fig. 1B). The protein of the main signal began to accumulate 2 h after zygote formation, reached a maximum level at 6 h, and persisted over 22 h.
Figure 6.
Temporal expression pattern of Zys3 proteins. Total protein samples of C. reinhardtii vegetative mt+ (V+) and mt− (V−) cells, mt+ (G+) and mt− (G−) gametes, and zygotes at 0, 10, and 30 min and 1, 2, 4, 6, and 22 h were fractionated, blotted, and probed with α-Zyspept3. Strong signals were detected in the vicinity of 40 kD at 2, 4, 6, and 22 h. A slightly heavier minor band that appeared at the same time may be the Zys3 precursor containing the signal peptide.
Subcellular Localization of Zys3 Gene Products
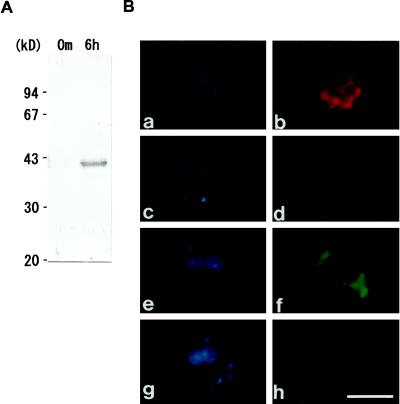
Figure 7 shows the results of western analysis of C. reinhardtii total proteins and α-Zysfuse3 antibody and the localization of Zys3 products in cells by epifluorescence microscopy. The specificity of α-Zysfuse3, an antibody raised against the fusion protein, was tested in the same way (Fig. 5). This antibody also reacted with the overexpressed 38-kD protein of E. coli (data not shown), as well as with the endogenous 40-kD protein and its putative precursor of C. reinhardtii zygotes at 6 h (Fig. 7A), but did not react with total protein from C. reinhardtii zygotes at 0 min (Fig. 7A). The α-Zysfuse3 was used to analyze the subcellular localization of Zys3 proteins to increase the sensitivity for signal detection.
Figure 7.
A, Specificity of α-Zysfuse3, an antibody raised against affinity-purified, bacterially synthesized fusion protein of Zys3 and a 6× His tag. The fractionated total protein of the C. reinhardtii zygote at 0 min and 6 h after mating by SDS-PAGE was blotted onto the nitrocellulose membrane. The α-Zysfuse3 antibody was shown by western-blot analysis to specifically react with the 40-kD Zys3 proteins and the precursor in the zygote at 6 h. B, Indirect fluorescence microscopy for analysis of localization of Zys3 products. α-Zysfuse3 was used as the primary antibody in the mt− gamete (a and b), and in the zygotes at 1 h (c and d) and 6 h (e and f) after fertilization to detect its selective reaction against Zys3 proteins. Preimmune serum was also applied to zygotes at 6 h (g and h). Cells were observed under UV light by staining with DAPI (a, c, e, and g) and under blue light to detect the FITC signal (b, d, f, and h). Strong fluorescence of FITC was observed only around the cell nucleus from zygotes 6 h after fertilization. Bar = 5 μm. Red autofluorescence in b and d resulted from remnant chlorophyll molecules of the specimens.
Specificity of α-Zysfuse3 was enough for the immunocytochemical analyses. As shown in Figure 7B, the α-Zysfuse3 antibody bound strongly to a protein in the C. reinhardtii zygotes at 6 h after fertilization, but did not react with anything in gametes or very young zygotes at 1 h. Preimmune serum of α-Zysfuse3 did not detect anything in zygotes at 6 h, confirming the specificity of this antibody. Visualization of the cell and chloroplast nuclei with DAPI (Fig. 7B, a, c, e, and g) indicated that Zys3 gene products exist within the cytoplasm and encompass the fused cell nucleus.
