Abstract
(Macro)autophagy encompasses both an unselective, bulk degradation of cytoplasmic contents as well as selective autophagy of damaged organelles, intracellular microbes, protein aggregates, cellular structures and specific soluble proteins. Selective autophagy is mediated by autophagic adapters, like p62/SQSTM1 and NBR1. p62 and NBR1 are themselves selective autophagy substrates, but they also act as cargo receptors for degradation of other substrates. Surprisingly, we found that homologs of NBR1 are distributed throughout the eukaryotic kingdom, while p62 is confined to the metazoans. As a representative of all organisms having only an NBR1 homolog we studied Arabidopsis thaliana NBR1 (AtNBR1) in more detail. AtNBR1 is more similar to mammalian NBR1 than to p62 in domain architecture and amino acid sequence. However, similar to p62, AtNBR1 homo-polymerizes via the PB1 domain. Hence, AtNBR1 has hybrid properties of mammalian NBR1 and p62. AtNBR1 has 2 UBA domains, but only the C-terminal UBA domain bound ubiquitin. AtNBR1 bound AtATG8 through a conserved LIR (LC3-interacting region) motif and required co-expression of AtATG8 or human GABARAPL2 to be recognized as an autophagic substrate in HeLa cells. To monitor the autophagic sequestration of AtNBR1 in Arabidopsis we made transgenic plants expressing AtNBR1 fused to a pH-sensitive fluorescent tag, a tandem fusion of the red, acid-insensitive mCherry and the acid-sensitive yellow fluorescent proteins. This strategy allowed us to show that AtNBR1 is an autophagy substrate degraded in the vacuole dependent on the polymerization property of the PB1 domain and of expression of AtATG7. A functional LIR was required for vacuolar import.
Keywords: NBR1, p62/SQSTM1, ATG8, autophagy, evolution, plants
Introduction
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved, lysosomal degradation route for organelles and macromolecules. The process involves sequestration of a part of the cytoplasm by the formation of a double membrane (the phagophore) that expands and closes upon itself to form the autophagosome. The autophagosome subsequently fuses with the lysosome to form an autolysosome whose inner membrane and content are subsequently degraded.1,2 Autophagy is important both for protein and organelle quality control, for restoring intracellular nutrients and energy during starvation and for innate immunity responses against intracellular microbes.1,3–5 Since the ubiquitin-proteasome system cannot handle large substrates such as organelles and protein aggregates, autophagy helps clear the cell of toxic aggregates and damaged organelles. In mammals there is both a constitutive, basal autophagy and a starvation-induced autophagy. In broad terms, the basal autophagy cleans the cell of damaged organelles and protein aggregates, while the induced autophagy is called upon during periods of nutrient and energy deprivation.
In yeast, the process involves 34 identified AuTophaGy (ATG) proteins, of which 15 constitute the core machinery.6–8 Likely, even more components are involved in mammalian autophagy processes as indicated from the size of the autophagy interaction network recently determined by proteomics analyses.9 Six different protein complexes are involved in the formation and expansion of the phagophore.8 These include two ubiquitin (Ub)-like protein conjugation systems involving the two Ub-like modifiers Atg8 and Atg12. Yeast has only a single Atg8 while mammals have seven ATG8 proteins divided into two subfamilies consisting of three MAP1 light chain 3 (LC3A, B and C) and four gamma-aminobutyrate receptor-associated protein (GABARAP) and GABARAP-like proteins (ATG8L/GEC-1/GABARAPL1, GATE-16/GABARAPL2 and GABARAPL3).10,11 The higher plant, Arabidopsis thaliana, has nine ATG8 proteins.12,13 The C-termini of ATG8 proteins are specifically cleaved by ATG4 family proteases and conjugated to phosphatidylethanolamine.14,15 The lipidated form is bound to the autophagosomal membranes and acts as an autophagic marker protein.14–16 The lipidated ATG8 is required for hemifusion of lipid membranes and also is suggested to drive the expansion of the autophagosome.17 In mammals, LC3B is regarded as the main ATG8 protein acting in starvation-induced autophagy. However, it seems clear that both the LC3 and GABARAP subfamilies of ATG8s are required for autophagy and that LC3 is more important for expansion while GABARAPL2 acts more at the closure step during formation and maturation of the autophagosome.18
Starvation-induced autophagy is largely a nonselective, bulk degradation process, but mounting evidence suggests that selective autophagy can target a variety of substrates.19–21 Selective autophagy is mediated by autophagic cargo receptors or adapters such as p62, which has been shown to facilitate docking of ubiquitinated substrates to the autophagosome.22 Recently, the mammalian protein NBR1 (neighbor of BRCA1 gene) was also characterized as a cargo receptor that cooperates with p62 in the autophagic clearance of ubiquitinated substrates.23,24
Other recently characterized mammalian autophagy cargo receptors or adapters are NDP52 and Nix/Bnip3L.25,26 As recently reviewed in reference 19, p62 acts as a cargo receptor for autophagic degradation of protein aggregates, soluble proteins, midbody rings, damaged mitochondria, peroxisomes, intracellular bacteria, phagocytic membrane remnants, bacteriocidal precursor- and viral capsid proteins. NBR1 has not yet been implicated in the selective autophagy of other substrates than ubiquitinated protein aggregates, but this is most likely because it is much less studied than p62. Both proteins share a similar overall domain organization with an N-terminal PB1 (Phox and Bem1p) domain followed by a ZZ-type zinc finger domain, a LIR (LC3-interacting region) motif and a C-terminal UBA (ubiquitin-associated) domain that mediates interaction with monoand polyubiquitin.19,27 The LIR motifs mediate direct interaction with LC3B and this interaction is required for the autophagic degradation of p62/NBR1-containing structures.22,23,28 The X-ray and solution structures of complexes of the LIR peptide of p62 and LC3B have been solved and show that the core sequence DDDWTHL of the p62 LIR is an extended β-strand bound at the interface between the N-terminal arm and the C-terminal Ub-like domain of LC3B.28–30 A recent analysis we made of verified LIR motifs suggests the LIR motif is eight amino acids long. Furthermore, a consensus LIR motif may be expressed as D/E-D/E-D/E-W/F/Y-X-X-L/I/V where there is not an absolute requirement for acidic residues at all three indicated positions, but usually there is at least one at these positions.19 We mapped two LIRs in human NBR1, but it is LIR1, with the core sequence SEDYIII, which is most important.23
Both p62 and NBR1 contain a C-terminal UBA domain that can bind mono- and polyubiquitin.23,31 The isolated UBA domain of p62 binds ubiquitin with low affinity,32,33 while full-length p62 binds much more strongly.23 Contrary to p62, the isolated UBA domain of NBR1 binds strongly to ubiquitin.23
The PB1 domain of p62 mediates polymerization of p62 via front-to-back interactions through electrostatic forces between appropriately positioned basic and acidic charge clusters.34,35 Among the human PB1 domain proteins, only p62 has this ability. The related human NBR1 protein is not able to polymerize via PB1 because its PB1 domain lacks the most N-terminal basic charge cluster. Instead, human NBR1 self-interacts via a coiled-coil domain.23 However, NBR1 interacts with p62 via its PB1 domain in a heterodimeric fashion.34 The polymerization property of the PB1 domain is required for efficient degradation of p62 by autophagy.28 It was recently shown that p62 and NBR1 can be recruited to ER-located autophagosome formation sites in fibroblasts before the phagophore with LC3 is recruited, and that this recruitment requires the homo-oligomerization property of p62.36
Autophagy has been extensively studied in budding yeast, mammals, Drosophila, and to some extent also in plants, but research on autophagy in nonmetazoan organisms is limited. To our knowledge, there is only one report suggestive of selective autophagy occurring in plants.37 We have undertaken a study of the evolutionary distribution of the autophagic adapters p62 and NBR1. We find that most organisms belonging to nonmetazoan kingdoms harbor a single conserved NBR1-homolog while metazoan organisms possess the p62 protein either exclusively or in addition to NBR1. A more detailed analysis of a plant representative, Arabidopsis thaliana NBR1 (At4g24690), showed that despite possessing structural similarity to the NBR1 group of proteins, it shares some distinct functional properties with mammalian p62. We show, moreover, that the Arabidopsis homolog of NBR1 is an autophagic substrate in the plant cell that polymerizes via an N-terminal PB1-domain, binds ubiquitin through a C-terminal UBA-domain, and interacts with the Arabidopsis homologs of ATG8 via an evolutionarily conserved LIR motif. Our results support the notion that nonmetazoan homologs of NBR1, such as AtNBR1, represent proteins with ancestral characteristics which in metazoans have undergone gene duplication, specific deletion of domains, and introduction of point mutations in the PB1 and UBA domains, to become the present metazoan p62 and NBR1.
Results
Evolutionary distribution of p62/SQSTM1 and NBR1: homologs of NBR1 are found throughout the eukaryotic kingdom while p62 is confined to the metazoans.
