Abstract
The significance of the widespread downregulation of Rap1GAP in human tumors is unknown. In previous studies we demonstrated that silencing Rap1GAP expression in human colon cancer cells resulted in sustained increases in Rap activity, enhanced spreading on collagen and the weakening of cell-cell contacts. The latter finding was unexpected based on the role of Rap1 in strengthening cell-cell adhesion and reports that Rap1GAP impairs cell-cell adhesion. We now show that Rap1GAP is a more effective inhibitor of cell-matrix compared to cell-cell adhesion. Overexpression of Rap1GAP in human colon cancer cells impaired Rap2 activity and the ability of cells to spread and migrate on collagen IV. Under the same conditions, Rap1GAP had no effect on cell-cell adhesion. Overexpression of Rap1GAP did not enhance the dissociation of cell aggregates nor did it impair the accumulation of β-catenin and E-cadherin at cell-cell contacts. To further explore the role of Rap1GAP in the regulation of cell-cell adhesion, Rap1GAP was overexpressed in non-transformed thyroid epithelial cells. Although the formation of cell-cell contacts required Rap1, overexpression of Rap1GAP did not impair cell-cell adhesion. These data indicate that transient, modest expression of Rap1GAP is compatible with cell-cell adhesion and that the role of Rap1GAP in the regulation of cell-cell adhesion may be more complex than is currently appreciated.
Key words: Rap1GAP, cell adhesion, matrix adhesion, Rap, E-cadherin, β-catenin
Introduction
Rap1GAP (RapGTPase activating protein) is one member of a family of negative regulators of Rap proteins (Rap1a/b, Rap2a/b/c in mammalian cells). The expression of Rap1GAP is decreased in tumors of the pancreas, skin, thyroid and colon.1–6 Rap1GAP expression is further decreased in invasive compared to benign lesions, suggesting that depletion of Rap1GAP enhances tumor progression.1,2,4,5 The advantages conferred to tumor cells by the downregulation of Rap1GAP are unknown. Overexpression of Rap1GAP in vitro impaired tumor cell proliferation, migration and invasion.3–5,7,8 Overexpression of Rap1GAP elicited variable effects on tumor formation,5,8–10 but consistently impaired metastasis.5,9,10 We previously reported that silencing Rap1GAP expression in human colon cancer cells impaired cell-cell adhesion and enhanced spreading on collagen.6 The weakening of cell-cell contacts together with alterations in matrix adhesion are hallmarks of tumor progression. These data suggest that loss of Rap1GAP endows cells with the ability to disseminate and provide a potential rationale for the progressive downregulation of Rap1GAP in human tumors.
The notion that loss of Rap1GAP enhances matrix adhesion is not surprising. Silencing Rap1GAP induced sustained increases in Rap activity.6 Activated Rap enhances cell-matrix adhesion by regulating integrin affinity and avidity.11–13 However, Rap activity promotes cell-cell adhesion (reviewed in refs. 14–17), a finding that is difficult to reconcile with the observation that silencing Rap1GAP weakened cell-cell adhesion.6 Given the importance of alterations in cellular adhesion in tumor progression together with the widespread downregulation of Rap1GAP in human tumors, we further explored the role of Rap1GAP in the regulation of cell adhesion. Surprisingly, transient and modest overexpression of Rap1GAP in human colon cancer cells and in thyroid epithelial cells impaired matrix adhesion in the absence of effects on cell-cell adhesion. These data indicate that the role of Rap1GAP in the regulation of Rap signaling is more complex than is currently appreciated.
Results
Rap1GAP impairs cell-matrix adhesion.
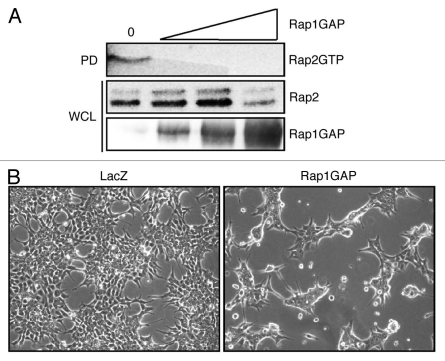
To analyze the role of Rap1GAP in the regulation of cell adhesion, an adenovirus was used to transiently overexpress Rap1GAP in HCT116 cells. Dose response experiments revealed that infection with the Rap1GAP adenovirus at 5 IU/cell was sufficient to block endogenous Rap2 activity (Fig. 1A, lane 2). Rap1 activity was undetectable, most likely due to the low level of Rap1 expression in these cells (data not shown). Cells overexpressing Rap1GAP exhibited dramatic alterations in cell morphology. Rap1GAP-expressing cells were more compact and less spread than LacZ-infected cells (Fig. 1B). There was no difference in the morphology of LacZ and mockinfected cells (data not shown).
Figure 1.
Overexpression of Rap1GAP impairs Rap 2 activity and induces morphological changes in colon cancer cells. (A) HCT116 cells were infected with Rap1GAP adenovirus at 0, 5, 10 or 21 infectious units (IU)/cell and Rap2 activity monitored in pull-down assays (PD). Whole cell lysates (WCL) were subjected to protein gel blotting for Rap2 and Rap1GAP. (B) Growing HCT116 cells were imaged (100x) at 48 h post-infection with Rap1GAP or LacZ adenoviruses.