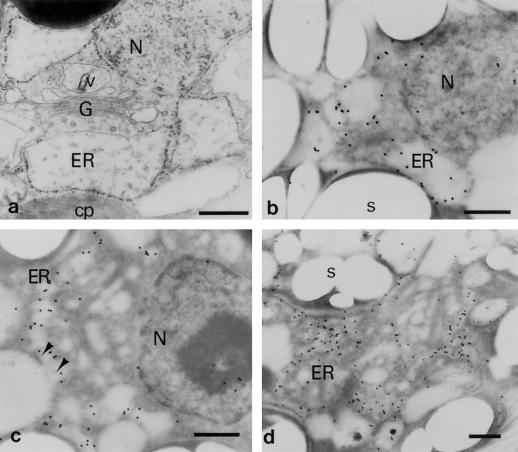
A general ultrastructural image of a C. reinhardtii zygote is presented in Figure 8a. The nucleus is surrounded by the rough ER, the Golgi apparatus, and a large chloroplast. The vesicular derivatives from the Golgi apparatus can be distinguished from the ER sac by the filamentous appearance of their contents (Fig. 8a; Minami and Goodenough, 1978). Ribosomes also attach to the outer surface of the nuclear envelope. Detection of Zys3 proteins was performed according to the method of Nozaki et al. (1994) using young (Figs. 8, b–d, and 9a) and premature zygotes (Fig. 9, b–d) that had been embedded in London Resin White and sectioned. The signals were localized on the surface of the ER membrane (Fig. 8b) or on the invaginated protrusions from the cytosol (Fig. 8c) at the early stages of zygote development (3 and 6 h after mating; Fig. 8, b and c, respectively). They were then localized specifically on the continuous, anastomosing ER membrane system, which spread throughout the C. reinhardtii zygotes 6 h after fertilization (Fig. 8d).
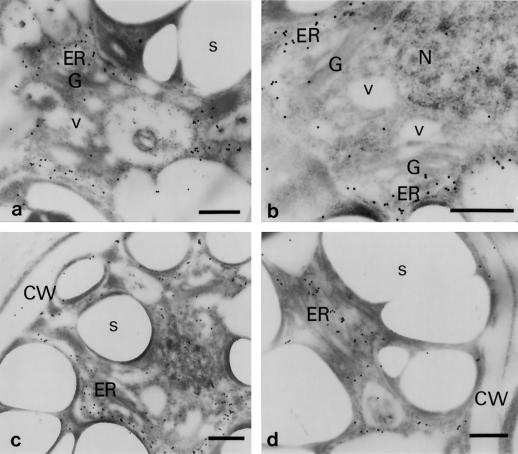
Figure 8.
Immunoelectron microscopic analyses with the anti-Zys3 protein antibody α-Zysfuse3. A general ultrastructural image of a very young zygote of C. reinhardtii is represented first as a control (a), and then at 3 h (b) and 6 h (c and d) after being reacted with α-Zysfuse3. N, Nucleus; cp, chloroplast; s, starch grain; G, Golgi apparatus; v, Golgi-related vesicle. Arrowheads indicate undefined structures protruding from the cytosolic side into the ER. Bars = 500 nm.
Figure 9.
Localization patterns of Zys3 proteins were investigated along the protein-secretion pathway of C. reinhardtii zygotes at 6 h (a) and 22 h (b–d). CW, Cell wall. Other abbreviations are as in Figure 8. Bars = 500 nm.
As for the distribution of signals along the protein-secretion pathway, the Zys3 proteins migrated to the vicinity of the cis side of the Golgi stacks by 6 h (Fig. 9a). Only negligible gold particles were detected in the Golgi sac and Golgi-derived vesicles at this stage (Fig. 9a). Even at 22 h after fertilization, none of particles moved to the Golgi apparatus, remaining instead on the membrane of the ER (Fig. 9b). This organelle appeared to extend toward the cell surface, but was distinct from the plasma membrane (Fig. 9c). Signal was barely seen on the plasma membrane or in the cell wall even at this later stage (Fig. 9d). In addition, there was no regularity in the distribution of signal in the gametes and zygotes 1 h after fertilization (data not shown). This localization pattern was supported by previous results with epifluorescence microscopy, in which signals were detected in the cytoplasm of premature zygotes, but no signal was evident in gametes and very young zygotes (Fig. 7). The fluorescent structure in Figure 7B, f, corresponded to that of the ER of a C. reinhardtii zygote.
DISCUSSION
C. reinhardtii is one of the simplest experimental organisms for the study of sexual reproduction (Goodenough et al., 1995). The mt+ and mt− cells are thought to differ only within a narrow region of the nuclear genome (Ferris and Goodenough, 1994), and the experimental control of gametogenesis and fertilization is far easier than in most other organisms. Gametes can be induced separately by deprivation of a nitrogen source, and the efficiency of mating can be elevated to more than 90% within 10 min after mixing masses of homogeneous mating types. To understand the fundamental features of fertilization and subsequent zygote development, the central part of sexual reproduction, it is most effective to employ this simple and efficient system.