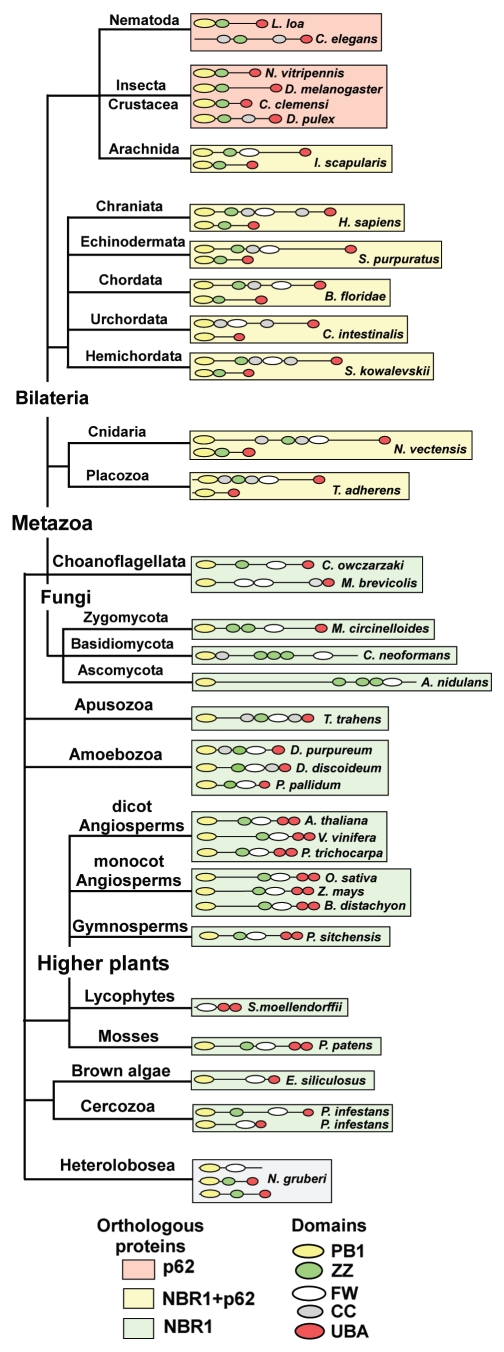
Both NBR1 and p62 share a conserved domain architecture, with an N-terminal PB1 domain, a ZZ-type zinc finger and a C-terminal UBA-domain. NBR1 contains an evolutionary conserved globular domain, not found in p62, characterized by the presence of 4 highly conserved tryptophan (W) residues. We have therefore named this domain the FW (4 tryptophan) domain. The FW domain is highly conserved in homologs of NBR1, and is also found in homologs of the human protein MGC4614 and in some bacterial proteins (Fig. S1). Since the PB1-ZZ-UBA domain combination is shared by p62 and NBR1, we find it reasonable to distinguish between p62- and NBR1-homologs by the presence of the FW domain. Using this distinction we hereafter refer to all homologs containing the FW domain as NBR1 homologs. We used BLAST searches and multiple sequence alignment tools to identify NBR1 and p62 homologs throughout the eukaryotic kingdom, and to analyze their evolutionary distribution. A representative selection of homologs and their distribution is shown in a phylogenetic tree in Figure 1. The tree is built on a phylogenetic model of eukaryotes presented in the tree of life web project,38 but does not aim to represent the true evolution of eukaryotes. Rather, it serves as a graphic representation of the conservation of NBR1 and p62 homologs in eukaryotes. Strikingly, most nonmetazoan organisms contain only a single NBR1 homolog and no p62 homolog. However, metazoans harbor both p62 and NBR1 (or only p62), suggesting that a duplication has occurred early in the metazoan lineage. To further test the relationship between the metazoan and nonmetazoan proteins, we constructed a phylogenetic tree based on alignments of the PB1, ZZ, FW and UBA domain sequences using the maximum likelihood method.39 Amoebozoa are thought to have diverged from the animal/fungal lineage after the plant/animal split, but the amoebae have maintained more of their ancestral genome diversity than plants and animals.40 The results show that when rooting the tree with NBR1 homologs of amoebae, NBR1 homologs of plants and metazoans branch out as a sister group to metazoan p62 homologs (Fig. S2). This supports our definition of NBR1 homologs based on the presence of the FW domain.
Figure 1.
Homologs of NBR1 are found throughout the eukaryotic kingdom while p62 is confined to the metazoans. The evolutionary distribution of NBR1- and p62-homologs in selected species from the eukaryotic kingdoms of opisthokonts (fungi, metazoa and choanozoa), apusozoa (flagellate protozoa), amoebozoa, archeaplastida (plantae) and chromalveolata (brown algae) is shown. The domain architectures of the proteins are shown. Background color coding is used to indicate species which possess both p62 and NBR1 (yellow), species containing only p62 (orange) and species harboring only NBR1 (green). Protein sequence accession numbers are listed in Table S1.
The choanoflagellates Monosiga brevicollis and Capsaspora owczarzaki, considered to be the closest living unicellular relatives of metazoa,41,42 both contain a single homolog of NBR1. Insects, crustaceans and nematodes seem to have lost NBR1 while keeping the p62 homolog. One could also suspect that this line diverged before the duplication event, but the finding of NBR1- and p62 homologs in the arthropod species Ixodes scapularis (deer tick) contradicts this assumption. Two homologs of NBR1 can be found in the organism Phytophthora infestans, the causative agent of late blight of potato, belonging to the cercozoan phylum of chromalveolates. This is not a general feature of chromalveolates since Ectocarpus siliculosus, a brown algae belonging to the stramemopile phylum, has only one NBR1 homolog (Fig. 1). The amoeboflagellate Naegleria gruberi belongs to the heteroboloseans, representing one of the earliest diverging eukaryotic lineages. It is the only nonmetazoan organism analyzed containing p62-like proteins (no FW domain). Naegleria also has a protein which appears to be a truncated NBR1-homolog. Some basal lineages of fungi have retained a clear homolog of NBR1 with a UBA domain, while many of the derived fungal lineages contain a more diverged homolog of NBR1 that has additional zinc fingers and lacks the C-terminal UBA-domain (Fig. 1). This apparent ‘loss’ of the traditional NBR1-homolog in fungi has been presented in a review by Kraft et al.21
Plant NBR1 homologs contain a duplicated C-terminal UBA domain but lack the coiled coil domain of mammalian NBR1 (Fig. 1). These features are highly conserved among the angiosperms and gymnosperms of the higher plant lineage and are also found in the moss Physcomitrella patens. A duplication of NBR1 has presumably occurred in the monocot lineage of angiosperms (data not shown), giving rise to a nearly identical NBR1 homolog that contains only one predicted C-terminal UBA-domain. We also identified a truncated NBR1-homolog in the spike moss Selaginella moellendorffii that contains the FW domain and the tandem C-terminal UBA domains (Fig. 1).
As a representative of nonmetazoan NBR1, we decided to analyze the Arabidopsis thaliana homolog of NBR1, hereafter referred to as AtNBR1. The process of autophagy is conserved in higher plants,43 and Arabidopsis represents a robust and versatile model system for in vivo studies.
Similar to p62, AtNBR1 homo-oligomerizes via the PB1 domain.
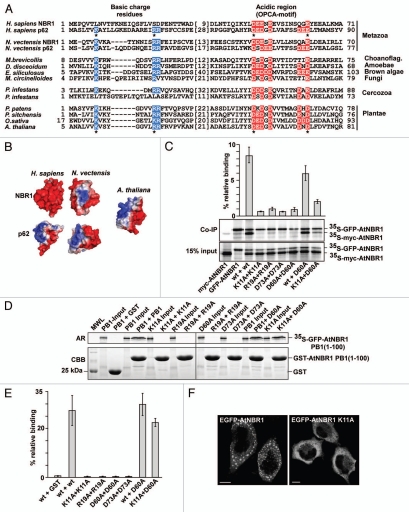
The PB1 domains of NBR1-homologs in most non-metazoan organisms harbor the N-terminal basic charge cluster and the C-terminal, acidic OPCA motif characteristic of the p62-type PB1 domain which has the ability to polymerize (Fig. 2A). The sea anemone Nematostella vectensis is considered to have a genome that reflects the early origins of multicellular metazoans.44 The predicted homologs of NBR1 and p62 in Nematostella both have p62-type PB1 domains (Fig. 2A). Molecular modeling revealed that the PB1-domain in the NBR1-homologs of Nematostella and Arabidopsis share a similar basic/acidic surface structure (Fig. 2B). This suggests that after the putative duplication of NBR1 in the metazoan lineage, p62 has kept the basic/acidic PB1-domain, while NBR1 has lost the basic surface cluster of the PB1 domain. In Phytophthora infestans we found a similar pattern of PB1-domain differentiation as in the metazoan p62 and NBR1 homologs since one of the 2 NBR1 homologs has lost the basic charge cluster (Fig. 2A). Studies have shown that p62 strongly depends on PB1-mediated homo-polymerization to be degraded by autophagy.28 We therefore asked whether AtNBR1 also has the ability to homo-polymerize via the PB1-domain. Four residues in the PB1-domain of p62 are especially important for polymerization, two residues located in the basic charge cluster (K7 and R21), and two acidic residues in the OPCA-motif (D69 and E82).34 Hence, we made single point mutants where the corresponding four residues in the PB1-domain of AtNBR1 (K11, R19, D60 and D73) were mutated to alanines. We then performed co-immunoprecipitation experiments with full-length, wt or mutant AtNBR1, using 35S-labeled, co-in vitro translated proteins tagged with GFP or a myc epitope tag. GFP-tagged AtNBR1 was immunoprecipitated with a GFP antibody, and the efficiency of co-precipitation of myc-tagged wt and mutant proteins was analyzed. The results show that AtNBR1 is able to self-interact, and that a functional PB1 domain is required for this interaction (Fig. 2C). All four point mutants lost the ability to self-interact. These mutations do not cause major structural changes in the domain, as an interaction could still be obtained between the wt protein and an acidic charge point mutant, as well as between two point mutants together (acidic mutation and basic mutation). The quantification of co-precipitated protein shows that the amount of co-precipitation was reduced in these point-mutant interactions compared with the wt interaction (Fig. 2C). This is consistent with the notion that wt AtNBR1 can polymerize via PB1 domains to form homo-polymers whereas a basic charge point mutant is only able to dimerize with an acidic charge point mutant.
Figure 2.