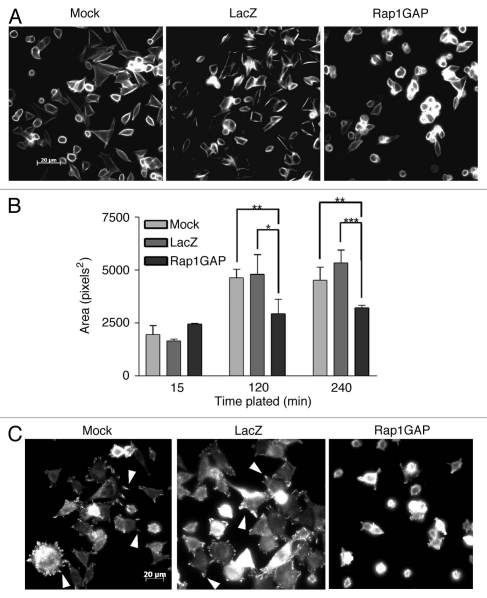
To determine if Rap1GAP impaired cell spreading, Rap1GAP-expressing cells were plated on collagen IV for various times (ranging from 15–240 min) and cell area measured by morphometry. Rap1GAP-infected cells were less spread than mock- or LacZ-infected cells (Fig. 2A). Measurements of cell area in rhodamine phalloidin-stained cells confirmed that Rap1GAP significantly impaired cell spreading (Fig. 2B). When plated on poly-L-lysine, Rap1GAP-expressing cells were similar in size to mock- and LacZ-infected cells, suggesting that Rap1GAP selectively impairs integrin-mediated spreading (data not shown).
Figure 2.
Rap1GAP impairs spreading on collagen. (A) Mock-, LacZ- and Rap1GAP-infected cells were plated on collagen IV for various times (240 min shown here), fixed and stained for F-actin with rhodamine phalloidin. (B) Cell area was measured and quantified using morphometry. At least three random fields (approximately 100 cells) were measured for each time point. The graph illustrates results from a representative experiment (*p < 0.05, **p < 0.01, ***p < 0.001). (C) Mock-, LacZ- and Rap1GAP-infected cells were plated on collagen IV for 90 min, fixed and stained for phospho-FAK (Y397). Arrows indicate examples of focal adhesions.
To explore consequences on integrin signaling, focal adhesions were analyzed by immunostaining for autophosphorylated FAK (phospho-FAKY397). Overexpression of Rap1GAP dramatically reduced the appearance of focal adhesions (Fig. 2C). Taken together, these results demonstrate that Rap1GAP overexpression impairs cell-matrix adhesion in human colon cancer cells.
Rap1GAP inhibits cell migration.
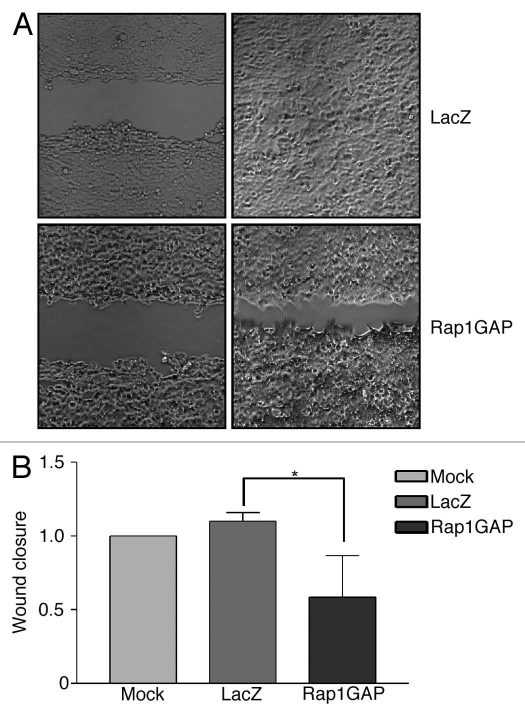
Stable overexpression of Rap1GAP inhibited cell migration in pancreatic carcinoma and melanoma cell lines.3–5 To assess whether acute expression of Rap1GAP is sufficient to inhibit migration, Rap1GAP was transiently overexpressed in HCT116 cells. Confluent monolayers of Rap1GAP- and LacZ-infected cells were wounded with a pipet tip and migration across the wounded area monitored after 24 h (Fig. 3A). Increased expression of Rap1GAP significantly impaired wound closure (Fig. 3B). These findings confirm that Rap1GAP expressed from the adenovirus is functional. More importantly, they demonstrate that transient overexpression of Rap1GAP is sufficient to inhibit Rap2 activity, cell spreading and cell migration.
Figure 3.
Rap1GAP impairs cell migration. (A) Confluent mock-, LacZ- and Rap1GAP-infected cells were wounded and images acquired immediately (left parts) and 24 h later (right parts). The magnification used was 100x. (B) Quantitation of wound closure from three independent experiments is shown (see materials and methods for details). The distance migrated by mock-infected cells was set to 1. Rap1GAP significantly decreased cell migration (p < 0.05).
Cell-cell adhesion is not disrupted by Rap1GAP.