This experimental system has enabled us to analyze many zygote-specific genes. At least 16 such genes have been isolated so far from C. reinhardtii by differential screening (Ferris and Goodenough, 1987; Wegener and Beck, 1991; Uchida et al., 1993). The message of the zymB and zymC genes does not significantly accumulate until zygote development has progressed 24 h after fertilization (Wegener and Beck, 1991), suggesting that these genes may participate in sporulation or later events. The Class V gene was expressed from 2.5 h after cell fusion, but was dependent on other products for its transcriptional activation (Matters and Goodenough, 1992). The function of this protein is unknown, although it contains sequences homologous to cell-surface receptors and protein kinases (Matters and Goodenough, 1992). In contrast, mRNAs of Classes I to IV, VI, and zys1–4 began to accumulate from a very early stage and required no zygote-specific protein synthesis (Ferris and Goodenough, 1987; Uchida et al., 1993). Woessner and Goodenough (1989) suggested a structural role in the architecture of the zygote cell wall for Class IV and VI genes. Armbrust et al. (1993) reported that transcription of ezy-1 (Class III) was sensitive to UV light and that its product had an almost identical electrophoretic mobility to a protein previously detected by Nakamura et al. (1988). This ezy-1 product is thought to bind directly to chloroplast nuclei to preferentially digest those derived from the mt− gamete. Some of these zygote-specific genes are likely to be involved in transcriptional regulation (Uchida et al., 1993). C. reinhardtii begins to express the Zys3 gene from the moment of cell fusion, suggesting its involvement in zygote development from an early stage.
Sequence analysis indicated that the zygote-specific gene, Zys3, encoding novel proteins with two kinds of repeat sequences, ankyrin repeats and WW domains, was probably involved in protein-to-protein interactions. Both yeast CDC10/SWI4 and SWI6 genes and the C. reinhardtii Zys3 gene contain the two ankyrin repeats, but in the yeast genes they are located separately, in the middle of the sequence, and in C. reinhardtii they are located tandemly, near the 5′ end (Breeden and Nasmyth, 1987; Fig. 1B). Transmembrane receptors have six ankyrin repeats on their internal side (Wharton et al., 1985; Yochem et al., 1988; Yochem and Greenwald, 1989), and cytoplasmic IκB and premature NF-κB have five and six repeats, respectively (Bours et al., 1990; Ghosh et al., 1990; Kieran et al., 1990; Haskill et al., 1991). In contrast, some ankyrins have more than 20 repeats, which is the largest number of all known genes containing this motif (Lux et al., 1990; Benett, 1992). This variety in the number of ankyrin repeats does not reflect the number of binding sites but, rather, the proper conformation for binding (Devarajan et al., 1996). Up to four WW domains have been detected in a single gene (human Nedd4; Sudol et al., 1995). Each WW domain has its specific ligand, XPPXY (Einbond and Sudol, 1996), so the number of binding sites harbored in the T106 and T91 transcripts can be estimated to be at least three and two, respectively.
Ankyrin repeats and WW domains are present in a variety of cytoskeletal, regulatory, and signaling protein molecules (Bennet, 1992; Bork and Sudol, 1994), but to date have never been reported together in a single gene. One WW-domain protein, dystrophin, also has repeats that are characteristic of spectrin, a cytoskeletal protein that ties together an actin filament and an ankyrin at the plasma membrane of animal cells (Ahn and Kunkel, 1993). Recently, the WW domain of this protein was proven to bind to a dystroglycan through its Pro-rich motif and to help stabilize the plasma membrane. Some mutations preclude this contact and thus cause muscular dystrophy (Einbond and Sudol, 1996). Organization of proteins by multiple functional protein-to-protein interaction domains helps to establish the complicated network of the membrane skeleton (Ahn and Kunkel, 1993).
Computer analyses suggested that Zys3 proteins have a signal peptide at their N terminus that targets the outside of the cell and therefore could be directed to the ER, the Golgi apparatus, and the Golgi-derived vesicles. We speculate that Zys3 proteins play a role in the organization of intracellular membrane conformation, in particular, in the organization of membranous organelles working in the protein-secretion system of the zygote through the protein-to-protein interaction.