AtNBR1 polymerizes via the N-terminal PB1-domain. (A) Alignment of PB1 domain sequences from p62- and NBR1 homologs of selected metazoan- and nonmetazoan species. Blue background color denotes basic residues and red background color denotes acidic residues of the charged clusters important for PB1 domain interactions. The OPCA-motif is indicated. Gaps are indicated with dashes and the numbers of amino acid residues not shown are specified in brackets. Residues crucial for the self-interaction of p62,34 and which are mutated in AtNBR1 in the analyses shown in (C–E) are indicated with asterisks. (B) Electrostatic surface potentials of the PB1 domains of NBR1 (PDB:2BKF) and p62 (PDB:2KKC) from H. sapiens and N. vectensis as well as the AtNBR1 PB1 domain. The N. vectensis and AtNBR1 PB1 domains were modeled using Swissmodel. (C) Co-immunoprecipitation experiments using full-length AtNBR1. Myc- and GFP-tagged AtNBR1 (wt or the indicated mutants) were co-translated in vitro in the presence of 35S-methionine and precipitated using an anti-GFP antibody. Immunoprecipitated and co-precipitated proteins as well as in vitro translated proteins corresponding to 15% of the input were resolved by SDS-PAGE and detected by autoradiography. The upper band corresponds to 35SGFP-AtNBR1, the lower to 35Smyc-AtNBR1. Quantifications of the interaction data are shown above the gel parts. Band intensity was measured using ImageJ (Fuji) and Y-axis values were calculated employing the following formula; (IP(myc/GFP)/input(myc/GFP)) × 100. (D) GST pulldown assays using in vitro translated 35S-labeled GFP-AtNBR1 PB1 (amino acids 1–100) (with the indicated point mutations) and GST or GST-PB1 (with indicated point mutations) constructs. Precipitated proteins were detected by autoradiography. (E) Quantitative representation of the interaction data shown in (D). Y-axis values are set to percent total binding protein; (pulldown/input) × 100. (F) AtNBR1 forms cytosolic aggregates when overexpressed with an N-terminal GFP-tag in HeLa cells, while the K11A point-mutant of AtNBR1 loses the ability to form aggregates. Results in (C and E) are mean values of three independent experiments with standard deviations indicated as bars. Bars represent 10 µm.
To confirm the role of the PB1 domain in polymerization we also performed GST-pulldown experiments using the isolated PB1-domain of AtNBR1. The wt-domain and all 4 point mutants were expressed as GST-PB1 fusion proteins and used in GST-pulldown experiments with in vitro translated, 35S-labeled, GFP-tagged PB1 domain constructs. As shown in Figure 2D, the wt PB1 domain showed self-interaction, while all 4 point mutants completely lost the ability to self-interact. However, in this experiment the interaction between wt and point mutated PB1, as well as between 2 point mutants, showed no significant decrease in binding compared with the wt-wt interaction (Fig. 2D and E). This is due to polymerization of the wt PB1-domains before they are subjected to the pulldown assays in vitro, causing a reduction in the interaction with GST-tagged PB1. Consistent with the results from the in vitro interaction experiments, we found wt AtNBR1 to form large, punctated, cytoplasmic structures when overexpressed as a GFP-fusion protein in HeLa cells (Fig. 2F). This suggests that AtNBR1 also has the ability to form aggregate structures in vivo, and when we expressed a GFP-tagged PB1-point mutant of AtNBR1 (K11A) the ability to aggregate was lost (Fig. 2F). Taken together, all these results show that AtNBR1 has a p62-like PB1 domain able to polymerize in vitro and in vivo.
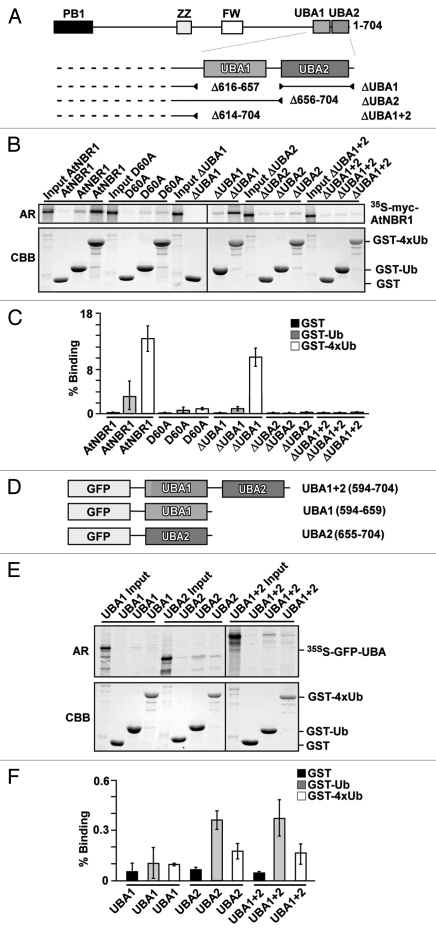
Only the C-terminal UBA domain of the twin UBA domains of AtNBR1 binds ubiquitin.
The UBA domain is conserved in the identified NBR1-homologs, and the C-terminal region of NBR1 in higher plants and in Physcomitrella contains two juxtaposed UBA-domains. Both domains harbor the conserved residues that are needed to create a hydrophobic ubiquitin binding patch. A secondary structure prediction shows that both domains consist of three predicted α-helixes (Fig. S3). To test if one or both UBA domains could bind to ubiquitin we made deletion constructs of full-length myc-AtNBR1 that lacked either the UBA1 or UBA2 or both UBA-domains (Fig. 3A). GST-pulldown assays using GST-tagged mono-ubiquitin and tetra-ubiquitin (4xUb) demonstrated that AtNBR1 has the ability to bind ubiquitin and that the binding is mediated by the C-terminal UBA2 domain. Deletion of the UBA2-domain completely eliminated the interaction with ubiquitin, while deletion of the UBA1 domain had no significant effect (Fig. 3B and C). It has previously been shown for the mammalian homologs that ubiquitin binding is strongly induced by PB1-mediated polymerization of p62 or coiled-coil-mediated oligomerization of NBR1, respectively.23,27,33 In line with these studies, the binding between AtNBR1 and 4xUb was reduced almost 10-fold when testing a monomeric point-mutant (D60A) (Fig. 3C). We next tested the binding capacity of the isolated UBA-domains fused to GFP (Fig. 3D). The isolated UBA2 domain interacted with ubiquitin while the UBA1 domain did not show significant binding. Compared with full-length AtNBR1, the observed binding capacity of UBA2 was considerably lower, and approximately similar to the binding of monomeric AtNBR1 (not shown). Another interesting observation was that the UBA2-domain interacted more strongly with mono-Ub than with 4xUb (Fig. 3E and F). In conclusion, AtNBR1 binds Ub only via its C-terminal UBA2 domain, and similar to p62, its binding efficiency is strongly dependent on polymerization of the full-length protein.
Figure 3.
AtNBR1 binds ubiquitin through the most C-terminal UBA domain (UBA2). (A) Schematic illustration of AtNBR1 UBA deletion constructs. (B) GST pull-down assays using in vitro translated 35S-labeled myc-AtNBR1 (indicated deletions) and immobilized GST or indicated GST-Ub and GST-4xUb constructs. Precipitated proteins were detected by autoradiography. (C) Quantitative representation of the interaction data shown in (B). Y-axis values are set to percent total binding protein; (pulldown/input) × 100. (D) AtNBR1 GFP-UBA domain fusion constructs used for pull-down experiments. (E) GST pull-down assays using in vitro translated 35S-labeled GFP-UBA constructs and GST or indicated GST-Ub and GST-4xUb constructs. Precipitated proteins were detected by autoradiography. (F) Quantitative representation of the interaction data shown in (E). Results in (C and F) are mean values of three independent experiments with standard deviations indicated as bars.
AtNBR1 binds AtATG8 through a conserved LIR-motif.
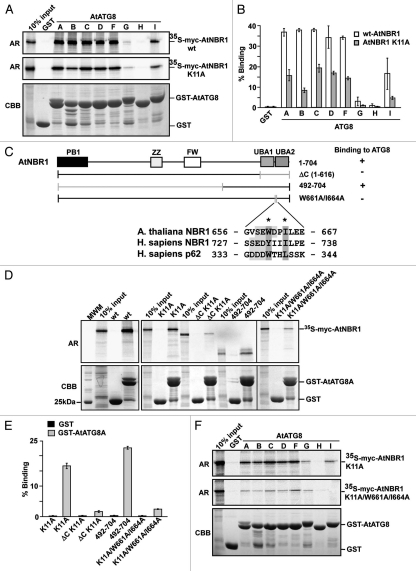
ATG8 is highly conserved throughout the eukaryotic kingdom and Arabidopsis contains a family of nine homologs of ATG8 (AtATG8A-I).12,13 AtATG8 is present in autophagosomes that are transported by autophagy to the central vacuole,13 and is used as a marker for autophagy in plants.45 To test if AtNBR1 had the ability to bind AtATG8 in vitro, we first cloned and expressed GST-fusions of eight of the nine homologs of AtATG8 in E. coli. Full-length, myc-tagged AtNBR1 and monomeric myc-AtNBR1 (K11A) were subsequently tested in GST-pulldown assays with the eight recombinant AtATG8 homologs (ATG8E was not tested). AtNBR1 bound to six of these eight AtATG8 family members (Fig. 4A). The binding was equally strong for AtATG8A, -C, -D and -F, while reduced binding was observed for AtATG8B and -I. AtATG8H did not interact and AtATG8G interacted very weakly with AtNBR1 (Fig. 4B). The interaction was not dependent on polymerization as shown for the monomeric AtNBR1 K11A mutant, but polymerization of the wt AtNBR1 resulted in a more efficient co-precipitation with the AtATG8 proteins (Fig. 4B).