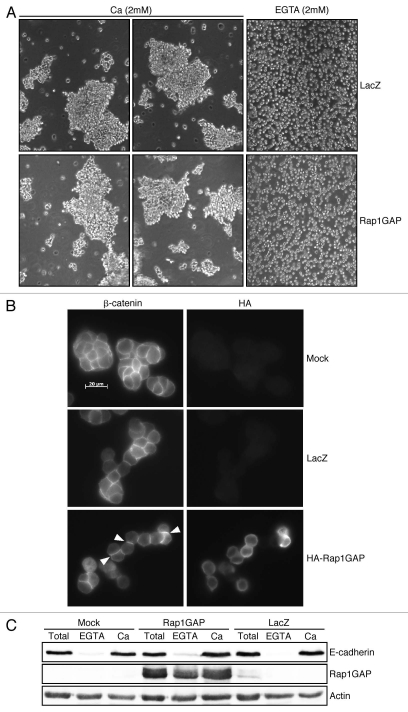
To assess the functional consequences of Rap1GAP overexpression on cell-cell adhesion, cell dissociation assays were conducted. Cells were released from tissue culture dishes and subject to dissociation by pipetting in the presence of calcium, which maintains E-cadherin-mediated cell-cell contacts or in EGTA to disrupt calcium-dependent contacts. Although expressed at levels sufficient to inhibit Rap2 activity (Fig. 1A), spreading on collagen IV (Fig. 2A) and cell migration (Fig. 3), Rap1GAP did not render cells more sensitive to dissociation (Fig. 4A).
Figure 4.
Rap1GAP does not impair cell-cell adhesion. (A) LacZ- and Rap1GAP-infected cells were trypsinized in the presence of calcium or EGTA for 10 min, pipetted 10x, plated and images acquired immediately (100x magnification). (B) At 24 h post-infection, mock-, LacZ- and Rap1GAP-infected cells were plated overnight, fixed and stained for β-catenin and HA-Rap1GAP. (C) Mock-, LacZ- and Rap1GAP-infected cells were disrupted directly on tissue culture dishes (total cell lysates) or trypsinized in the presence of calcium or EGTA prior to lysis. Lysates were subjected to protein gel blotting for E-cadherin, Rap1GAP and actin to confirm equal protein loading.
To further investigate effects on cell-cell adhesion, we examined whether Rap1GAP induced the loss of adherens junction proteins from cell-cell junctions. Cells were fixed and stained for HA to identify cells expressing HA-Rap1GAP and with antibodies to β-catenin or E-cadherin to label adherens junctions. The proportion of cells with β-catenin (Fig. 4B) or E-cadherin at cell-cell contacts was similar in Rap1GAP-expressing and control cells (β-catenin: mock-85.3%, LacZ-84.6%, Rap1GAP-87.7%; E-cadherin: mock-77.8%, LacZ-71.8%, Rap1GAP-76.1%). At least 50 pairs of cells were analyzed for each condition.
To further explore the effects of RapGAP on the accumulation of E-cadherin at the plasma membrane, cell surface expression of E-cadherin was analyzed in trypsin sensitivity assays. Cells were disrupted directly on tissue culture dishes (total in Fig. 4C) or trypsinized in the presence of EGTA or calcium prior to disruption. As expected, treatment with EGTA removed E-cadherin from the cell surface, while calcium protected cell surface E-cadherin (Fig. 4C). In agreement with the results obtained in the immunostaining experiments, overexpression of Rap1GAP did not reduce cell surface expression of E-cadherin. Collectively, these data show that transient, modest overexpression of Rap1GAP is not sufficient to weaken cell-cell adhesion or to disrupt E-cadherin-mediated cell-cell contacts in human colon cancer cells.
Effects of Rap1GAP on the formation of cell-cell contacts.
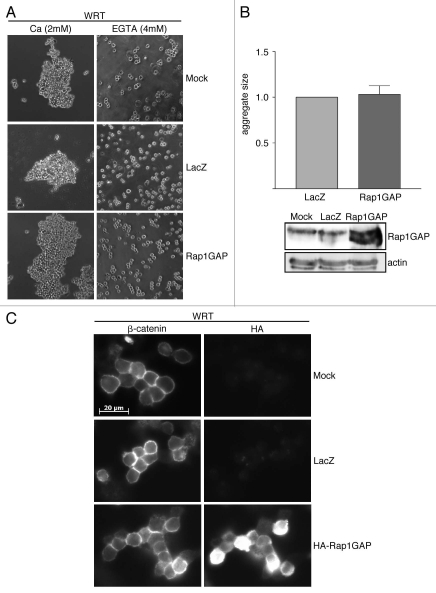
We detected only Rap2 activity in HCT116 cells. As the role of Rap2 in E-cadherin-mediated cell adhesion is unknown, we conducted similar experiments in Wistar rat thyroid (WRT) epithelial cells, which express abundant levels of Rap1.19,24 Rap1GAP was transiently overexpressed in these cells at the lowest dose sufficient to inhibit Rap1 activity.3,18 At 48 h post-infection, cell association assays were conducted. Rap1GAP-expressing cells formed aggregates in a manner indistinguishable from mock- and LacZ-infected cells (Fig. 5A). To confirm that there was no effect of Rap1GAP on cell association, we compared the sizes of the aggregates formed in three independent experiments (Fig. 5B). There was no significant difference in the size of the aggregates formed by Rap1GAP- versus LacZ-infected cells.
Figure 5.
Rap1GAP does not impair cell-cell adhesion in thyroid epithelial cells. (A) Cell association assays were conducted in mock-, LacZ- and Rap1GAP-infected WRT cells. Images were acquired at 150x magnification. (B) Aggregate size (LacZ set to 1.0) from three independent experiments is shown. There was no significant difference in the size of aggregates formed by Rap1GAP-infected versus LacZ-infected cells (Rap1GAP 1.03 ± 0.09). Replicate plates were collected and subjected to protein gel blotting for Rap1GAP. Actin was analyzed to document equal protein loading. (C) Mock-, LacZ- and Rap1GAP-infected cells were plated on laminin-coated coverslips overnight. Cells were fixed and stained for β-catenin and HA-Rap1GAP.