Peters et al. (1995) studied the mouse ankyrin gene Ank3, a major isoform of three ankyrins in kidney cells. Various mRNA species are transcribed from a single gene, Ank3, some of which completely lack the ankyrin repeat. Transcripts were alternatively spliced depending on the tissue in which they were expressed. In addition, proteins lacking the repeat domain were localized in the cytoplasm, whereas those containing the repeats were attached to a structure located directly below the plasma membrane, suggesting their functional differentiation within a single cell. In Zys3 an exact region existent only in one mRNA species did not cover the total repeat domain (Fig. 1B); consequently, no complete ankyrin repeat is present in the putative amino acid sequence of the other. The sequence of the deleted region included the consensus characteristics of introns that begin with “gt” and end with “ag” (Fig. 1B). These characteristics are also apparent in the intron of another C. reinhardtii gene (Matters and Goodenough, 1992). It is most likely that T106 and T91 mRNAs are transcribed from the same template via alternative splicing (Figs. 1B and 4). The entire nucleotide sequence, except for the ankyrin-repeat region and the 5′ end of the 5′ untranslated region of T106, was identical between the two species, suggesting that differential posttranscriptional regulation of this molecule occurs in C. reinhardtii. Since these species accumulate within a single cell in the same pattern (Fig. 3D), they may differ in their functions in vivo based on their ability to interact with other proteins.
Genomic Southern-blot analysis (Fig. 4) indicated that Zys3 is a single-copy gene. The numbers and lengths of each band were identical between mt+ and mt− genomic DNA (data not shown), so we can conclude that the Zys3 gene was not specific to either mating type.
Western blotting with the α-Zyspept3 antibody confirmed that these proteins exhibited zygote-specific expression (Fig. 6), and that Zys3 proteins began to accumulate 2 h after fertilization and remained in the cells for about 1 d. Nuclear fusion, flagellar degeneration, zygote wall formation, and chloroplast fusion are known to occur 4 to 6 h after fertilization, when these proteins accumulate to maximum levels. The fact that the Zys3 products remained for more than 22 h suggests a role in the regulation of the progression of long-term zygote development. Since RNase protection analysis revealed identical expression patterns of the two related mRNA species, we expected multiple species of proteins of different molecular masses. However, such signals could not be detected by western-blot analysis, possibly due to the scantiness of the smaller species as suggested by their mRNA levels (Fig. 3C).
Cytoplasmic localization of Zys3 products was demonstrated by indirect immunofluorescence microscopy applied to whole-cell specimens. Figure 7B shows that the signal was detected exclusively in the zygote at 6 h after fertilization. Strong fluorescence was concentrated around the nucleus. However, the structure of a C. reinhardtii cell is marked by its single, large chloroplast that occupies nearly half the cell, and it was therefore not clear whether Zys3 products were distributed uniformly throughout the cytoplasm or in particular organelles. This problem was resolved by observation of sectioned specimens via electron microscopy (below). DAPI fluorescence (Fig. 7B, a, c, e, and g) confirmed previous reports (Kuroiwa et al., 1982); 1 h after cell fusion the zygote contained cell and chloroplast nuclei derived from both parents, but at 6 h it lacked the male-derived chloroplast nuclei and had a single, fused cell nucleus.
Electron microscopy revealed that most signals of gold particles were first detected on the surface of the membrane of the ER sac, then near the cis side of the Golgi apparatus (Figs. 8 and 9a). Later, they continued to gather distinctly at the ER, but the form of this organelle changed into the anastomosing type, which could not have been recognized without this antibody (Figs. 8d and 9). As shown in Figure 8a, this form of ER was not discernible, even in the micrograph of cells fixed with both OsO4 and glutaraldehyde soon after mating, and has never been reported at other haploid stages; therefore, it must be formed in parallel with zygote development or synthesis of this protein.
Many studies have shown that there are close relationships between ER and actin filament organization (e.g. Quader et al., 1987; Staehelin, 1997; Zhou et al., 1997), strongly suggesting that some mechanism that integrates the cytoskeleton with the membrane system exists on the ER, as has been discovered at the plasma membrane of animal cells (Bennet, 1992; Zhou et al., 1997). Recently, one of the human ankyrin isoforms was shown to colocalize with the ER and Golgi-matrix components βIΣ* spectrin and β-COP (Devarajan et al., 1996), suggesting its functional relationship in the organization of intracellular membrane systems. Although the total sizes of Zys3 proteins were far smaller than those of mammalian, nematode, or Drosophila ankyrins, the domain organization and localization pattern support the idea that they function in the membrane skeleton for the regulation of ER configuration (Figs. 2B, 8, and 9). Large, immunogenic, spectrin-like molecules were also discovered in the vegetative C. reinhardtii cell (Lorenz et al., 1995), but it is not known whether these proteins are also expressed in zygotes, or if there is any relationship between them and other proteins such as the ankyrins.