Figure 4.
AtNBR1 binds to Arabidopsis ATG8 family proteins via a LIR-motif located between the twin UBA domains. (A) GST pull-down assays using in vitro translated 35S-labeled myc-AtNBR1 (polymeric and monomeric K11A mutants) and immobilized GST or GST-ATG8 (indicated isoforms) constructs. Precipitated proteins were detected by autoradiography. (B) Quantitative representation of the interaction data shown in (B) (polymeric and monomeric AtNBR1). Y-axis values are set to percent total binding protein; (input/pulldown) × 100. (C) Constructs used and a summary of GST pull-down assays between full-length ATG8A fused to GST and deletion mutants of AtNBR1 (upper part). The lower part shows an alignment of the LIR in AtNBR1 to the corresponding sequences in human p62 and NBR1. The W661 and I664 residues mutated to A are indicated with asterisks. (D) GST pull-down assays using in vitro translated 35S-labeled myc-AtNBR1 (indicated deletions and mutations) and GST or indicated GST-ATG8A constructs. Precipitated proteins were detected by autoradiography. (E) Quantitative representation of the interaction data shown in (B). (F) GST pull-down assays using in vitro translated 35S-labeled, monomeric (K11A mutant) myc-AtNBR1 (indicated mutations) and immobilized GST or indicated GST-ATG8 (indicated isoforms) constructs. Precipitated proteins were detected by autoradiography. Results in (B and E) are mean values of three independent experiments with standard deviations indicated as bars.
We have previously shown that both p62 and NBR1 interact with ATG8 homologs via a LIR motif.22,23 To map the interaction region and look for a potential LIR motif in AtNBR1 a series of deletion constructs were made. Only the most informative ones are shown in Figure 4C. When mapping this region, it became apparent that the polymerization of AtNBR1 masked the effect of specific deletions, and it was therefore optimal to use monomeric AtNBR1 in combination with the desired deletion/mutation. Upon deletion of the C-terminal region containing both UBA-domains (amino acids 617–704), the interaction was significantly reduced (Fig. 4D and E). Within this region, a short stretch of residues encompassing amino acids 660–675 was identified as the most likely interaction region based on similarity to the LIR in p62 and NBR1. The consensus LIR motif W/YXXL/I is found between the two UBA domains in AtNBR1 as WDPI, and this region is highly conserved among all the NBR1 homologs of higher plants (Fig. S3). A double-point mutation substituting the core hydrophobic residues tryptophan and isoleucine in the LIR motif with alanines (W661A/I664A) displayed complete loss of binding similar to deletion of the entire region (ΔC) (Fig. 4E). However, the short C-terminal region alone did not bind sufficiently well to be tested in GST-pulldown assays, and the shortest construct to show significant binding was the one containing amino acids 492–704. Finally, we also performed a pulldown assay with the LIR mutant of AtNBR1 against eight isoforms of AtATG8 (Fig. 4F). The double LIR-point mutation (W661A/I664A) severely reduced the interaction with all six isoforms of AtATG8 that bound strongly to AtNBR1. These results show that the binding of AtNBR1 to AtATG8 isoforms is mediated by a conserved LIR motif located in the junction of UBA1 and UBA2 overlapping with the first predicted α-helix of UBA2 (Fig. S3).
AtNBR1 requires co-expression of AtATG8 or human GABARAPL2 to be recognized as an autophagic substrate in HeLa cells.
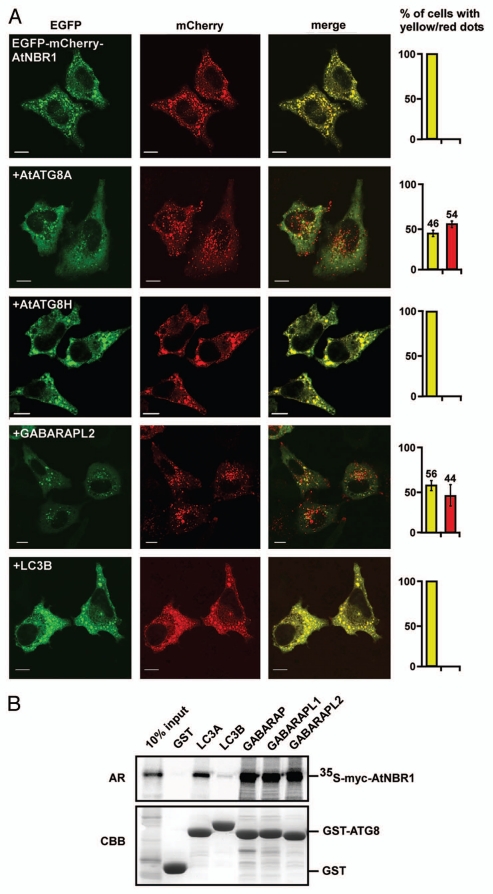
Previous studies have shown that ATG-proteins from Arabidopsis can complement homologs in yeast.46 Having established that AtNBR1 shares biochemical properties with p62 and NBR1, we next asked whether AtNBR1 could be recognized as an autophagic substrate and imported to acidified compartments in a mammalian system. To test this, we expressed AtNBR1 in HeLa cells and used the double tag-strategy to differ between localization in neutral and acidified compartments. The double-tag strategy was previously used to demonstrate that p62 and NBR1 are sequestered to acidic lysosomes in mammalian cells.22,23 We utilized a double tag consisting of EGFP and the acid stable red fluorescent protein mCherry. When overexpressed in HeLa cells, EGFP-mCherry-AtNBR1 accumulated in large cytosolic aggregates (Fig. 5A). However, in all transfected cells the red and green fluorescent signals were completely overlapping. This implies that AtNBR1-containing aggregates are not acidified and therefore not recognized as autophagic substrates in a mammalian system. We reasoned that AtATG8 was possibly required to link AtNBR1 to the autophagic system. To address this possibility, we tested whether co-expression of myc-tagged AtATG8A could rescue HeLa cells for the inability to degrade AtNBR1 by autophagy. As seen in Figure 5A, structures containing EGFP-mCherry-AtNBR1 were efficiently acidified when co-transfected with AtATG8A. As a negative control we co-expressed AtATG8H and here we found no acidified structures. We also co-expressed human LC3B and as expected, this also failed to induce an acidification of the structures containing AtNBR1. This indicates that AtNBR1 does not recognize mammalian LC3B and is therefore not sequestered into autophagosomes. However, when we co-expressed human GABARAPL2 (GATE-16) we saw acidification of AtNBR1-containing structures. Although GABARAPL2 is expressed in HeLa cells, the level of endogenous GABARAPL2 is likely too low to enable efficient selective degradation of overexpressed AtNBR1. To test this further we performed a pulldown experiment using in vitro translated AtNBR1 with the mammalian homologs of ATG8 (Fig. 5B). The results show that AtNBR1 binds strongly to the mammalian GABARAP subfamily members (GABARAP, GABARAPL1 and GABARAPL2), weakly to LC3A, and not at all to LC3B, supporting the in vivo specificities observed.
Figure 5.
AtNBR1 is not recognized as an autophagic substrate in HeLa cells unless AtATG8 or GABARAPL2 are co-expressed. (A) Overexpression of GFP-mCherry-AtNBR1 in HeLa cells causes accumulation of cytosolic aggregates. When co-expressed with myc-tagged AtATG8A, red punctated structures appear and the amount of cytosolic aggregates is reduced. This pattern is also found upon co-expression with myc-GABARAPL2. No red structures are found upon co-expression with myc-AtATG8H or myc-LC3B. The graphs to the right illustrate the percentage of transfected cells containing only yellow structures (yellow bar) or a mix of both yellow and red (red bar). Each graph represents the mean of three separate transfections (>100 cells counted per transfection) with standard deviation indicated. Picture bars represent 10 µm. (B) GST pull-down assays using in vitro translated 35S-labeled myc-AtNBR1 and GST or indicated GST-Ub and GST-ATG8 constructs. Precipitated proteins were detected by autoradiography.
AtNBR1 forms cytosolic bodies in plant cells and is localized to the central vacuole by autophagy.
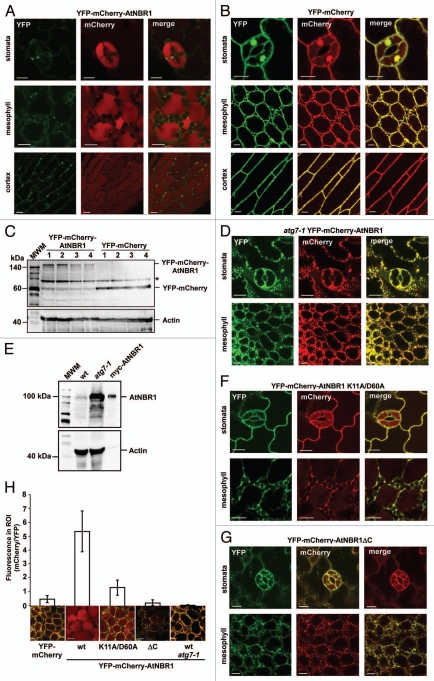
Whereas animal cells have many small acidic vacuoles (lysosomes), plant cells have one acidified vacuole that fills almost the entire cell, and autophagosomes fuse with the central vacuole to deliver their contents for degradation. RFP has previously been used to tag proteins targeted to the central vacuole of plant cells.37 To test whether AtNBR1 is targeted to the central vacuole in plants we created transgenic Arabidopsis lines using the double tag to distinguish between cytosolic and vacuolar localization. When looking at seedlings of Arabidopsis Col-0 expressing the double-tagged, YFP-mCherry-AtNBR1 (Fig. 6A), we found that AtNBR1 forms punctated structures that could be observed as yellow (green + red) dots in the cytosol lining the periphery of the cells. The central vacuole exhibited strong red fluorescence, indicating that AtNBR1 is imported into the central vacuole of the cell. This pattern could be found within different tissues of the plant (stomata, mesophyll and cortex are shown). Transgenic Arabidopsis expressing YFP-mCherry was included as control protein that can be recruited to the vacuole by nonselective autophagy. The YFP-mCherry exhibited diffuse cytosolic and nuclear localization, and only low levels of red fluorescence could be detected in the central vacuole (Fig. 6B). Protein gel blots of extracts prepared from 4 different lines of transgenic plants expressing either YFP-mCherry or YFP-mCherry-AtNBR1, respectively, showed that proteins of the expected sizes were expressed at relatively similar levels (Fig. 6C). When double-tagged AtNBR1 was expressed in the autophagy-defect atg7-1 line, the cells contained large amounts of cytosolic AtNBR1-containing aggregates and no red fluorescence could be detected in the central vacuole (Fig. 6D). ATG7 is essential for delivery of autophagic substrates to the vacuole in Arabidopsis.47 When crude extracts from two-week-old Arabidopsis seedlings (Col-0 and atg7-1) were subjected to protein gel blot analysis with an anti-AtNBR1 antibody, we found that AtNBR1 had accumulated in the atg7-1 line (Fig. 6E). This clearly demonstrates that vacuolar import of endogenous AtNBR1 is mediated by autophagy.