To further assess the effects of Rap1GAP on cell-cell contacts, the localization of β-catenin was analyzed. β-catenin accumulated at cell-cell contacts in both Rap1GAP- and LacZ-infected cells (Fig. 5C). The proportion of Rap1GAP-expressing cells with β-catenin at cell-cell contacts was similar to that in mock- and LacZ-infected cells (experiment 1: mock-92.0%, LacZ-94.9%, Rap1GAP-93.8%; experiment 2: mock-83.6%, LacZ-83.3%, Rap1GAP-78.6%). At least 90 pairs of cells were analyzed per condition.
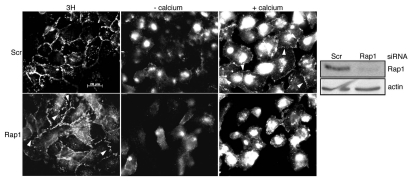
To exclude the possibility that cell-cell contact formation was Rap1-independent in these cells, the expression of Rap1 was silenced using RNA interference. Depletion of Rap1 disrupted the integrity of cell-cell contacts in growing cells (Figs. 6 and 3H). The junctions between cells were more irregular and disorganized in the absence of Rap1. β-catenin was lost from cell-cell contacts when cells were deprived of calcium (Fig. 6, -calcium). Interestingly, under these conditions, Rap1-depleted cells were consistently more dissociated from one another compared to control cells. Within 1 h of transfer to calciumsupplemented medium, control cells spread and began to form cell-cell contacts that labeled for β-catenin (Fig. 6, +calcium). The reformation of cell-cell contacts was delayed in Rap1-depleted cells (Fig. 6, +calcium). These results indicate that the absence of inhibitory effects of Rap1GAP on cell-cell adhesion is not because cell-cell contact formation is independent of Rap1.
Figure 6.
Cell-cell junction formation requires Rap1. WRT cells transfected with scrambled or Rap1-directed siRNAs were maintained in 3H growth medium (left parts), transferred to low calcium medium for 1 h (middle parts) or deprived of calcium for 1 h and then transferred to calcium-containing medium for 1 h (right parts). Cells were fixed and stained for β-catenin. Total cell lysates were subjected to protein gel blotting to confirm Rap1 depletion. The same filter was reprobed for actin to confirm equal protein loading.
Discussion
Although widely used as a tool to inhibit Rap activity, little is known about the cellular functions regulated by Rap1GAP. Genetic silencing of Rap1GAP in human HT29 colon cancer cells increased Rap1 and Rap2 activity, confirming that endogenous Rap1GAP is an essential negative regulator of both proteins in vivo.6 Despite the increase in Rap activity, downregulation of Rap1GAP impaired cell-cell adhesion.6 This finding was difficult to reconcile with reports that Rap1 promotes E-cadherin-mediated cell adhesion (reviewed in refs. 14 and 17). Additionally, previous reports demonstrated that overexpression of Rap1GAP impaired VE-cadherin- and E-cadherin-mediated cell-cell adhesion.25–28 This prompted us to further explore the role of Rap1GAP in the regulation of cell adhesion.
We selected human HCT116 colon cancer cells for this analysis, as these cells retain E-cadherin-mediated cell-cell contacts. Transient overexpression of Rap1GAP at the lowest level sufficient to inhibit Rap2 activity, which comprised all of the detectable Rap activity in these cells, impaired cell spreading on collagen, decreased focal adhesions and inhibited migration. However, under these conditions, Rap1GAP did not render cells more sensitive to dissociation, nor did it impair the accumulation of β-catenin and E-cadherin at cell-cell junctions. The absence of inhibitory effects on cell-cell adhesion could indicate that E-cadherin-mediated adhesion is independent of Rap2 activity. Rap1 and Rap2 mediate overlapping and divergent functions (reviewed in ref. 29). Both Rap proteins promote integrin activation, cell spreading and cell motility.30 However, their localizations differ31 and specific effectors for Rap2 have been identified (reviewed in ref. 32). Rap2 promotes cell-cell adhesion via integrin activation30 and stabilizes β-catenin in Xenopus embryos.33 It is not yet known whether Rap2 regulates E-cadherin-mediated cell-cell adhesion. Hence, we conducted similar experiments in rat thyroid epithelial cells, which express abundant levels of Rap1.19,24 As previously reported, overexpression of Rap1GAP blocked Rap1 activity.3,17 Under the same conditions, Rap1GAP overexpression had no effect on cell-cell adhesion or the accumulation of β-catenin at cell-cell junctions. Silencing the expression of Rap1 in these cells disrupted cell-cell contacts and delayed the formation of cell-cell contacts in calcium switch experiments, confirming that Rap1 is required for cell-cell adhesion. The fact that modest, transient overexpression of Rap1GAP selectively impaired cell-matrix over cell-cell adhesion in two different cell lines suggests that the roles of cellular Rap1GAP are more complex than is presently appreciated.