The Zys3 proteins continued to associate with the membrane system for a relatively long time. Figures 6 and 9 indicate that these proteins could still be detected in the ER of zygotes 22 h after fertilization. As pointed out by Minami and Goodenough (1978), young C. reinhardtii zygotes have increasing secretory activity through which glycoprotein fibers are transported through the Golgi apparatus. Therefore, these Zys3 molecules may be synthesized for the activation of ER functions in processing, sorting, and targeting of rigid, zygote-specific cell wall materials and/or mucilaginous extracellular matrices.
As noted by Cavalier-Smith (1976), a marked increase in the volume or number of ER, Golgi, and related vesicles can be observed in the zygote. Given that sexual reproduction was originally established against adverse environments (Goodenough et al., 1995), the formation of a rigid, desiccation-resistant cell wall may accompany the early stage of zygospore formation. The development of a synthetic secretory system represented by the ER and Golgi apparatus and related genes underlies this wall construction. In this case a specific modification of ER composition, and thus a modification of ER properties, was controlled solely by the stimulus of fertilization (Arnburst et al., 1993; Uchida et al., 1993) and was confined to the young zygote of C. reinhardtii. A similar ER form can be seen in other cells, such as those of the higher plants (e.g. Jones, 1980; Quader et al., 1987), but their distribution and kinetics seem unlikely to be directly related to sexual reproduction. For C. reinhardtii, a drastic increase in secretory systems may be required only at the time of zygote formation.
Although the origin and evolution of sexual reproduction remain controversial (Graham, 1993), many scientists believe that the oogamous reproductive system evolved from an isogamous system of ancestral flagellates. C. reinhardtii should exhibit many fundamental aspects of fertilization and zygote development. Analyses of the regulatory mechanisms of the morphological change and activation of the ER in phylogenically related organisms may help to clarify the evolution of this structure. Further studies, including a search for Zys3 homologs in related species, promoter analysis, and the determination of proteins that interact with Zys3, will reveal the mechanisms of regulation of the intricate intracellular membrane system during sexual reproduction of C. reinhardtii.
ACKNOWLEDGMENTS
We thank Dr. Soichi Nakamura of the University of Ryukyus for his kind gift of C. reinhardtii strain 137c, and Drs. Shigeyasu Tanaka and Takeshi Suzuki of Gunma University for their kind advice on the design of the antigen region to which the peptide antibody was raised. We are also very grateful to Drs. Sachihiro Matsunaga and Chieko Saito of the University of Tokyo for their technical instruction in many of these experiments.
Abbreviations:
- DAPI
4′,6-diamidino-2-phenylindole
- FITC
fluorescein isothiocyanate
- IPTG
isopropyl-1-thio-β-d-galactopyranoside
Footnotes
This work was supported by a Grant-in-Aid for Special Promoted Research (project no. 06101002 to T.K.) from the Ministry of Education, Science and Culture of Japan.