Figure 6.
AtNBR1 forms cytosolic, punctated bodies and is imported to the central vacuole by autophagy in vivo. (A) Transgenic Arabidopsis expressing YFP-mCherry-AtNBR1. Scattered punctate structures containing AtNBR1 can be found along the rim of the cells and the central vacuole of the cells is exhibiting diffuse red fluorescence. This pattern can be found in all tissues of the plant. The emitted fluorescence of YFP has been converted to green for visual purposes. (B) Transgenic Arabidopsis expressing YFP-mCherry. (C) Protein gel blot using an mCherry antibody to visualize the expression of YFP-mCherrry-AtNBR1 and YFP-mCherry in four different transgenic plant lines, respectively. The actin levels are shown in the lower part. The asterisk denotes an unspecific band. (D) The atg7-1 knockout mutant line of Arabidopsis expressing YFP-mCherry-AtNBR1. (E) Protein gel blot of crude protein extract from 2-week-old seedlings of Arabidopsis Col-0 and Arabidopsis atg7-1, using anti-AtNBR1 antibody. In vitro translated myc-AtNBR1 is included as positive control. Actin is included as loading control. (F and G) Transgenic Arabidopsis expressing monomeric YFP-mCherry-AtNBR1 K11A D60A construct (F) and YFP-mCherry-AtNBR1ΔC with UBA1, -2 and LIR deleted (G). (H) Quantification of vacuolar import of AtNBR1. Quantification was performed by dividing the average gray value in the cytosol (YFP) with the average gray value in vacuole (mCherry), using split channel images in ImageJ. The graphical representation is based on three separate lines per construct, >30 counted cells from three separate plants of each line (total of 100 cells), with standard deviation indicated as bars.
When expressing mutated forms of AtNBR1 we observed a reduction in the intensity of vacuolar mCherry. Point mutations in the basic and acidic motifs of the PB1 domain or deletion of the C-terminal region (LIR + UBA), caused disruption of aggregate-formation and resulted in increased cytosolic accumulation of AtNBR1 (Fig. 6F and G). To quantify the amount of vacuolar import we compared the intensity of mCherry in the central vacuole of the different lines (Fig. 6H). Due to differences in expression levels between lines, the intensity of vacuolar mCherry was compared with the level of cytosolic YFP and the quantification revealed that wild-type AtNBR1 is very efficiently transported to the central vacuole of the plant cell. Upon inhibition of polymerization (YFP-mCherry-AtNBR1 K11A) the import to the vacuole fell to near-control levels (YFP-mCherry). No vacuolar import of AtNBR1 was detected when both LIR and UBA domains were deleted or when AtNBR1 was expressed in the atg7-1 line. Taken together, our results with the transgenic double-tag plants and endogenous NBR1, show that AtNBR1 is an autophagy substrate degraded in the vacuole of Arabidopsis in a manner dependent on the presence of the polymerizing PB1 domain and the C-terminal region containing both LIR and UBA domains.
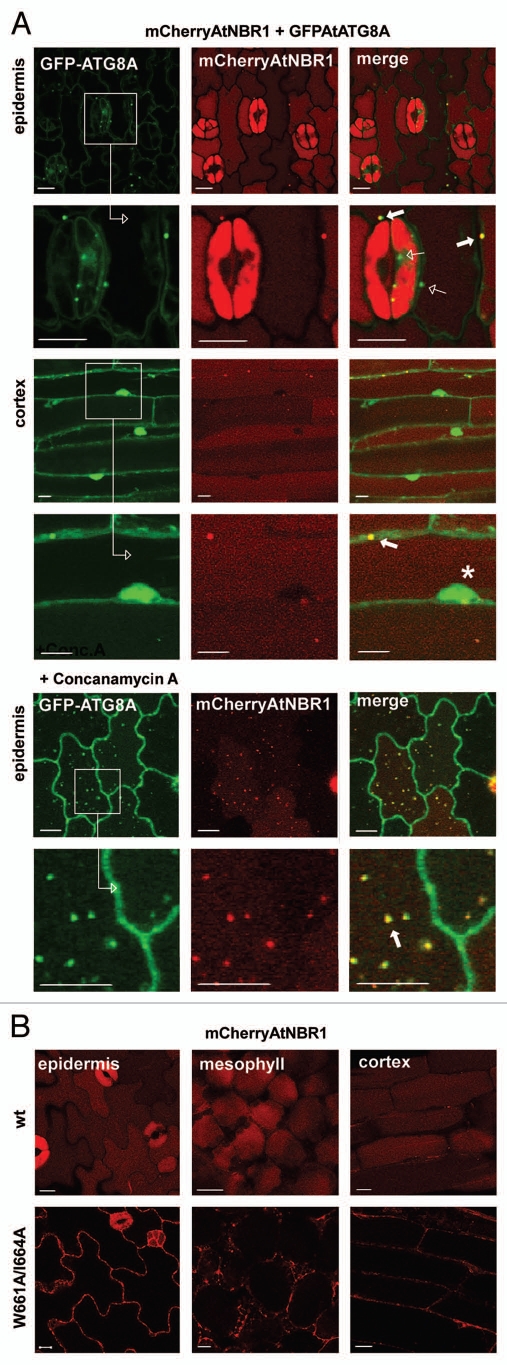
AtNBR1 co-localizes with AtATG8A in vivo and a functional LIR is required for vacuolar import.
Arabidopsis ATG8 has previously been shown to colocalize with autophagic substrates such as cytochrome b5 and Rubisco-containing bodies.37,48 Under nutrient-rich conditions, GFP-ATG8A can be found throughout the cytosol and nucleus of the cell. Upon treatment with concanamycin A, which neutralizes the vacuole and inhibits degradation, punctate bodies containing ATG8 appeared in the central vacuole.49 To check the interaction between AtNBR1 and ATG8 in vivo, we transformed a stably expressing GFP-ATG8A line with a mCherry-AtNBR1 construct. We also performed the same transformation with a double LIR point mutant of AtNBR1. The results showed that AtNBR1 colocalized with ATG8A in punctated cytosolic bodies with overlapping fluorescence (Fig. 7A). Cytosolic bodies containing only AtATG8A could also be found, but all AtNBR1-containing bodies colocalized with AtATG8A. The central vacuole exhibited diffuse red fluorescence while the cytosol and nucleus displayed diffuse green fluorescence. Upon treatment with concanamycin A, we observed a gradual neutralization of the vacuole, accompanied by the appearance of punctated bodies containing both ATG8A and AtNBR1 within the vacuole (Fig. 7A). When overexpressing AtNBR1 with a C-terminal deletion we previously saw that vacuolar import was severely reduced (Fig. 6F). However, this deletion included both the C-terminal UBA domains and the characterized LIR. To verify whether a functional LIR alone is required for vacuolar import of ATNBR1 we also analyzed plants expressing mCherry-AtNBR1 with two point mutations in the LIR motif (W661A/I664A). When comparing the cellular localization of wt mCherry-AtNBR1 and LIR-mutated mCherry-AtNBR1, it was clearly seen that the LIR mutations caused cytosolic accumulation of AtNBR1 (Fig. 7B). Hence, a functional LIR motif is required for autophagic degradation of AtNBR1.
Figure 7.
AtNBR1 colocalizes with AtATG8A in vivo and a functional LIR is required for vacuolar import. (A) Stable co-expression of mCherry-AtNBR1 and GFP-AtATG8A in Arabidopsis. Images collected from two different tissues with enlarged insets shown below. The central vacuole exhibits diffuse red fluorescence, while the cytosol and nucleus (marked with asterisk) exhibits diffuse green fluorescence. AtNBR1 co-localizes with AtATG8A in punctated cytosolic bodies with overlapping fluorescence (filled arrows). Punctate structures containing only GFP-AtATG8A are also found (open arrows). Upon treatment with Concanamycin A, an accumulation of punctated bodies of overlapping fluorescence can be seen within the central vacuole. (B) Transgenic Arabidopsis expressing mCherry-AtNBR1 (upper inset) and mCherry-AtNBR1 W661A I664A mutant (lower inset). Wild-type (wt) mCherryAtNBR1 is mostly localized to the central vacuole of the plants cells while the AtNBR1 LIR-mutant accumulated in the cytosol.