There are only a handful of reports in the literature that have analyzed the consequences of Rap1GAP overexpression on cadherin-mediated adhesion. Overexpression of Rap1GAPII impaired the ability of endothelial cells to plate on immobilized Fc-VE-cadherin.25,27 Rap1GAPII contains a GoLoco domain in the N-terminus that mediates high affinity interaction with heterotrimeric G protein α subunits.34–36 As Rap1GAP contains only a partial GoLoco domain, it is possible that it fails to localize to cell-cell contacts. We do not favor this explanation as it implies that pools of active Rap remain in HCT116 cells overexpressing Rap1GAP, which we were unable to detect. In addition, overexpression of Rap1GAP in Rap1GAP-depleted HT29 colon cancer cells restored the accumulation of E-cadherin and p120-catenin at cell-cell contacts, further supporting a role for Rap1GAP in the regulation of cell-cell adhesion.6 Overexpression of Rap1GAP disrupted the accumulation of VE-cadherin at cell-cell contacts in confluent HUVECs.26 Whether this was due to impaired matrix adhesion, higher levels of Rap1GAP overexpression or differing roles of Rap1GAP in the regulation of VE-cadherin- versus E-cadherin-mediated adhesion remains to be determined. Microinjection of Rap1GAP into MCF7 cells had no effect on cell-cell contacts in growing cells, but delayed the accumulation of E-cadherin at cell-cell contacts following a calcium switch.28 The significance of this delay is unclear in that microinjection of activated Rap1 elicited similar effects. It is noteworthy that other investigators have reported aberrant cell-cell junctions in cells expressing high levels of activated Rap1.37 Moreover, two reports showed that the disruption of cell-cell contacts activates Rap1 in epithelial cells.22,38 In both studies, trans-ligation of E-cadherin was required to inactivate Rap1, suggesting that sustained Rap1 activity may be incompatible with cell-cell adhesion under some circumstances.
The mechanism through which Rap1GAP selectively impairs cell-matrix adhesion remains to be elucidated. It is conceivable that at low levels of Rap1GAP expression, Rap1GAP is targeted to cell-matrix attachment sites, enhancing the inactivation of Rap1 at these sites. At high levels of Rap1GAP expression, Rap1GAP might then be targeted to cell-cell contact sites, leading to the disruption of cell-cell adhesion. Interestingly, the scaffolding protein AF6 has been shown to co-localize Rap1GAP and Rap1 at sites of cell-matrix attachment.39 AF6 also enhances the stability of membrane E-cadherin.40 It is tempting to speculate that in addition to limiting the duration of Rap activity, Rap1GAP may play a role in determining the cellular sites of Rap signaling. There are precedents for such a role. Rap1GAP has been proposed to limit Rap activation to intracellular membranes.41 In Saccharomyces cerevisiae, Bud2p (RapGAP) recruits Rsr1p/Bud1p (Rap1) to the incipient bud site (reviewed in ref. 42).
Based on previous findings,6 we suggest that loss of Rap1GAP in human tumors renders tumor cells more sensitive to dissociation, thereby facilitating tumor cell dissemination. In support of that notion, we now show that Rap1GAP overexpression is compatible with cell-cell adhesion in tumor cells and in non-transformed epithelial cells. Further studies are required to determine how Rap1GAP contributes to the regulation of cell-cell adhesion, and whether loss of Rap1GAP in other tumor cells facilitates their dissemination.
Materials and Methods
Reagents.
Rap1GAP (sc-28189), β-catenin (sc-7199), HA polyclonal (sc-805) and HRP secondary antibodies were from Santa Cruz Biotechnology. Monoclonal HA antibody was kindly provided by Dr. Jeffrey Field (Department of Pharmacology, University of Pennsylvania). Glutathione sepharose beads (17-0756-01) were from GE Healthcare. E-cadherin antibody was from EMD Biosciences (205601). Phospho-FAK (Y397) antibody (44624G), rhodamine-conjugated phalloidin (R415) and Alexa-fluor conjugated secondary antibodies (A21202, A21203 and A21207) were from Invitrogen. For Amaxa transfections, Cell Line Nucleofector Kit V was used (VCA-1003). Scrambled (1027280), Rap1 (SI01968722, SI03090010) and Rap1GAP (SI01737050) siRNAs were from Qiagen. Rap2 antibody (610215), Matrigel (356230) and collagen IV (354233) were from BD Biosciences. Laminin was from Collaborative Biomedical Products (CB40232EA). Low-calcium medium was from Invitrogen (SMEM, 11380).
Cell lines and reagents.
HCT116 human colon carcinoma cells were a generous gift from Dr. John Lynch (Department of Medicine, University of Pennsylvania). Cells were propagated in McCoy's 5A medium supplemented with 10% fetal bovine serum (FBS). Wistar rat thyroid (WRT) cells were propagated in Coon's modified Ham's F-12 medium supplemented with calf serum (5%), insulin (10 µg/ml), TSH (1 mU/ml) and transferrin (5 µg/ml) (referred to as 3H growth medium). WRT cells were starved in basal medium (Coon's modified Ham's F-12 medium devoid of growth factors and serum).
Adenovirus infection.
Rap1GAP and LacZ adenoviruses were constructed and purified as described previously in reference 3. Control (LacZ) and Rap1GAP viruses were infected at an equal MOI (multiplicity of infection) of 5 infectious units (IU)/cell in HCT116 cells and 10 IU/cell in WRT cells. HCT116 cells were infected overnight in growth medium. WRT cells were infected overnight in basal medium and transferred to 3H growth medium the following day.
Protein gel blotting.
Protein gel blotting was performed as described previously in reference 18. Proteins were detected using the FUJI LAS-3000 system and Multi Gauge 3.0 software (Fuji).
Rap activation.
Rap activation was assessed as described previously using the RalGDS Rap-binding domain fused to GST to selectively retrieve GTP-bound Rap1 or Rap2.19
Cell spreading.