LITERATURE CITED
- Ahn AH, Kunkel LM. The structural and functional diversity of dystrophin. Nat Genet. 1993;3:283–291. doi: 10.1038/ng0493-283. [DOI] [PubMed] [Google Scholar]
- Altshul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andrews BJ, Herskowits I. The yeast SWI4 protein contains a motif present in developmental regulators and is part of a complex involved in cell-cycle-dependent transcription. Nature. 1989;342:830–833. doi: 10.1038/342830a0. [DOI] [PubMed] [Google Scholar]
- Armbrust EV, Ferris PJ, Goodenough UW. A mating type-linked gene cluster expressed in Chlamydomonas zygotes participates in the uniparental inheritance of the chloroplast genome. Cell. 1993;74:801–811. doi: 10.1016/0092-8674(93)90460-8. [DOI] [PubMed] [Google Scholar]
- Aves SJ, Durkacz BW, Carr A, Nurse P. Cloning, sequencing and transcriptional control of the Shizosaccharomyces pombe cdc 10 ‘start’ gene. EMBO J. 1985;4:457–463. doi: 10.1002/j.1460-2075.1985.tb03651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett V. Ankyrins. J Biol Chem. 1992;267:8703–8706. [PubMed] [Google Scholar]
- Bork P, Sudol M. The WW domain: a signalling site in dystrophin? Trends Biochem Sci. 1994;19:531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- Bours V, Franzoso G, Azarenko V, Park S, Kanno T, Brown K, Siebenlist U. The oncoprotein Bcl-3 directly transactivates through κB motifs via association with DNA-binding p50B homodimers. Cell. 1993;72:729–739. doi: 10.1016/0092-8674(93)90401-b. [DOI] [PubMed] [Google Scholar]
- Bours V, Villalobos J, Bord PR, Kelly K, Siebenlist U. Cloning of a mitogen-inducible gene encoding a κB DNA-binding protein with homology to the rel oncogene and to cell-cycle motifs. Nature. 1990;348:76–80. doi: 10.1038/348076a0. [DOI] [PubMed] [Google Scholar]
- Breeden L, Nasmyth K. Similarity between cell-cycle genes of budding yeast and fission yeast and the Notch gene of Drosophila. Nature. 1987;329:651–654. doi: 10.1038/329651a0. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Nature. 1970;228:333–335. doi: 10.1038/228333a0. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Protoplasma. 1976;87:297–315. doi: 10.1007/BF01624002. [DOI] [PubMed] [Google Scholar]
- Chan DC, Bedford MT, Leder P. Formin binding proteins bear WWP/WW domains that bind proline-rich peptides and functionally resemble SH3 domains. EMBO J. 1996;15:1045–1054. [PMC free article] [PubMed] [Google Scholar]
- Devarajan P, Stabach PR, Mann AS, Ardito T, Kashgarian M, Morrow JS. Identification of a small cytoplasmic ankyrin (AnkG119) in the kidney and muscle that binds βIΣ* spectrin and associates with the Golgi apparatus. J Cell Biol. 1996;133:819–830. doi: 10.1083/jcb.133.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einbond A, Sudol M. Towards prediction of cognate complexes between the WW domain and proline-rich ligands. FEBS Lett. 1996;384:1–8. doi: 10.1016/0014-5793(96)00263-3. [DOI] [PubMed] [Google Scholar]
- Ferris PJ, Goodenough UW. Transcription of novel genes, including a gene linked to the mating-type locus, induced by Chlamydomonas fertilization. Mol Cell Biol. 1987;7:2360–2366. doi: 10.1128/mcb.7.7.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Goodenough UW. The mating-type locus of Chlamydomonas reinhardtii contains highly rearranged DNA sequences. Cell. 1994;76:1135–1145. doi: 10.1016/0092-8674(94)90389-1. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Gifford AM, Riviere LR, Tempest P, Nolan GP, Baltimore D. Cloning of the p50 DNA binding subunit of NF-κB: homology to rel and dorsal. Cell. 1990;62:1019–1029. doi: 10.1016/0092-8674(90)90276-k. [DOI] [PubMed] [Google Scholar]
- Gilbert SF (1988) Developmental Biology, Ed 2. Sinauer Associates, Sunderland, MA, pp 522–561
- Goodenough UW, Armbrust EV, Campbell AM, Ferris PJ. Molecular genetics of sexuality in Chlamydomonas. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:21–44. [Google Scholar]
- Graham LE. The Origin of Land Plants. New York: John Wiley & Sons; 1993. [Google Scholar]
- Hart MJ, Callow MG, Souza B, Polakis P. IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. EMBO J. 1996;15:2997–3005. [PMC free article] [PubMed] [Google Scholar]