Discussion
Phylogenetic analyses based on conservation of domain organization and sequence similarity suggest that At4g24690 is the Arabidopsis homolog of mammalian NBR1, hence we named it AtNBR1. However, based on biochemical properties such as PB1 domain polymerization and UBA domain-mediated binding properties to ubiquitin, AtNBR1 has functional characteristics more similar to mammalian p62 than mammalian NBR1. AtNBR1 and other nonmetazoan NBR1 homologs, appear as functional hybrids that share some properties with metazoan p62 and some with mammalian NBR1. It is tempting to speculate that NBR1 represents the ancestral gene and that metazoan p62 arose via gene duplication of NBR1 followed by loss of the FW- and coiled coil domains. Metazoan NBR1 subsequently lost the basic charge cluster of its PB1 domain and thereby its ability to polymerize via the PB1 domain, whereas this property was retained by metazoan p62. Interestingly, some groups of animals have experienced a loss of NBR1 and only kept p62, as indicated from our analyses of available sequences from nematodes, insects and crustaceans.
We found the p62-type PB1 domain to be conserved in the NBR1-homologs of non-metazoan species. AtNBR1 uses the N-terminal PB1 domain to polymerize and a functional PB1 domain is required for aggregate formation and autophagic degradation of AtNBR1 in transgenic plants. This represents a functional link between AtNBR1 and mammalian p62, which also requires a functional PB1 domain to form aggregates,50 and for efficient degradation by autophagy.28 Mammalian NBR1 uses its coiled-coil domain to self-interact.23 A putative coiled-coil can be found in some NBR1 homologs in other monophyletic lineages of eukaryotes, but is absent in plants. The oligoor polymerization property seems to be a requirement for efficient sequestration of autophagic adapters into phagophores (forming autophagosomes) whether it is in the yeast cytoplasm-to-vacuole-targeting pathway or in selective autophagy mediated by p62 or NBR1.19
In addition to the PB1 domain polymerization properties, also the ubiquitin binding properties of AtNBR1 and human p62 show striking similarities distinct from human NBR1. Only the most C-terminal UBA-domain (UBA2) of AtNBR1 has the ability to bind ubiquitin in vitro. Similar to p62, full-length, polymeric AtNBR1 binds very well to ubiquitin while the monomeric PB1 domain mutant binds much more poorly. For monomeric AtNBR1 or the isolated UBA2-domain, the interaction between AtNBR1 and ubiquitin is reduced almost 10-fold compared with polymeric AtNBR1. Monomeric ubiquitin interactions are commonly weak, but enhanced physiologically to high-affinity interactions via other interactions or polymerization of the ubiquitin binding proteins.51 This correlates well with the observed effect of PB1 polymerization on the ubiquitin binding capacity of AtNBR1. Furthermore, monomeric AtNBR1 and the isolated UBA2 domain have the same binding capacity, which confirms that apart from PB1-mediated polymerization, no other part of AtNBR1 contributes to the binding. The reason why NBR1 orthologs in higher plants, lycophytes and mosses have twin UBA domains is unknown, particularly since UBA1 cannot bind ubiquitin.
We found AtNBR1 to bind selectively to six of the eight Arabidopsis ATG8 isoforms. Differential expression patterns in tissues of Arabidopsis seedlings suggest a functional redundancy of the AtATG8 isoforms,49 a notion that is supported by our experimental results. AtNBR1 binds to the AtATG8 isoforms using a conserved LIR motif (SEWDPILE). Based on what is known so far about LIR-ATG8 protein interactions,19,30 we can surmise that the W and I residues will dock into the hydrophobic pockets and the acidic E and D residues will contribute by electrostatic interactions to the binding to the AtATG8s.
When overexpressed alone in human HeLa cells, AtNBR1 accumulated in cytosolic aggregates, but upon co-expression with AtATG8A AtNBR1 was sequestered into acidified compartments. Human GABARAPL2, but not human LC3B or LC3A (data not shown), was able to complement the function of AtATG8. The inability of LC3A and -B to complement the function of AtATG8 is most likely due to weak and no binding to AtNBR1, respectively. We found that endogenous AtNBR1 accumulates in autophagy-deficient plants. Furthermore, in transgenic plants AtNBR1 acts as a selective autophagy substrate dependent on PB1 domain polymerization and the LIR-ATG8 interaction. In mammals p62 and NBR1 act as cargo receptors for selective autophagic degradation of ubiquitinated targets.22,23 Both the ability of AtNBR1 to interact with ubiquitin and the UBA domain conservation in plants as well as in the majority of nonmetazoans, indicates that ubiquitin may have an important role in selective autophagy in these organisms as well. A study performed with the cellular slime mold Dictyostelium discoideum showed that autophagy-deficient cells accumulated protein aggregates containing ubiquitin and the putative Dictyostelium homolog of p62.52 In a recent review on selective autophagy it is suggested that fungi have retained a homolog of NBR1 that lacks the C-terminal UBA-domain, and that the yeast Atg19 protein, a cargo receptor in the Cvt-pathway, is a divergent homolog of NBR1.21 As mentioned above, we also find that the fungi have a NBR1 homolog without UBA domain, but we were not able to detect significant sequence homology between NBR1 or p62 proteins to yeast Atg19 to support an evolutionary relationship. However, a functional relationship is undoubtedly evident. The lack of UBA domains in fungal NBR1 homologs fits with the fact that there is no involvement of ubiquitin in the Cvt-pathway, mitophagy or pexophagy in yeast species.19,21
The presence of two p62/NBR1 homologs is not unique to the metazoan lineage as we found two p62-like sequences (containing PB1-, ZZ- and UBA, but lacking FW domains) in Naegleria gruberi and two NBR1-like sequences in Phytophthora infestans. Also, monocots contain two NBR1 proteins, one with a single UBA domain and the other with twin UBA domains. This demonstrates that nomenclature is not straightforward. However, we feel that our distinction of NBR1 homologs and p62-like proteins based on the presence or absence of the FW domain is a logical solution. In fact, Naegleria is the only non-metazoan organism analyzed containing p62-like proteins. The conservation of ATG-related genes in unicellular eukaryotes (protists) have been addressed in two recent reviews,53,54 and p62/NBR1 homologs have been noted in Dictyostelium, Monosiga, Thalassiosira, Phytophthora and Naegleria.54 In case of the putative Dictyostelium homolog of p62 mentioned above,52 we found that it contains the FW domain and should therefore be classified as an NBR1 ortholog (see Fig. 1 and Fig. S2).
To our knowledge this is the first report describing a selective autophagy substrate in plants. Presently, there are several insertion mutant lines of AtNBR1 available, but we found that none of these offered efficient knockout of AtNBR1. It is possible that a complete and constitutive knockout of the AtNBR1 gene leads to plant lethality. Future studies might therefore have to be directed toward generating inducible knockout/knockdown plants in order to unravel the functional role of AtNBR1 in more detail than is currently possible.
Materials and Methods
Plasmids used in this study.
Plasmids used in this study are listed in Table 1. They were made by conventional restriction enzyme-based cloning or by use of the Gateway recombination system (Invitrogen). Point mutants were made using the QuickChange site-directed mutagenesis kit (Stratagene, 200523). Gateway LR reactions were performed as described in the Gateway cloning technology instruction manual (Invitrogen). Oligonucleotides for mutagenesis, PCR and DNA sequencing reactions were obtained from Invitrogen and Sigma. All plasmid constructs were verified by restriction digestion and/or DNA sequencing (BigDye, Applied Biosystems, 4337455). Details of their construction are available upon request.
Table 1.