HCT116 cells were plated in serum-containing growth medium on collagen IV for various times, fixed and stained with rhodamine phalloidin. Cell area was measured using a Zeiss Axiophot fluorescence microscope and Zeiss Axiovision software.
Wound assays.
Cells were infected with LacZ or Rap1GAP virus overnight. Cells were plated in 35 mm dishes with a line made down the center. At 24–48 h post-infection, confluent cell monolayers were wounded perpendicularly to the line (5–6 wounds/dish). Images were captured immediately and after 24 h using a Nikon Eclipse TE2000 microscope and analyzed using Image J software. The area measured after 24 h was subtracted from the area measured at 0 h. The area closed by mockinfected control cells was set to 1.
Cell dissociation assays.
Cell dissociation assays were performed as described previously in reference 20. Cells were treated with 0.01% trypsin in the presence of EGTA (2 mM) or calcium (2 mM) for 10 min. Cells were triturated 15 times, plated and images acquired immediately using a Nikon Eclipse TE2000 microscope.
Cell association assays.
Cell association assays were performed as described previously in references 21 and 22. WRT cells were dissociated into single cells in 0.01% trypsin/EGTA (2 mM). Two × 106 cells were collected by centrifugation, washed and suspended in 3H growth medium containing EGTA (2 mM) or calcium (2 mM). After incubation at 37°C for 30 min, cells were plated on agarose-coated dishes to prevent matrix attachment and rocked for 16 h. Images were acquired using a Nikon Eclipse TE2000 microscope. Aggregate size was measured and quantified using morphometry.
Trypsin sensitivity assays.
Experiments were performed as described previously in reference 23. Cells were lysed directly on tissue culture dishes (total cell lysates) or trypsinized (0.01%) in the presence of calcium (2 mM) or EGTA (2 mM) prior to lysis in RIPA buffer 50 mM Tris-HCl, pH 8.0; 150 mM NaCl; 0.5% deoxycholate; 1% NP-40; 0.1% sodium dodecyl sulfate (SDS); 10 mM NaF; 2 mM Na3VO4; aprotinin, leupeptin and Pefabloc (each at 10 µg/mL). Lysates (equal protein content) were subjected to protein gel blotting for E-cadherin.
Immunostaining.
For cell spreading and immunostaining for focal adhesions, HCT116 cells were plated on collagen IV for 15, 90, 120 and 240 min. For analysis of adherens junction proteins, HCT cells were plated on matrigel and WRT cells on laminin. Cells were fixed in MeOH:acetone (1:1) for 15 min at room temperature, stained with primary antibodies diluted in PBS, 5 mg/ml bovine serum albumin, 0.2% Triton-X-100 for 1 h at 37°C and then with Alexa-fluor-conjugated secondary antibodies for 1 h at 37°C. Images were captured using a Zeiss Axiophot fluorescence microscope and Zeiss Axiovision software. All images for a given antibody within an experiment were captured for the same times.
Silencing experiments.
WRT cells were transfected with Rap1 (400 nM) or scrambled siRNAs (400 nM) using Amaxa electroporation and plated onto glass slips. At 72 h post-transfection, cells were transferred to low calcium medium (SMEM) for 1 h and then refed with 3H growth medium for various times. Cells were fixed and stained for β-catenin.
Statistics.
All experiments were performed at least two times with similar results. Data are presented as means ± SD and significance was assessed by t-test. A p value < 0.05 was considered to be statistically significant.
Acknowledgments
This work was funded by PHS grant CA127986 to J.M. L.V. is funded by grant R25 CA101871-07 from the National Cancer Institute.
References
- 1.Nellore A, Paziana K, Ma C, Tsygankova OM, Wang Y, Puttaswamy K, et al. Loss of Rap1GAP in papillary thyroid cancer. J Clin Endocrinol Metab. 2009;94:1026–1032. doi: 10.1210/jc.2008-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zuo H, Gandhi M, Edreira MM, Hochbaum D, Nimgaonkar VL, Zhang P, et al. Downregulation of Rap1GAP through epigenetic silencing and loss of heterozygosity promotes invasion and progression of thyroid tumors. Cancer Res. 2010;70:1389–1397. doi: 10.1158/0008-5472.CAN-09-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsygankova OM, Prendergast GV, Puttaswamy K, Wang Y, Feldman MD, Wang H, et al. Downregulation of Rap1GAP contributes to Ras transformation. Mol Cell Biol. 2007;27:6647–6658. doi: 10.1128/MCB.00155-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng H, Gao L, Feng Y, Yuan L, Zhao H, Cornelius LA. Downregulation of Rap1GAP via promoter hypermethylation promotes melanoma cell proliferation, survival and migration. Cancer Res. 2009;69:449–457. doi: 10.1158/0008-5472.CAN-08-2399. [DOI] [PubMed] [Google Scholar]