- Haskill S, Beg A, Tompkins SM, Morris JS, Yurochko AD, Sampson-Johannes A, Mondal K, Ralph P, Baldwin AS., Jr Characterization of an immediate-early gene induced in adherent monocytes that encodes IκB-like activity. Cell. 1991;65:1281–1289. doi: 10.1016/0092-8674(91)90022-q. [DOI] [PubMed] [Google Scholar]
- Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1991;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- Itadani H, Sugita M, Sugiura M. Structure and expression of a DNA encoding an RNA helicase-like protein in tobacco. Plant Mol Biol. 1994;24:249–252. doi: 10.1007/BF00040593. [DOI] [PubMed] [Google Scholar]
- Johnson KA, Rosenbaum JL. The basal bodies of Chlamydomonas do not contain immunologically detectable DNA. Cell. 1990;62:615–619. doi: 10.1016/0092-8674(90)90105-n. [DOI] [PubMed] [Google Scholar]
- Jones RL. Quantitative and qualitative changes in the endoplasmic reticulum of barley aleurone layers. Planta. 1980;150:70–81. doi: 10.1007/BF00385617. [DOI] [PubMed] [Google Scholar]
- Kieran M, Blank V, Logeat F, Vandekerckhove J, Lottspeich F, Le Bail O, Urban MB, Kourilsky P, Baeuerle PA, Israel A. The DNA binding subunit of NF-κB is identical to factor KBF 1 and homologous to the rel oncogene product. Cell. 1990;62:1007–1018. doi: 10.1016/0092-8674(90)90275-j. [DOI] [PubMed] [Google Scholar]
- Kirk MM, Kirk DL. Transcriptional regulation of protein synthesis, in response to light, at a critical stage of Volvox development. Cell. 1985;41:419–428. doi: 10.1016/s0092-8674(85)80015-5. [DOI] [PubMed] [Google Scholar]
- Kuroiwa T. The replication, differentiation, and inheritance of plastids with emphasis on the concept of organelle nuclei. Int Rev Cytol. 1991;128:1–62. [Google Scholar]
- Kuroiwa T, Kawano S, Nishibayashi S, Sato C. Epifluorescent microscopic evidence for maternal inheritance of chloroplast DNA. Nature. 1982;298:481–483. doi: 10.1038/298481a0. [DOI] [PubMed] [Google Scholar]
- Kuroiwa T, Kawano S, Sato C. Mechanisms of maternal inheritance. II. RNA synthesis involved in preferential destruction of chloroplast DNA of male origin. Proc Jpn Acad Ser B Phys Biol Sci. 1983;59:182–185. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamarco K, Thompson CC, Byers BP, Walton EM, McKnight SL. Identification of Ets- and Notch-related subunits in GA binding protein. Science. 1991;253:789–792. doi: 10.1126/science.1876836. [DOI] [PubMed] [Google Scholar]
- Lorenz M, Biskirska B, Hanus-Lorenz B, Strzalka K, Shikorski AF. Proteins reacting with anti-spectrin antibodies are present in Chlamydomonas cells. Cell Biol Int. 1995;19:625–632. doi: 10.1006/cbir.1995.1110. [DOI] [PubMed] [Google Scholar]
- Lux SE, John KM, Bennett V. Analysis of cDNA for human erythrocyte ankyrin indicates a repeated structure with homology to tissue-differentiation and cell-cycle control proteins. Nature. 1990;344:36–62. doi: 10.1038/344036a0. [DOI] [PubMed] [Google Scholar]
- Matters GL, Goodenough UW. A gene/pseudogene tandem duplication encodes a cysteine-rich protein expressed during zygote development in Chlamydomonas reinhardtii. Mol Gen Genet. 1992;232:81–88. doi: 10.1007/BF00299140. [DOI] [PubMed] [Google Scholar]
- Minami S, Goodenough UW. Novel glycopolypeptide synthesis induced by gametic cell fusion in Chlamydomonas reinhardtii. J Cell Biol. 1978;77:165–181. doi: 10.1083/jcb.77.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Sato C, Kuroiwa T. Polypeptides related to preferential digestion of male chloroplast nucleoids in Chlamydomonas. Plant Sci. 1988;56:129–136. [Google Scholar]
- Nozaki H, Kuroiwa H, Kroiwa T. Light and electron microscopic characterization of two types of pyrenoids in Gonium (Goniaceae, Chrolophyta) J Phycol. 1994;30:279–290. [Google Scholar]
- Ohno H, Takimoto G, McKeithan TW. The candidate proto-oncogene bcl-3 is related to genes implicated in cell lineage determination and cell cycle control. Cell. 1990;60:991–997. doi: 10.1016/0092-8674(90)90347-h. [DOI] [PubMed] [Google Scholar]
- Ohta N, Nagashima H, Kawano S, Kuroiwa T. Isolation of the chloroplast DNA and the sequence of the trn K gene from Cyanidium cardarium strain RK-1. Plant Cell Physiol. 1992;33:657–661. [Google Scholar]