Plasmids used in this study
| Plasmid | Description | Reference |
| Gateway cloning vectors | ||
| pENTR 1A | Gateway® Entry Vector, KanR | Invitrogen |
| pENTR 2B | Gateway® Entry Vector, KanR | Invitrogen |
| pENTR 3C | Gateway® Entry Vector, KanR | Invitrogen |
| pDest-EGFP-C1 | Mammalian EGFP fusion expression vector, CMV promoter, AmpR | Ref. 50 |
| pDest-myc | Mammalian myc-tag fusion expression vector, CMV & T7 promoters, AmpR | Ref. 34 |
| pDest-mCherry-C1 | Mammalian mCherry fusion expression vector, backbone aspDestEGFP-C1, AmpR | Ref. 22 |
| pcDNA-Dest53 | Mammalian GFP fusion expression vector CMV and T7 promotors, AmpR | Invitrogen |
| pDest15 | Mammalian GST fusion expression vector CMV and T7 promotors, AmpR | Invitrogen |
| pENTR-AtNBR1 | Gateway® PENTR/SD-DTOPO vector with At NBR1 | SSP Consortium |
| pENTR-AtATG8G | Gateway® PENTR/SD-DTOPO vector with Arabidopsis ATG8G | SSP Consortium |
| pUNI51 cloning vector | ||
| pUNI51 | pUNI51 Cloning Vector, Universal cloning vector used for ORF clones | SSP Consortium |
| Plant expression vectors | ||
| pEarleygate104 | Plant YFP fusionvector, Binary, 35S promoter, KanR, Bar | Ref. 58 |
| pK2GW7 | Plant overexpression vector, 35 promotor, Binary, SmR, Kanamycin | Ref. 59 |
| pUNI51 cloning vectors | ||
| pUNI51-ATG8A | pUNI51 Cloning Vector with AtATG8A | SSP Consortium |
| pUNI51-ATG8B | pUNI51 Cloning Vector with AtATG8B | SSP Consortium |
| pUNI51-ATG8C | pUNI51 Cloning Vector with AtATG8C | SSP Consortium |
| pUNI51-ATG8D | pUNI51 Cloning Vector with AtATG8D | SSP Consortium |
| pUNI51-ATG8F | pUNI51 Cloning Vector with AtATG8F | SSP Consortium |
| pUNI51-ATG8H | pUNI51 Cloning Vector with AtATG8H | SSP Consortium |
| pUNI51-ATG8I | pUNI51 Cloning Vector with AtATG8I | SSP Consortium |
| Entry clones made by subcloning and/or site-directed mutagenesis | ||
| pENTR- AtNBR1 K11A | Gateway® Entry Vector with AtNbr1 point mutantK11A | This study |
| pENTR- AtNBR1 R19A | Gateway® Entry Vector with AtNbr1 point mutant R19A | This study |
| pENTR- AtNBR1 D60A | Gateway® Entry Vector with AtNbr1 point mutant D60A | This study |
| pENTR- AtNBR1 D73A | Gateway® Entry Vector with AtNbr1 point mutant D73A | This study |
| pENTR- AtNBR1 PB1 (aa 1–100) | Gateway® Entry Vector with AtNbr1 PB1 (aa 1–100) | This study |
| pENTR- AtNBR1 PB1 K11A | Gateway® Entry Vector with AtNbr1 PB1 K11A | This study |
| pENTR- AtNBR1 PB1 R19A | Gateway® Entry Vector with AtNbr1 PB1 R19A | This study |
| pENTR- AtNBR1 PB1 D60A | Gateway® Entry Vector with AtNbr1 PB1 D60A | This study |
| pENTR- AtNBR1 PB1 D73A | Gateway® Entry Vector with AtNbr1 PB1 D73A | This study |
| pENTR- AtNBR1ΔUBA1 (616–657) | Gateway® Entry Vector with AtNbr1ΔUBA1 | This study |
| pENTR- AtNBR1ΔUBA2 (656–704) | Gateway® Entry Vector with AtNbr1ΔUBA2 | This study |
| pENTR-AtNBR1ΔUBA1+2 (614–704) | Gateway® Entry Vector with AtNbr1ΔUBA3 | This study |
| pENTR- AtNBR1UBA1 (594–655) | Gateway® Entry Vector with AtNbr1UBA1 | This study |
| pENTR- AtNBR1UBA2 (655–704) | Gateway® Entry Vector with AtNbr1UBA1 | This study |
| pENTR- AtNBR1UBA1+2 (594–704) | Gateway® Entry Vector with AtNbr1UBA1 | This study |
| pENTR- AtNBR1Δ1-492 | Gateway® Entry Vector with AtNbr1Δ1-412 | This study |
| pENTR- AtNBR1 W661A I664A | Gateway® Entry Vector with AtNbr1 point mutantW661A I664A | This study |
| pENTR- AtNBR1 K11A W661A I664A | Gateway® Entry Vector with AtNbr1 point mutantK11A W661A I664A | This study |
| pENTR-ATG8A | Gateway® Entry Vector with AtATG8A | This study |
| pENTR-ATG8B | Gateway® Entry Vector with AtATG8B | This study |
| pENTR-ATG8C | Gateway® Entry Vector with AtATG8C | This study |
| pENTR-ATG8D | Gateway® Entry Vector with AtATG8D | This study |
| pENTR-ATG8F | Gateway® Entry Vector with AtATG8F | This study |
| pENTR-ATG8H | Gateway® Entry Vector with AtATG8H | This study |
| pENTR-ATG8I | Gateway® Entry Vector with AtATG8I | This study |
| pENTR- mCherry-AtNbr1 | Gateway® Entry Vector with AtNbr1 containing anN-terminal cherry-tag | This study |
| cDNA constructs, AmpR, made by gateway LR reactions (this study) | ||
| pDest-myc-AtNBR1 | pDest-myc- AtNBR1ΔUBA1 | pDest-myc-GABARAPL2 |
| pDest-myc-AtNBR1 K11A | pDest-myc- AtNBR1ΔUBA2 | pDest-myc-LC3B |
| pDest-myc-AtNBR1 R19A | pDest-myc- AtNBR1ΔUBA1+2 | pDestEarleygate104-mCherry-AtNbr1 |
| pDest-myc-AtNBR1 | D60A pDest-myc- AtNBR1Δ1-492 | pDestEarleygate104-mCherry-AtNBR1 K11A D60A |
| pDest-myc-AtNBR1 D73A | pDest-myc- AtNBR1 K11A ΔUBA2 | pDestEarleygate104-mCherry-AtNBR1 ΔUBA1+2 |
| pDest53 -AtNBR1 | pDest15-AtATG8A | pDestEarleygate104-mCherry |
| pDest53 -AtNBR1 K11A | pDest15-AtATG8B | pK7GW2-mCherry-AtNBR1 |
| pDest53 -AtNBR1 R19A | pDest15-AtATG8C | pK7GW2-mCherry-AtNBR1 W661A I664A |
| pDest53 -AtNBR1 D60A | pDest15-AtATG8D | pDest15-AtNBR1-UBA2 |
| pDest53 -AtNBR1 D73A | pDest15-AtATG8F | |
| pDest53 -AtNBR1 PB1 (1–100) | pDest15-AtATG8G | |
| pDest53 -AtNBR1 PB1 (1–100) K11A | pDest15-AtATG8H | |
| pDest53 -AtNBR1 PB1 (1–100) R19A | pDest15-AtATG8I | |
| pDest53 -AtNBR1 PB1 (1–100) D60A | pDest-myc-AtNBR1 K11A W661A I664A | |
| pDest53 -AtNBR1 PB1 (1–100) D73A | pDestEGFP-C1-AtNBR1 | |
| pDest15-AtNBR1-PB1 (1–100) | pDestEGFP-C1-AtNBR1 K11A | |
| pDest15-AtNBR1-PB1 (1–100) K11A | pDestEGFP-C1-AtNBR1 K11A W661A I664A | |
| pDest15-AtNBR1-PB1 (1–100) R19A | pDestEGFP-C1-mCherry-AtNBR1 | |
| pDest15-AtNBR1-PB1 (1–100) D60A | pDest-myc-AtATG8A | |
| pDest15-AtNBR1-PB1 (1–100) D73A | pDest-myc-AtATG8H | |
| Other cDNA-constructs | ||
| pDest15-Ub | Mammalian GST-fusion expression vector with Ubiquitin | T. Lamark |
| pDest15-4xUb | Mammalian GST-fusion expression vector with 4x-Ubiquitin | T. Lamark |
Cell culture and transfections.
HeLa cells were grown in Eagle's minimum essential medium supplemented with 10% fetal bovine serum (Biochrom AG, S0615), nonessential amino acids, 2 mm l-glutamine and 1% streptomycin-penicillin (Sigma, P4333). Subconfluent cells were transfected with plasmids using TransIT-LT1 (Mirus, MIR2300) following the supplier's instructions. Twenty four h after transfection cells were fixed in 4% paraformaldehyde and analyzed by confocal fluorescence microscopy.
Transformation and growth of Arabidopsis thaliana.
Agrobacterium tumefaciens strain GV3101 was transformed with the binary plant expression vectors and then used to transform plants (ecotype Col-0) using the flower drip method (modified from Clough and Bent).55 Basta-resistant plants expressing the YFP-mCherry-fusion constructs were then selected using a fluorescence stereomicroscope (SteREO LUMAR, Zeiss), screening for seedlings with good expression of YFP. Seeds of at least three high-expressing lines were used for further studies. Kanamycin-resistant plants were selected by growth on Murashige and Skoog (MS) basal medium (Sigma, M9274) containing 50 ug/ml kanamycin, whereafter 40 transformants were allowed to seed and then each individual line was screened by confocal microscopy. Seeds of at least three well-expressing lines were used for further studies.
For growth on plates, seeds of Arabidopsis were surface-sterilized in 5% chlorine (commercial) bleach and spread on solid MS medium (Sigma). The plates were stratified for 2 d at 6°C and then incubated in a growth chamber under a standard light regime of 16 h light/8 h dark cycle at 24°C. All media were purchased from Sigma. Transgenic seedlings for microscopy were germinated and grown in liquid cultures using Hoagland solution under normal growth conditions (16 h light/8 h dark cycle, room-temp.). All plants were examined within a period of 4–7 d after germination.
Fluorescence confocal microscopy analysis.
Cultured HeLa-cells and Arabidopsis seedlings were examined using a Zeiss Axiovert 200 microscope with a 40 × 1.2 W C-Apochromat objective, equipped with an LSM510-META confocal module using LSM 5 software version 3.2. Images were processed using Canvas version 9 and 10 (ACD Systems).
Co-immunoprecipitation.
Expression vectors (0.5 µg) for GFP- and myc-tagged proteins were in vitro co-transcribed/co-translated in a total volume of 25 µl using the TNT T7 coupled rabbit reticulocyte lysate system (Promega, L4610) according to the manufacturer's protocol, with added 35S-labeled methionine (Perkin-Elmer, NEG009A001MC). Twenty microliters of the in vitro translated 35S-labeled proteins were immediately diluted in 200 µl of ice-cold NET-N buffer (20 mM TRIS-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.5% Nonidet P-40) containing one tablet per 10 ml of Complete Mini, EDTA-free protease inhibitor mixture (Roche, 11 836 170001). The samples were preincubated with a 50% solution of Protein A-agarose beads (Santa Cruz; sc-2001) in NET-N buffer for 10 min at 4°C on a rotating wheel and then incubated with 0.1 µg of a custom made anti-GFP polyclonal antibody for 1 h and for another 30 min in the presence of bovine serum albumin-saturated Protein A-agarose beads. The complexes were washed five times with 400 µl NET-N and resuspended in 15 µl of 2 × SDS-polyacrylamide gel load buffer and boiled for 5 min. The samples were resolved on SDS-polyacrylamide gels. 35S-Labeled proteins were detected using a Fujifilm bioimaging analyzer BAS-5000 (Fuji).