- 5.Zhang L, Chenwei L, Mahmood R, van Golen K, Greenson J, Li G, et al. Identification of a putative tumor suppressor gene Rap1GAP in pancreatic cancer. Cancer Res. 2006;66:898–906. doi: 10.1158/0008-5472.CAN-05-3025. [DOI] [PubMed] [Google Scholar]
- 6.Tsygankova OM, Ma C, Tang W, Korch C, Feldman MD, Lv Y, et al. Downregulation of Rap1GAP in human tumor cells alters cell/matrix and cell/cell adhesion. Mol Cell Biol. 2010;30:3262–3274. doi: 10.1128/MCB.01345-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey CL, Kelly P, Casey PJ. Activation of Rap1 promotes prostate cancer metastasis. Cancer Res. 2009;69:4962–4968. doi: 10.1158/0008-5472.CAN-08-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z, Mitra RS, Henson BS, Datta NS, McCauley LK, Kumar P, et al. Rap1GAP inhibits tumor growth in oropharyngeal squamous cell carcinoma. Am J Pathol. 2006;168:585–596. doi: 10.2353/ajpath.2006.050132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin KB, Tan P, Freeman SA, Lam M, McNagny KM, Gold MR. The Rap GTPases regulate the migration, invasiveness and in vivo dissemination of B-cell lymphomas. Oncogene. 2010;29:608–615. doi: 10.1038/onc.2009.345. [DOI] [PubMed] [Google Scholar]
- 10.Freeman SA, McLeod SJ, Dukowski J, Austin P, Lee CC, Millen-Martin B, et al. Preventing the activation or cycling of the Rap1 GTPase alters adhesion and cytoskeletal dynamics and blocks metastatic melanoma cell extravasation into the lungs. Cancer Res. 2010;70:4590–4601. doi: 10.1158/0008-5472.CAN-09-3414. [DOI] [PubMed] [Google Scholar]
- 11.Reedquist KA, Ross E, Koop EA, Wolthuis RM, Zwartkruis FJ, van Kooyk Y, et al. The small GTPase, Rap1, mediates CD31-induced integrin adhesion. J Cell Biol. 2000;148:1151–1158. doi: 10.1083/jcb.148.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caron E, Self AJ, Hall A. The GTPase Rap1 controls functional activation of macrophage integrin alphaM-beta2 by LPS and other inflammatory mediators. Curr Biol. 2000;10:974–978. doi: 10.1016/s0960-9822(00)00641-2. [DOI] [PubMed] [Google Scholar]
- 13.Bos JL, de Bruyn K, Enserink J, Kuiperij B, Rangarajan S, Rehmann H, et al. The role of Rap1 in integrin-mediated cell adhesion. Biochem Soc Trans. 2003;31:83–86. doi: 10.1042/bst0310083. [DOI] [PubMed] [Google Scholar]
- 14.Kooistra MR, Dube N, Bos JL. Rap1: a key regulator in cell-cell junction formation. J Cell Sci. 2007;120:17–22. doi: 10.1242/jcs.03306. [DOI] [PubMed] [Google Scholar]
- 15.Pannekoek WJ, Kooistra MR, Zwartkruis FJ, Bos JL. Cell-cell junction formation: the role of Rap1 and Rap1 guanine nucleotide exchange factors. Biochim Biophys Acta. 2009;1788:790–796. doi: 10.1016/j.bbamem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 16.Boettner B, Van Aelst L. Control of cell adhesion dynamics by Rap1 signaling. Curr Opin Cell Biol. 2009;21:684–693. doi: 10.1016/j.ceb.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Retta SF, Balzac F, Avolio M. Rap1: a turnabout for the crosstalk between cadherins and integrins. Eur J Cell Biol. 2006;85:283–293. doi: 10.1016/j.ejcb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Vuchak LA, Tsygankova OM, Prendergast GV, Meinkoth JL. Protein kinase A and B-Raf mediate extracellular signal-regulated kinase activation by thyrotropin. Mol Pharmacol. 2009;76:1123–1129. doi: 10.1124/mol.109.060129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsygankova OM, Saavedra A, Rebhun JF, Quilliam LA, Meinkoth JL. Coordinated regulation of Rap1 and thyroid differentiation by cyclic AMP and protein kinase A. Mol Cell Biol. 2001;21:1921–1929. doi: 10.1128/MCB.21.6.1921-1929.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagafuchi A, Ishihara S, Tsukita S. The roles of catenins in the cadherin-mediated cell adhesion: functional analysis of E-cadherin-alpha catenin fusion molecules. J Cell Biol. 1994;127:235–245. doi: 10.1083/jcb.127.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urushihara H, Ozaki HS, Takeichi M. Immunological detection of cell surface components related with aggregation of Chinese hamster and chick embryonic cells. Dev Biol. 1979;70:206–216. doi: 10.1016/0012-1606(79)90017-4. [DOI] [PubMed] [Google Scholar]
- 22.Balzac F, Avolio M, Degani S, Kaverina I, Torti M, Silengo L, et al. E-cadherin endocytosis regulates the activity of Rap1: a traffic light GTPase at the crossroads between cadherin and integrin function. J Cell Sci. 2005;118:4765–4783. doi: 10.1242/jcs.02584. [DOI] [PubMed] [Google Scholar]
- 23.Miyashita Y, Ozawa M. Increased internalization of p120-uncoupled E-cadherin and a requirement for a dileucine motif in the cytoplasmic domain for endocytosis of the protein. J Biol Chem. 2007;282:11540–11548. doi: 10.1074/jbc.M608351200. [DOI] [PubMed] [Google Scholar]