- Peters LL, John KM, Lu FM, Eicher EM, Higgins A, Yialamas M, Turtzo LC, Otsuka AJ, Lux SE. Ank3 (epithelial ankyrin), a widely distributed new member of ankyrin gene family and the major ankyrin in kidney, is expressed in alternatively spliced forms, including forms that lack the repeat domain. J Cell Biol. 1995;130:313–330. doi: 10.1083/jcb.130.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quader H, Hofmann A, Schnepf E. Shape and movement of the endoplasmic reticulum in onion bulb epidermis cells: possible involvement of actin. Eur J Cell Biol. 1987;44:17–26. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F. Determining of nucleotide sequences in DNA. Science. 1981;214:1205–1210. doi: 10.1126/science.7302589. [DOI] [PubMed] [Google Scholar]
- Snell WJ. Study of the release of cell wall degrading enzymes during adhesion of Chlamydomonas gametes. Exp Cell Res. 1982;138:109–119. doi: 10.1016/0014-4827(82)90096-9. [DOI] [PubMed] [Google Scholar]
- Spence AM, Coulson A, Hodgkin J. The product of fem-1, a nematode sex-determining gene, contains a motif found in cell cycle control proteins and receptors for cell-cell interactions. Cell. 1990;60:981–990. doi: 10.1016/0092-8674(90)90346-g. [DOI] [PubMed] [Google Scholar]
- Spurr AR. A low viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Staehelin LA. The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J. 1997;11:1151–1165. doi: 10.1046/j.1365-313x.1997.11061151.x. [DOI] [PubMed] [Google Scholar]
- Sudol M, Bork P, Einbond A, Kastury K, Druck T, Negrini M, Huebner K, Lehman D. Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J Biol Chem. 1995;270:14733–14741. doi: 10.1074/jbc.270.24.14733. [DOI] [PubMed] [Google Scholar]
- Thompson CC, Brown TA, McKnight SL. Convergence of Ets- and Notch-related structural motifs in a heteromeric DNA binding complex. Science. 1991;253:762–768. doi: 10.1126/science.1876833. [DOI] [PubMed] [Google Scholar]
- Uchida H, Kawano S, Sato N, Kuroiwa T. Isolation and characterization of novel genes which are expressed during the very early stage of zygote formation in Chlamydomonas reinhardtii. Curr Genet. 1993;24:296–300. doi: 10.1007/BF00336779. [DOI] [PubMed] [Google Scholar]
- Wegener D, Beck CF. Identification of novel genes specifically expressed in Chlamydomonas reinhardtii zygotes. Plant Mol Biol. 1991;16:937–946. doi: 10.1007/BF00016066. [DOI] [PubMed] [Google Scholar]
- Wharton KA, Johansen KM, Xu T, Arvatanis-Tsakonas S. Nucleotide sequence from the neurogenic locus Notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43:567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- Woessner JP, Goodenough UW. Molecular characterization of a zygote wall protein: an extensin-like molecule in Chlamydomonas reinhardtii. Plant Cell. 1989;1:901–911. doi: 10.1105/tpc.1.9.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley KC, Wiese BA, Smith RF. BEAUTY: an enhanced BLAST-based search tool that integrates multiple biological in-formation resources into sequence similarity search results. Genome Res. 1995;5:173–184. doi: 10.1101/gr.5.2.173. [DOI] [PubMed] [Google Scholar]
- Yochem J, Greenwald I. Glp-1 and Lin-12, genes implicated in distinct cell-cell interactions in C. elegans, encode similar transmembrane proteins. Cell. 1989;58:553–563. doi: 10.1016/0092-8674(89)90436-4. [DOI] [PubMed] [Google Scholar]
- Yochem J, Weston K, Greenwald I. The Caenorhabditis elegans lin-12 gene encodes a transmembrane protein with overall similarity to Drosophila Notch. Nature. 1988;335:547–550. doi: 10.1038/335547a0. [DOI] [PubMed] [Google Scholar]
- Youngblom J, Schloss JA, Silflow CD. The two β-tubulin genes of Chlamydomonas reinhardtii code for identical proteins. Mol Cell Biol. 1984;4:2686–2696. doi: 10.1128/mcb.4.12.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Scheirer DC, Fowle WH, Goodman HM. Expression of antisense or sense RNA of an ankyrin repeat-containing gene blocks chloroplast differentiation in Arabidopsis. Plant Cell. 1992;4:1575–1588. doi: 10.1105/tpc.4.12.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Birkenmeier CS, Williams MW, Sharp JJ, Barker JE, Bloch RJ. Small, membrane-bound, alternatively spliced forms of ankyrin 1 associated with the sarcoplasmic reticulum of mammalian skeletal muscle. J Cell Biol. 1997;136:621–631. doi: 10.1083/jcb.136.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]