GST-pulldown experiments.
All GST-tagged proteins were expressed in Escherichia coli BL21(DE3). GST fusion proteins were purified on glutathione-Sepharose 4 Fast Flow beads (GE Healthcare, 17-5132-01). 35S-labeled GFP- and myc-tagged proteins were synthesized in vitro using the TnT T7 coupled reticulocyte lysate system. Translation reaction products from 0.5 µg of plasmid were pre-incubated with 10 ul glutathione-Sepharose beads with 100 µl of NETN-E buffer (50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.5% Nonidet P-40) supplemented with Complete Mini EDTA-free protease inhibitor cocktail (Roche) for 30 min at 4°C to reduce unspecific binding. The supernatant from the pre-incubation was then incubated with 3–20 µl of GST-labeled proteins on glutathione-Sepharose beads for 1 h at 4°C and then the beads were washed five times with 400 µl of NETN-E buffer, boiled with 2× SDS-PAGE gel loading buffer, and subjected to SDS-PAGE. Gels were stained with Coomassie brilliant blue and vacuum-dried. 35S-labeled proteins were detected using a Fujifilm bioimaging analyzer BAS-5000 (Fuji).
Immunological analysis of AtNBR1.
For antibody production, the C-terminal-UBA domain of AtNBR1 was expressed with an N-terminal GST-tag in E. coli BL21(DE3) and purified using the GST SpinTrap™ (GE Healthcare, 28-9523-59). A polyclonal antibody was produced in rabbit against the most C-terminal UBA domain of AtNBR1 by Agrisera AB, Sweden. Before use in protein gel blots, the serum was depleted of antibodies with affinity for GST using recombinant GST coupled to Glutathione Sepharose 4 Fast Flow beads. For protein gel blots, 100 mg of seedlings (roots included) were collected from plates and immediately frozen in liquid nitrogen. All indicated wt plants were ecotype Col-0, atg7-1 line is ecotype WS. Five-mm stainless steel beads (Qiagen, 69989) were used to grind the material in the Qiagen TissueLyser. Two-hundred µl 1× SDS-PAGE gel loading buffer was added to each sample followed by boiling for 5 min, bead removal, and then 5 min centrifugation at 13,000 rpm in a microcentrifuge. The supernatant was removed and used for protein gel blotting.
Bioinformatics.
All sequences were collected using BLAST searches on genome-portals available online.56 The alignments were performed using ClustalW,57 and aligned sequences were analyzed using phyML (maximum likelihood).39 PB1-domains of the N. vectensis and A. thaliana proteins were modeled using the Swissmodel database (automated mode) and solvent accessible electrostatic surface potentials were calculated and visualized using the APBS plugin in PyMOL.
Acknowledgements
We are grateful to Leidulf Lund at Tromso University Phytotron for extensive help with growing and managing the transgenic plants. We thank Dr. Taijoon Chung at the Vierstra Laboratory for the generous gift of seeds from transgenic GFP-AtATG8 and the atg7-1 knockout line of Arabidopsis. This work was supported by grants from the FRIBIO and FUGE programmes of the Norwegian Research Council, the Norwegian Cancer Society, the Aakre Foundation and the Blix Foundation to T.J.
Abbreviations
- ATG
AuTophaGy-related
- Cvt
cytoplasm-to-vacuole-targeting
- GABARAP
gamma-aminobutyrate receptor associated protein
- GFP
enhanced green fluorescent protein
- (MAP)LC3
microtubule-associated protein 1 light chain 3
- LIR
LC3 interacting region
- NBR1
next to BRCA1 gene 1
- PB1 domain
Phox and Bem1 domain
- UBA domain
ubiquitin-associated domain
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 2.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deretic V. Autophagy in infection. Curr Opin Cell Biol. 2010;22:252–262. doi: 10.1016/j.ceb.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 7.Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584:1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-102. [DOI] [PubMed] [Google Scholar]
- 9.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He H, Dang Y, Dai F, Guo Z, Wu J, She X, et al. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J Biol Chem. 2003;278:29278–29287. doi: 10.1074/jbc.M303800200. [DOI] [PubMed] [Google Scholar]
- 11.Xin Y, Yu L, Chen Z, Zheng L, Fu Q, Jiang J, et al. Cloning, expression patterns and chromosome localization of three human and two mouse homologues of GABA(A) receptor-associated protein. Genomics. 2001;74:408–413. doi: 10.1006/geno.2001.6555. [DOI] [PubMed] [Google Scholar]
- 12.Slavikova S, Ufaz S, Avin-Wittenberg T, Levanony H, Galili G. An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J Exp Bot. 2008;59:4029–4043. doi: 10.1093/jxb/ern244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, et al. Processing of ATG8s, ubiquitin-like proteins and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16:2967–2983. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 16.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 18.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 21.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 22.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 23.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 24.Waters S, Marchbank K, Solomon E, Whitehouse C, Gautel M. Interactions with LC3 and polyubiquitin chains link nbr1 to autophagic protein turnover. FEBS Lett. 2009;583:1846–1852. doi: 10.1016/j.febslet.2009.04.049. [DOI] [PubMed] [Google Scholar]
- 25.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 27.Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 28.Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 29.Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 30.Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010;584:1379–1385. doi: 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 31.Vadlamudi RK, Joung I, Strominger JL, Shin J. p62, a phosphotyrosine-independent ligand of the SH2 domain of p56lck, belongs to a new class of ubiquitin-binding proteins. J Biol Chem. 1996;271:20235–20237. doi: 10.1074/jbc.271.34.20235. [DOI] [PubMed] [Google Scholar]
- 32.Long J, Gallagher TR, Cavey JR, Sheppard PW, Ralston SH, Layfield R, et al. Ubiquitin recognition by the ubiquitin-associated domain of p62 involves a novel conformational switch. J Biol Chem. 2008;283:5427–5440. doi: 10.1074/jbc.M704973200. [DOI] [PubMed] [Google Scholar]
- 33.Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 34.Lamark T, Perander M, Outzen H, Kristiansen K, Overvatn A, Michaelsen E, et al. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J Biol Chem. 2003;278:34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- 35.Wilson MI, Gill DJ, Perisic O, Quinn MT, Williams RL. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol Cell. 2003;12:39–50. doi: 10.1016/S1097-2765(03)00246-6. [DOI] [PubMed] [Google Scholar]
- 36.Itakura E, Mizushima N. p62 targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol. 2011;192:17–27. doi: 10.1083/jcb.201009067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toyooka K, Moriyasu Y, Goto Y, Takeuchi M, Fukuda H, Matsuoka K. Protein aggregates are transported to vacuoles by a macroautophagic mechanism in nutrient-starved plant cells. Autophagy. 2006;2:96–106. doi: 10.4161/auto.2.2.2366. [DOI] [PubMed] [Google Scholar]
- 38.Maddison DR, Schulz KS, Maddison WP. The Tree of Life Web Project. Zootaxa. 2007;1668:19–40. [Google Scholar]
- 39.Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:557–559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruiz-Trillo I, Inagaki Y, Davis LA, Sperstad S, Landfald B, Roger AJ. Capsaspora owczarzaki is an independent opisthokont lineage. Curr Biol. 2004;14:946–947. doi: 10.1016/j.cub.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 43.Bassham DC. Function and regulation of macro-autophagy in plants. Biochim Biophys Acta. 2009;1793:1397–1403. doi: 10.1016/j.bbamcr.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 45.Matsuoka KDJ. Chimeric fluorescent fusion proteins to monitor autophagy in plants. Methods Enzymol. 2008;451:541–555. doi: 10.1016/S0076-6879(08)03231-X. [DOI] [PubMed] [Google Scholar]
- 46.Fujiki Y, Yoshimoto K, Ohsumi Y. An Arabidopsis homologue of yeast ATG6/VPS30, a component of phosphatidylinositol-3-kinase, is essential for pollen development. Plant Cell Physiol. 2005;46:28. [Google Scholar]
- 47.Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. Autophagic Nutrient Recycling in Arabidopsis Directed by the ATG8 and ATG12 Conjugation Pathways. Plant Physiol. 2005;138:2097–2110. doi: 10.1104/pp.105.060673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, et al. Mobilization of Rubisco and Stroma-Localized Fluorescent Proteins of Chloroplasts to the Vacuole by an ATG Gene-Dependent Autophagic Process. Plant Physiol. 2008;148:142–155. doi: 10.1104/pp.108.122770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sláviková S, Shy G, Yao Y, Glozman R, Levanony H, Pietrokovski S, et al. The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J Exp Bot. 2005;56:2839–2849. doi: 10.1093/jxb/eri276. [DOI] [PubMed] [Google Scholar]
- 50.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calvo-Garrido J, Escalante R. Autophagy dysfunction and ubiquitin-positive protein aggregates in Dictyostelium cells lacking Vmp1. Autophagy. 2010;6:100–109. doi: 10.4161/auto.6.1.10697. [DOI] [PubMed] [Google Scholar]
- 53.Rigden DJ, Michels PA, Ginger ML. Autophagy in protists: Examples of secondary loss, lineage-specific innovations and the conundrum of remodeling a single mitochondrion. Autophagy. 2009;5:784–794. doi: 10.4161/auto.8838. [DOI] [PubMed] [Google Scholar]
- 54.Duszenko M, Ginger ML, Brennand A, Gualdron-Lopez M, Colombo MI, Coombs GH, et al. Autophagy in protists. Autophagy. 2011;7:127–158. doi: 10.4161/auto.7.2.13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 56.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 59.Karimi M, Inzé D, Depicker A. GATEWAY(TM) vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/S1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.