- 24.Tsygankova OM, Feshchenko E, Klein PS, Meinkoth JL. Thyroid-stimulating hormone/cAMP and glycogen synthase kinase 3beta elicit opposing effects on Rap1GAP stability. J Biol Chem. 2004;279:5501–5507. doi: 10.1074/jbc.M305824200. [DOI] [PubMed] [Google Scholar]
- 25.Sakurai A, Fukuhara S, Yamagishi A, Sako K, Kamioka Y, Masuda M, et al. MAGI-1 is required for Rap1 activation upon cell-cell contact and for enhancement of vascular endothelial cadherin-mediated cell adhesion. Mol Biol Cell. 2006;17:966–976. doi: 10.1091/mbc.E05-07-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wittchen ES, Worthylake RA, Kelly P, Casey PJ, Quilliam LA, Burridge K. Rap1 GTPase inhibits leukocyte transmigration by promoting endothelial barrier function. J Biol Chem. 2005;280:11675–11682. doi: 10.1074/jbc.M412595200. [DOI] [PubMed] [Google Scholar]
- 27.Fukuhara S, Sakurai A, Sano H, Yamagishi A, Somekawa S, Takakura N, et al. Cyclic AMP potentiates vascular endothelial cadherin-mediated cell-cell contact to enhance endothelial barrier function through an Epac-Rap1 signaling pathway. Mol Cell Biol. 2005;25:136–146. doi: 10.1128/MCB.25.1.136-146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogan C, Serpente N, Cogram P, Hosking CR, Bialucha CU, Feller SM, et al. Rap1 regulates the formation of E-cadherin-based cell-cell contacts. Mol Cell Biol. 2004;24:6690–6700. doi: 10.1128/MCB.24.15.6690-6700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spilker C, Kreutz MR. RapGAPs in brain: multipurpose players in neuronal Rap signalling. Eur J Neurosci. 2010;32:1–9. doi: 10.1111/j.1460-9568.2010.07273.x. [DOI] [PubMed] [Google Scholar]
- 30.McLeod SJ, Shum AJ, Lee RL, Takei F, Gold MR. The Rap GTPases regulate integrin-mediated adhesion, cell spreading, actin polymerization and Pyk2 tyrosine phosphorylation in B lymphocytes. J Biol Chem. 2004;279:12009–12019. doi: 10.1074/jbc.M313098200. [DOI] [PubMed] [Google Scholar]
- 31.Pizon V, Desjardins M, Bucci C, Parton RG, Zerial M. Association of Rap1a and Rap1b proteins with late endocytic/phagocytic compartments and Rap2a with the Golgi complex. J Cell Sci. 1994;107:1661–1670. doi: 10.1242/jcs.107.6.1661. [DOI] [PubMed] [Google Scholar]
- 32.Borland G, Smith BO, Yarwood SJ. EPAC proteins transduce diverse cellular actions of cAMP. Br J Pharmacol. 2009;158:70–86. doi: 10.1111/j.1476-5381.2008.00087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi SC, Kim GH, Lee SJ, Park E, Yeo CY, Han JK. Regulation of activin/nodal signaling by Rap2-directed receptor trafficking. Dev Cell. 2008;15:49–61. doi: 10.1016/j.devcel.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Jordan JD, Carey KD, Stork PJ, Iyengar R. Modulation of rap activity by direct interaction of Galpha(o) with Rap1 GTPase-activating protein. J Biol Chem. 1999;274:21507–21510. doi: 10.1074/jbc.274.31.21507. [DOI] [PubMed] [Google Scholar]
- 35.Mochizuki N, Ohba Y, Kiyokawa E, Kurata T, Murakami T, Ozaki T, et al. Activation of the ERK/MAPK pathway by an isoform of rap1GAP associated with G alpha(i) Nature. 1999;400:891–894. doi: 10.1038/23738. [DOI] [PubMed] [Google Scholar]
- 36.Meng J, Glick JL, Polakis P, Casey PJ. Functional interaction between Galpha(z) and Rap1GAP suggests a novel form of cellular cross-talk. J Biol Chem. 1999;274:36663–36669. doi: 10.1074/jbc.274.51.36663. [DOI] [PubMed] [Google Scholar]
- 37.Boettner B, Govek EE, Cross J, Van Aelst L. The junctional multidomain protein AF-6 is a binding partner of the Rap1A GTPase and associates with the actin cytoskeletal regulator profilin. Proc Natl Acad Sci USA. 2000;97:9064–9069. doi: 10.1073/pnas.97.16.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asuri S, Yan J, Paranavitana NC, Quilliam LA. E-cadherin dis-engagement activates the Rap1 GTPase. J Cell Biochem. 2008;105:1027–1037. doi: 10.1002/jcb.21902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su L, Hattori M, Moriyama M, Murata N, Harazaki M, Kaibuchi K, et al. AF-6 controls integrin-mediated cell adhesion by regulating Rap1 activation through the specific recruitment of Rap1GTP and SPA-1. J Biol Chem. 2003;278:15232–15238. doi: 10.1074/jbc.M211888200. [DOI] [PubMed] [Google Scholar]
- 40.Hoshino T, Sakisaka T, Baba T, Yamada T, Kimura T, Takai Y. Regulation of E-cadherin endocytosis by nectin through afadin, Rap1 and p120ctn. J Biol Chem. 2005;280:24095–24103. doi: 10.1074/jbc.M414447200. [DOI] [PubMed] [Google Scholar]
- 41.Ohba Y, Kurokawa K, Matsuda M. Mechanism of the spatio-temporal regulation of Ras and Rap1. EMBO J. 2003;22:859–869. doi: 10.1093/emboj/cdg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gulli MP, Peter M. Temporal and spatial regulation of Rho-type guanine-nucleotide exchange factors: the yeast perspective. Genes Dev. 2001;15:365–379. doi: 10.1101/gad.876901. [DOI] [PubMed] [Google Scholar]